The crystal structure of the hypothetical protein YqgQ from B. subtilis is reported at 2.1 Å resolution.
Keywords: YqgQ hypothetical protein, Se-SAD, novel fold, DUF910, Protein Structure Initiative, New York SGX Research Consortium for Structural Genomics
Abstract
The crystal structure of the hypothetical protein YqgQ from Bacillus subtilis has been determined to 2.1 Å resolution. The crystals belonged to space group P21, with unit-cell parameters a = 51.85, b = 41.25, c = 55.18 Å, β = 113.4°, and contained three protein molecules in the asymmetric unit. The structure was determined by the single-wavelength anomalous dispersion method using selenium-labeled protein and was refined to a final R factor of 24.7% (R free = 28.0%). The protein molecule mainly comprises a three-helical bundle. Its putative function is inferred to be single-stranded nucleic acid binding based on sequence and structural homology.
1. Introduction
The crystal structure of YqgQ, an 8.6 kDa protein of unknown function from Bacillus subtilis, has been determined at 2.1 Å resolution by the single-wavelength anomalous dispersion method (SAD). YqgQ represents a new protein fold and is a member of the DUF910 family in the Pfam database. This uncharacterized protein was selected by the New York Structural GenomiX Research Consortium (NYSGXRC) for structure determination (NYSGXRC target ID 10278a). This is the first structure to be reported for the DUF910 family. The protein fold is defined, according to SCOP, as a helical bundle of three α-helices forming a left-handed twist.
2. Experimental methods
2.1. Protein production and purification
The target gene for 10278a was amplified using the polymerase chain reaction (PCR) from B. subtilis genomic DNA using the forward primer AATACATTTTATGATGTGCAGC and the reverse primer AGCCTTTATAAAAATCTCTTCCG. The amplified gene was gel purified and cloned into a pSGX4(BS) vector designed to express the protein with a fusion tag, which was removed after purification. Protein expression and purification utilized previously published protocols, which are described in detail in PepcDB (http://pepcdb.pdb.org)
2.2. Crystallization, data collection and processing
Crystals were grown at room temperature by the sitting-drop vapor-diffusion technique against a reservoir containing 0.066 M sodium dihydrogen phosphate and 3.385 M dipotassium hydrogen phosphate (pH 8.2) as a precipitant (1 µl reservoir solution plus 1 µl protein at 7 mg ml−1 in 20 mM HEPES buffer pH 7.2, 200 mM NaCl, 5 mM l-methionine and 5 mM dithiothreitol). Diffraction-quality crystals were obtained by the microseeding technique and were flash-frozen in liquid nitrogen, using mother liquor containing glycerol [final concentration of 15%(v/v)] as a cryoprotectant. Single-wavelength Se-SAD diffraction data covering 360° rotation in ϕ were collected on NSLS beamline X25 (National Synchrotron Light Source, Brookhaven National Laboratory) to 2.1 Å resolution under standard cryogenic conditions using 1° oscillation per frame at the selenium absorption edge (λ = 0.9801 Å) and were processed, scaled and merged with HKL-2000 (Otwinowski & Minor, 1997 ▶). Details of the data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection, phasing and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Unit-cell parameters (Å, °) | a = 51.85, b = 41.25, c = 55.18, β = 113.4 |
| Space group | P21 |
| Data-collection statistics | |
| Wavelength (Å) | 0.9801 |
| Resolution range (Å) | 50–2.10 (2.18–2.10) |
| Unique reflections | 12344 (1447) |
| Completeness (%) | 97.5 (84.7) |
| Mean I/σ(I) | 12.2 (2.8) |
| Multiplicity | 6.5 (3.9) |
| Rmerge† | 0.08 (0.37) |
| Phasing statistics | |
| Phasing power‡ (ano) | 1.046 |
| FOM‡ (centric/acentric) | 0.040/0.293 |
| FOM after density modification | 0.88 |
| Refinement statistics | |
| Resolution range (Å) | 47.59–2.1 |
| No. of reflections (work) | 11871 |
| No. of reflections (test) | 959 |
| R factor/Rfree§ | 0.247/0.280 |
| B factor from Wilson plot (Å2) | 40.6 |
| R.m.s.d. bond lengths (Å) | 0.017 |
| R.m.s.d. bond angles (o) | 1.80 |
| Average B values (Å2) | |
| Main chain | 17.7 |
| Side chain | 38.1 |
| No. of non-H atoms | 1590 |
| No. of water molecules | 49 |
R
merge = 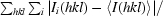
 , where I
i(hkl) is the intensity of the ith measurement and 〈I(hkl)〉 is the mean intensity for that reflection.
, where I
i(hkl) is the intensity of the ith measurement and 〈I(hkl)〉 is the mean intensity for that reflection.
As defined in SHARP.
R factor = 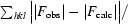
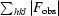 , where |F
calc| and |F
obs| are the calculated and observed structure-factor amplitudes, respectively. R
free is the same but for a subset of reflections that were chosen randomly as a test set.
, where |F
calc| and |F
obs| are the calculated and observed structure-factor amplitudes, respectively. R
free is the same but for a subset of reflections that were chosen randomly as a test set.
2.3. Structure determination and refinement
All nine possible Se-atom sites were identified with comparable occupancy using SOLVE (Terwilliger & Berendzen, 1999 ▶) and phases were refined using SHARP (de La Fortelle & Bricogne, 1997 ▶; Table 1 ▶). The final electron-density map after density modification was of high quality, allowing automated model building of ∼90% of the polypeptide chain with ARP/wARP (Perrakis et al., 1999 ▶). The atomic model was refined with CNS (Brünger et al., 1998 ▶) using data extending to 2.1 Å resolution. PROCHECK (Laskowski et al., 1993 ▶) shows ∼98% of residues in the most favorable region of the Ramachandran plot. The final refined atomic model contains three copies of the molecule containing residues 1–62 (the first residue Leu arises from a cloning artifact) and 47 water molecules (Table 1 ▶). The electron density for the ten C-terminal residues was missing. Unidentified continuous electron density has been modeled as water molecules. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB code 2nn4).
3. Results and discussion
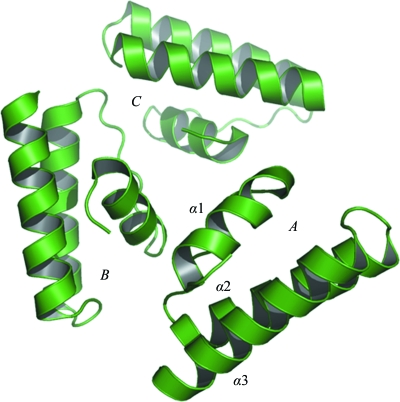
The asymmetric unit contains three structurally similar polypeptide chains, with a root-mean-square deviation (r.m.s.d.) of ∼0.6 Å for 62 common Cα-atom pairs for each pair of protomers. The protein has a novel fold, a three-helical bundle, with the helix order being left-handed and with the third helix flanked by a loop (Fig. 1 ▶). The buried interface surface area between molecules A and C is 755 Å2 (17.2% of the total surface area), while it is 294 Å2 (6.7% of the total surface area) between molecules B and C (Laskowski, 2001 ▶). There is no common interface between protomers A and B. Although the buried area between A and C is comparable to values found in biologically relevant dimers (Cavaille et al., 1999 ▶), the three protomers do not form a biologically relevant oligomer (Henrick & Thornton, 1998 ▶). The three protomers of YqgQ in the asymmetric unit are stabilized by ionic, hydrophobic and hydrogen-bond interactions. The interface between molecules A and C contains one salt bridge between Arg23 and Glu44, whereas the interface between molecules B and C contains two hydrogen bonds from His16 to Tyr5 and Gln9.
Figure 1.
The asymmetric unit of the YqgQ crystal structure with the three protomers labeled A, B and C. The three helices are labeled α1, α2 and α3 in protomer A.
3.1. Sequence homology of YqgQ
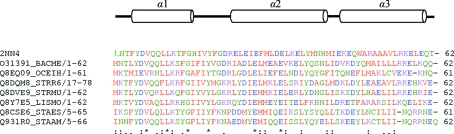
A search for conserved domains using BLAST (Altschul et al., 1997 ▶) revealed a set of hypothetical protein sequences with sequence identities ranging from 57 to 26%. The O31391 protein from B. megaterium shows 47% identity to YqgQ and has open reading frame protein 1 (ORF1) function. Generally, ORF1 proteins are single-stranded nucleic acid-binding proteins (Kolosha & Martin, 1997 ▶, 2003 ▶). They enhance the annealing of complementary oligonucleotides, participate in protein–protein interactions (Martin et al., 2000 ▶) and function as nucleic acid chaperones (Martin & Bushman, 2001 ▶). O31391 is a member of the DUF910 family of proteins; the family consists of 83 short bacterial proteins of unknown function (Fig. 2 ▶).
Figure 2.
Multiple sequence alignment of YqgQ and homologous proteins from the DUF910 family. Strictly invariant residues are indicated by ‘*’ and conserved residues by ‘:’ The secondary structure (top) is also aligned with the sequences.
3.2. Structural homology with YqgQ
A DALI (Holm & Sander, 1993 ▶) search was performed using the YqgQ model to identify three-dimensional structural homologs. The search revealed structures with significant similarity to YqgQ, including DNA-directed RNA polymerase α (PDB code 2a68, chain D) and RNA-directed RNA polymerase catalytic subunit (PDB code 3a1g, chain C), which have Z scores of 6.1 and 5.3 with r.m.s.d.s of 2.0 and 2.8 Å, respectively, despite having low sequence identity (5–14%) to YqgQ.
One of the aims of structural genomics is to annotate the functional aspects of a protein from its fold. Accordingly, the Protein Function Prediction webserver and DALI searches were used to obtain a putative function for the protein.
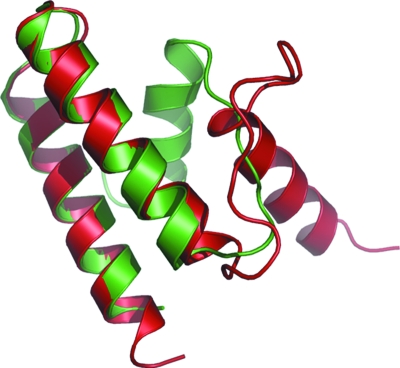
The Protein Function Prediction (PFP) webserver (http://dragon.bio.purdue.edu/pfp/) was used to identify the putative function of the protein. The database indicates that this protein may possess RNA-directed RNA polymerase activity. Structural superposition of RNA-directed RNA polymerase catalytic subunit PB-1 (PDB code 3a1g, chain C) and YqgQ (Fig. 3 ▶) shows that α-helices 2 and 3 superpose exactly with the two helices in the PB-1 domain, which plays a distinct role in viral RNA polymerase (Yuan et al., 2009 ▶) and is essential for viral RNA transcription initiation (He et al., 2008 ▶). The structural comparison gives an indication of the putative function of the protein. At this point there are no structural homologs available, hence it can only be speculated that the protein may be indirectly involved in an RNA polymerization reaction during bacterial cell growth.
Figure 3.
Structural superposition of YqgQ (green) with the RNA-directed RNA polymerase catalytic subunit of PB-1 from PDB entry 3a1g (red).
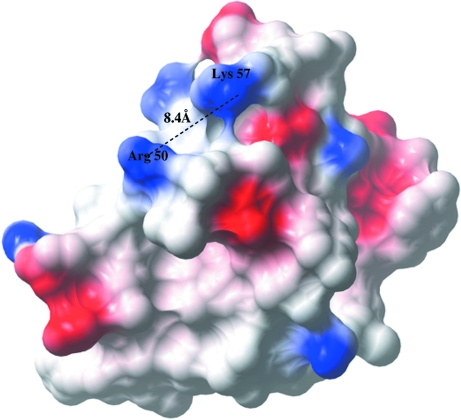
In addition to the structural comparison, protein-sequence comparison was taken into account to derive a putative function for YqgQ. The protein-sequence homology search shows that YqgQ is similar to an open reading frame 1 (ORF1) protein. ORF1 proteins interact with nucleic acids through positively charged Arg residues (Martin et al., 2005 ▶). The YqgQ structure contains positively charged residues Arg50 and Lys57 in helix 3. The distance between their side-chain N atoms is ∼8.4 Å, which is comparable to the distance between two consecutive phosphate groups (∼6.0 Å) in a nucleic acid. This suggests that YqgQ may also bind to single-stranded nucleic acids. The electrostatic potential surface calculated using CCP4mg (Potterton et al., 2004 ▶) is shown in Fig. 4 ▶.
Figure 4.
Electrostatic potential surface of YqgQ. Positively charged side-chain residues (Arg50 and Lys57) at the putative nucleic acid-binding pocket are present on the surface of the protein. The distance between the side chains of Arg50 and Lys57 is comparable to the distance between consecutive phosphate groups in a single-stranded nucleic acid.
In conclusion, we have determined the crystal structure of a conserved hypothetical protein comprising a three-helical bundle. The protein is a representative structure of a pool of short peptides of unknown function in B. subtilis (DUF910).
Supplementary Material
PDB reference: YqgQ, 2nn4, r2nn4sf
Acknowledgments
This research was supported by the National Institutes of Health (GM074945) under DOE Prime Contract No. DEAC02-98CH10886 with Brookhaven National Laboratory. We gratefully acknowledge data-collection support from beamline X25 (NSLS).
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res.25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Cavaille, J., Chetouani, F. & Bachellerie, J. P. (1999). RNA, 5, 66–81. [DOI] [PMC free article] [PubMed]
- He, X., Zhou, J., Bartlam, M., Zhang, R., Ma, J., Lou, Z., Li, X., Li, J., Joachimiak, A., Zeng, Z., Ge, R., Rao, Z. & Liu, Y. (2008). Nature (London), 454, 1123–1126. [DOI] [PubMed]
- Henrick, K. & Thornton, J. M. (1998). Trends Biochem. Sci.23, 358–361. [DOI] [PubMed]
- Holm, L. & Sander, C. (1993). J. Mol. Biol.233, 123–138. [DOI] [PubMed]
- Kolosha, V. O. & Martin, S. L. (1997). Proc. Natl Acad. Sci. USA, 94, 10155–10160. [DOI] [PMC free article] [PubMed]
- Kolosha, V. O. & Martin, S. L. (2003). J. Biol. Chem.278, 8112–8117. [DOI] [PubMed]
- La Fortelle, E. de & Bricogne, G. (1997). Methods Enzymol.276, 472–493. [DOI] [PubMed]
- Laskowski, R. A. (2001). Nucleic Acids Res.29, 221–222. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Martin, S. L. & Bushman, F. D. (2001). Mol. Cell. Biol.21, 467–475. [DOI] [PMC free article] [PubMed]
- Martin, S. L., Cruceanu, M., Branciforte, D., Li, P. W., Kwok, S. C., Hodges, R. S. & Williams, M. C. (2005). J. Mol. Biol.348, 549–561. [DOI] [PubMed]
- Martin, S. L., Li, J. & Weisz, J. A. (2000). J. Mol. Biol.304, 11–20. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Yuan, P., Bartlam, M., Lou, Z., Chen, S., Zhou, J., He, X., Lv, Z., Ge, R., Li, X., Deng, T., Fodor, E., Rao, Z. & Liu, Y. (2009). Nature (London), 458, 909–913. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: YqgQ, 2nn4, r2nn4sf
PDB reference: YqgQ, 2nn4, r2nn4sf