Malonyl-CoA:acyl-carrier protein transacylase (MCAT; FabD) from S. aureus has been cloned, overexpressed, purified and crystallized. The crystal belonged to space group P21, with unit-cell parameters a = 41.608, b = 86.717, c = 43.163 Å, α = γ = 90, β = 106.330°, and data were collected to 1.2 Å resolution using synchrotron radiation.
Keywords: fatty-acid synthesis, malonyl-CoA:acyl-carrier protein transacylase, FabD, antibacterial drug targets, Staphylococcus aureus
Abstract
Malonyl-CoA:acyl-carrier protein transacylase (MCAT), encoded by the fabd gene, is a key enzyme in type II fatty-acid biosynthesis. It is responsible for transferring the malonyl group from malonyl-CoA to the holo acyl-carrier protein (ACP). Since the type II system differs from the type I system that mammals use, it has received enormous attention as a possible antibiotic target. In particular, only a single isoform of MCAT has been reported and a continuous coupled enzyme assay has been developed. MCAT from Staphylococcus aureus was overexpressed in Escherichia coli and the protein was purified and crystallized. Diffraction data were collected to 1.2 Å resolution. The crystals belonged to space group P21, with unit-cell parameters a = 41.608, b = 86.717, c = 43.163 Å, α = γ = 90, β = 106.330°. The asymmetric unit contains one SaMCAT molecule.
1. Introduction
Fatty-acid biosynthesis, which consists of condensation, reduction, dehydration and reduction steps, is essential for the maintainance of homeostasis in all organisms. Mammals, fungi and some bacteria utilize type I fatty-acid biosynthesis, which involves a large multifunctional enzyme into which an acyl-carrier protein is integrated together with the catalytic domains. In contrast, the majority of bacteria and all plants use the type II system, in which discrete dissociated enzymes each carry out a single reaction in the pathway in a sequential manner (Campbell & Cronan, 2001 ▶; Heath et al., 2001 ▶; White et al., 2005 ▶). Owing to these differences, the enzymes of the type II system have been considered to be attractive antibacterial targets.
Malonyl-CoA is a coenzyme A derivative that plays a key role in chain elongation in fatty-acid biosynthesis as well as in polyketide biosynthesis. In the former, it provides two-carbon units to fatty acids and commits them to fatty-acid chain synthesis. It is formed by carboxylating acetyl-CoA using the enzyme acetyl-CoA carboxylase and is utilized in fatty-acid biosynthesis by the enzyme malonyl-coenzymeA:acyl-carrier protein transacylase (MCAT). MCAT serves to transfer malonate from malonyl-CoA to the terminal thiol of holo acyl-carrier protein (ACP). Crystal structures of MCAT from Escherichia coli (Serre et al., 1995 ▶; Oefner et al., 2006 ▶), Streptomyces coelicolor (Keatinge-Clay et al., 2003 ▶), Helicobacter pylori (Zhang et al., 2007 ▶) and Mycobacterium tuberculosis (Li et al., 2007 ▶) have been reported.
We have crystallized MCAT from Staphylococcus aureus, which is the most common cause of staphylococcal infections. It can cause a range of illnesses from life-threatening diseases such as pneumonia, meningitis, endocarditis, toxic shock syndrome and septicaemia to minor skin infections such as pimples, impetigo and boils (Kluytmans et al., 1997 ▶).
2. Experimental methods
2.1. Cloning
The MCAT gene (UniProtKB/TrEMBL accession No. Q99UN8) was amplified from S. aureus genomic DNA by polymerase chain reaction (PCR) using the forward primer 5′-GGGAATTCCATA TGGCAATTATTTTTCCGGGACAAG-3′ and the reverse primer 5′-CCGCTCGAGTTAGTCATTTTCATTCCATCCTTT-3′ (NdeI and XhoI restriction sites are indicated in bold). The PCR product was digested with NdeI and XhoI restriction endonucleases and ligated to digested pET-28a(+) (Novagen). The final construct encoded an N-terminally truncated MCAT protein consisting of residues 5–308 of MCAT fused to the N-terminal His tag MGSSHHHHHHSSGLVPRGSH.
2.2. Expression and purification
The recombinant plasmid was transformed into E. coli BL21 (DE3) strain and the cells were grown at 310 K in Luria–Bertani medium supplemented with kanamycin (50 mg ml−1). The transformants were grown in LB medium at 310 K until they reached an optical density of 0.6 at 600 nm; expression was then induced with 0.5 mM IPTG. The cells were grown for 16 h at 291 K after induction and were then harvested by centrifugation at 4000 rev min−1 for 20 min at 277 K. The cells were resuspended in lysis buffer (100 mM Tris–HCl, 50 mM NaCl, 1 mM MgCl2 pH 8.0, 2% glycerol and 0.2 mM PMSF) and lysed by sonication.
After centrifugation, the recombinant S. aureus MCAT (SaMCAT) protein was purified utilizing the N-terminal histidine tag on an Ni–NTA column (GE Healthcare, USA) equilibrated with buffer A (20 mM Tris–HCl, 100 mM NaCl pH 8.0 and 1 mM TCEP). The protein was eluted with a linear gradient to buffer A containing 1 M imidazole. The pooled fraction containing the MCAT protein was then loaded onto an anion-exchange Q-Sepharose column pre-equilibrated with buffer B (20 mM Tris–HCl, 1 mM DTT pH 8.0), eluted with a linear gradient of 0–1 M NaCl and identified by SDS–PAGE analysis. The final purification step was gel filtration on a HiLoad 26/60 Superdex-75 prep-grade column equilibrated with 20 mM Tris–HCl, 100 mM NaCl and 1 mM TCEP pH 7.5. The fraction containing SaMCAT was concentrated to 15 mg ml−1 using an YM10 membrane and an Amicon concentrator (Millipore) and the protein concentration was determined using the Bio-Rad protein assay with BSA as a standard.
2.3. Crystallization
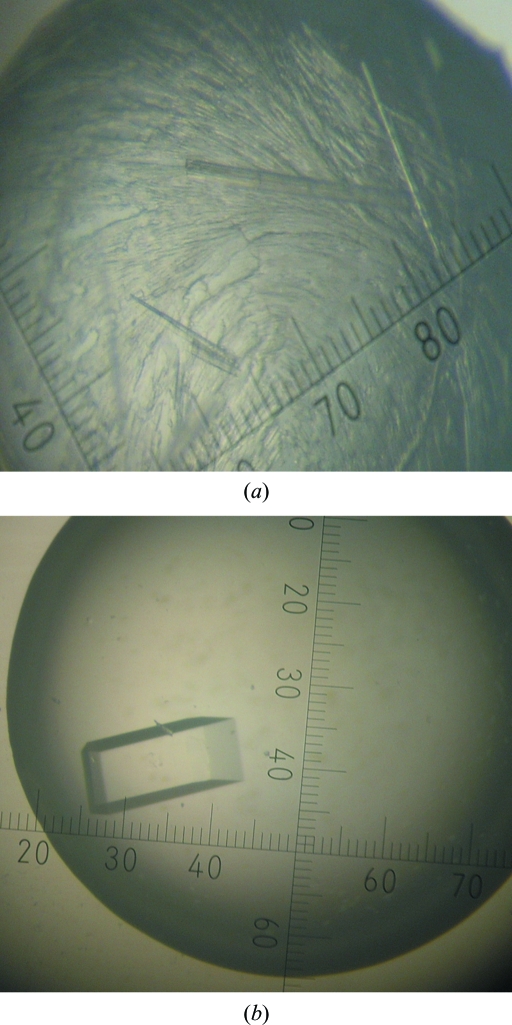
An initial search for crystallization conditions was made by screening against 1200 conditions using a Hydra II Plus One system (Matrix Technology). Microcrystals of SaMCAT were obtained after 3 d using Hampton Research PEG/Ion Screen condition No. 28 (20% PEG 3350, 0.2 M calcium acetate pH 7.3) as the reservoir solution. This condition was optimized by the hanging-drop method using 24-well tissue-culture VDX plates (Hampton Research) at 295 K. The PEG 3350 and calcium acetate concentrations were varied. Diffraction-quality SaMCAT crystals appeared after 5 d by mixing 1.5 µl protein solution at 15 mg ml−1 in 20 mM Tris–HCl pH 7.5, 100 mM NaCl and 1 mM TCEP with an equal volume of reservoir solution consisting of 21–23% PEG 3350 and 0.5–0.52 M calcium acetate (Fig. 1 ▶).
Figure 1.
A crystal of MCAT from S. aureus. (a) Microcrystals obtained using PEG/Ion Screen condition No. 28 consisting of 20% PEG 3350, 0.2 M calcium acetate pH 7.3. (b) Optimized crystal obtained after 5 d in 21–23% PEG 3350, 0.5–0.52 M calcium acetate. Its approximate dimensions are 0.15 × 0.05 × 0.05 mm.
2.4. Data collection and processing
For X-ray data collection, a native SaMCAT crystal was transferred to a solution consisting of 22% PEG 3350, 0.5 M calcium acetate and 30%(v/v) glycerol in four steps within a minute before being flash-cooled. X-ray diffraction data were collected using an ADSC Quantum CCD detector (ADSC, Madison, Wisconsin, USA) at 100 K on beamline 6C of Pohang Light Source (PLS), Republic of Korea. The wavelength was 1.12713 Å and the exposure time was 3 s per image. The crystal was rotated through a total of 305°, with 1.0° oscillation per frame. The crystal-to-detector distance was set to 70 mm. The raw data were processed and scaled using the HKL software package (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
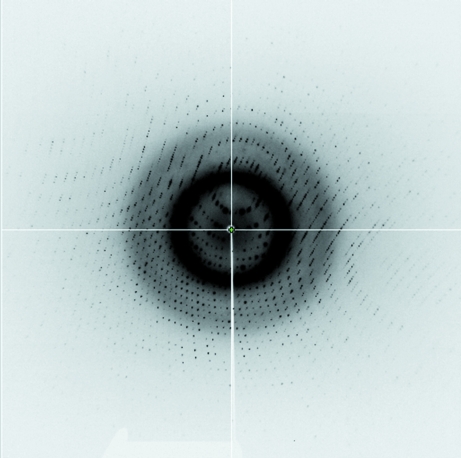
SaMCAT was successfully cloned in E. coli with a fused hexahistidine tag and was purified to homogeneity by Ni–NTA, anion-exchange and size-exclusion chromatography. Diffraction-quality crystals were obtained manually in hanging drops containing 21–23% PEG 3350 and 0.5–0.52 M calcium acetate as the well solution. An X-ray diffraction data set was collected on beamline 6C at PLS. The X-ray diffraction data were collected from a monoclinic SaMCAT crystal. A total of 1 211 660 measured reflections were merged into 91 645 unique reflections with an R merge (on intensity) of 3.7%. The merged data set was 97.9% complete to 1.2 Å resolution (Fig. 2 ▶). The crystal belonged to space group P21, with unit-cell parameters a = 41.608, b = 86.717, c = 43.163 Å, α = γ = 90, β = 106.330°. The asymmetric unit contained one molecule of SaMCAT, giving a crystal volume per protein mass (V M) of 2.22 Å3 Da−1 and a solvent content of 44.52% (Matthews, 1968 ▶). Data-collection statistics are given in Table 1 ▶. The structure was solved with the molecular-replacement program MOLREP in the CCP4 suite using E. coli MCAT (PDB code 1mla; Serre et al., 1995 ▶) as a search model; the structural details will be described in a separate paper.
Figure 2.
X-ray diffraction image of an SaMCAT crystal collected to 1.2 Å resolution.
Table 1. Data-collection statistics for SaMCAT.
Values in parentheses are for the highest resolution shell.
| Source | PLS beamline 6C |
| Wavelength (Å) | 1.12713 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 41.608, b = 86.717, c = 43.163, α = γ = 90, β = 106.330 |
| Temperature (K) | 100 |
| Redundancy | 5.1 |
| Resolution range (Å) | 50.0–1.2 (1.24–1.20) |
| Total/unique reflections | 1211660/91645 |
| Completeness (%) | 97.9 (88.9) |
| Mean I/σ(I) | 41.2 (6.6) |
| Rmerge† (%) | 3.7 (15.7) |
R
merge = 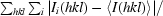
 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
Acknowledgments
We are grateful to the staff of beamline 6C at Pohang Light Source, Republic of Korea for assistance during data collection. This work was supported by the Functional Proteomics Center, 21C Frontier Program of the Korea Ministry of Science and Technology and a grant from the Korea Institute of Science and Technology.
References
- Campbell, J. W. & Cronan, J. J. (2001). Annu. Rev. Microbiol.55, 305–332. [DOI] [PubMed]
- Heath, R. J., White, S. W. & Rock, C. O. (2001). Prog. Lipid Res.40, 467–497. [DOI] [PubMed]
- Keatinge-Clay, A. T., Shelat, A. A., Savage, D. F., Tsai, S. C., Miercke, L. J., O’Connell, J. D., Khosla, C. & Stroud, R. M. (2003). Structure, 11, 147–154. [DOI] [PubMed]
- Kluytmans, J., van Belkum, A. & Verbrugh, H. (1997). Clin. Microbiol. Rev.10, 505–520. [DOI] [PMC free article] [PubMed]
- Li, Z., Huang, Y., Ge, J., Fan, H., Zhou, X., Li, S., Bartlam, M., Wang, H. & Rao, Z. (2007). J. Mol. Biol.371, 1075–1083. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Oefner, C., Schulz, H., D’Arcy, A. & Dale, G. E. (2006). Acta Cryst. D62, 613–618. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Serre, L., Verbree, E. C., Dauter, Z., Stuitje, A. R. & Derewenda, Z. S. (1995). J. Biol. Chem.270, 12961–12964. [DOI] [PubMed]
- White, S. W., Zheng, J., Zhang, Y. M. & Rock, C. O. (2005). Annu. Rev. Biochem.74, 791–831. [DOI] [PubMed]
- Zhang, L., Liu, W., Xiao, J., Hu, T., Chen, J., Chen, K., Jiang, H. & Shen, X. (2007). Protein Sci.16, 1184–1192. [DOI] [PMC free article] [PubMed]