Recombinant human CLEC5A was crystallized in the trigonal space group P31 and X-ray diffraction data were collected to 1.56 Å resolution.
Keywords: CLEC5A, MDL-1, C-type lectins, DAP12, dengue
Abstract
The human C-type lectin-like protein CLEC5A (also known as MDL-1) is expressed on the surface of myeloid cells and plays a critical role in dengue-virus-induced disease by signalling through the transmembrane adaptor protein DAP12. The C-type lectin-like domain of CLEC5A was expressed in Escherichia coli, refolded and purified. Recombinant CLEC5A crystals were grown by sitting-drop vapour diffusion using polyethylene glycol 6000 as a precipitant. After optimization, crystals were grown which diffracted to 1.56 Å using synchrotron radiation. The results presented in this paper suggest that crystals producing diffraction of this quality will be suitable for structural determination of human CLEC5A.
1. Introduction
The myeloid DAP12-associated lectin (MDL-1; also known as CLEC5A) is a type II transmembrane receptor that is expressed on the surface of monocytes and macrophages (Bakker et al., 1999 ▶). It is a member of the C-type lectin superfamily and plays a role in the activation of macrophages (Bakker et al., 1999 ▶). CLEC5A has a short cytoplasmic domain, but interacts in the plasma membrane with the signalling adaptor DAP12, which has an immunoreceptor tyrosine-based activation motif (ITAM) that can be phosphorylated in its cytoplasmic domain (Bakker et al., 1999 ▶; Yim et al., 2001 ▶). This interaction depends upon a charge–charge interaction between an aspartic acid in the transmembrane region of DAP12 and a lysine in the transmembrane region of CLEC5A (Bakker et al., 1999 ▶). Dengue virus is the most common cause of arboviral disease worldwide and is a major cause of mortality in endemic regions (Wilder-Smith & Schwartz, 2005 ▶). CLEC5A has very recently been shown to interact directly with the dengue virion, thereby causing phosphorylation of DAP12 and triggering a signalling cascade which results in the release of proinflammatory cytokines (Chen et al., 2008 ▶). These cytokines are responsible for many of the pathophysiological changes seen in dengue-virus-induced disease (Pang et al., 2007 ▶). Structural characterization of CLEC5A will provide a rational basis for understanding how dengue virus interacts with host immune cells and should provide insight into the role of CLEC5A in dengue-virus-induced lethal disease, which currently accounts for approximately 20 000 deaths per year (Chen et al., 2008 ▶).
2. Materials and methods
2.1. Cloning and expression
A portion of DNA encoding the extracellular domain of human CLEC5A from residue 64 to 188 was amplified from peripheral blood cDNA using the polymerase chain reaction (PCR), cut with the restriction enzymes NdeI and EcoRI and cloned into the T7 vector pGMT7 (O’Callaghan et al., 1998 ▶, 2001 ▶). This plasmid (pOC563) provided a template for PCR amplification using the primers CO1428 (5′-TATATTCATATGTGCCCCAAAGACTGGGAATTTTATC) and CO1429 (5′-ATAATAGAATTCCTAGGCATTCTTCTCACAGATCCTGCG). The amplified product was cut with NdeI and EcoRI and cloned into pGMT7 cut with the same enzymes. The design of the resulting construct was based on a BLASTP search against the Protein Data Bank (PDB) using the protein sequence of CLEC5A as a probe (Altschul et al., 1997 ▶) and also on a homology model of CLEC5A generated by the SWISS-MODEL comparative protein-modelling program (Schwede et al., 2003 ▶). The resulting plasmid pOC579 encoded the extracellular domain of human CLEC5A from residues 71 to 187 (Fig. 1 ▶). Both pOC563 and pOC579 encode protein sequences that contain an additional start methionine at their N-termini. All PCRs were performed with cloned Pfu polymerase (Stratagene, La Jolla, California, USA). The sequences of all constructs were verified using a 3730xl DNA Analyzer (Applied Biosystems, Foster City, California, USA).
Figure 1.
Sequences of the proteins expressed by plasmids pOC563 and pOC579. The truncated protein expressed by plasmid pOC579 is underlined and also contains a start methionine at its N-terminus. Predicted paired cysteines are displayed in the same colour.
For expression, plasmids pOC563 and pOC579 were separately transformed into Escherichia coli strain BL21 (DE3) pLysS. Cells were grown in Luria–Bertani medium supplemented with 100 µg ml−1 ampicillin (Sigma, Gillingham, England) and protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopryanoside (Sigma, Gillingham, England) in mid-log phase growth (A 600 = 0.6). Cells were harvested by centrifugation 4 h post-induction.
Harvested cells were resuspended in ice-cold phosphate-buffered saline and lysed by sonication. Inclusion bodies were purified by repeated washes in 0.5% Triton X-100, 50 mM Tris pH 8.0, 100 mM NaCl, 0.1% sodium azide. Detergent removal was facilitated by washing in 50 mM Tris pH 8.0, 100 mM NaCl. The resultant protein was solubilized overnight in 6 M guanidine hydrochloride, 50 mM MES pH 6.5, 10 mM ethylene diaminetetraacetic acid (EDTA), 2 mM dithiothreitol. Protein purity was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and quantified using the Bio-Rad protein assay (Bio-Rad, Hercules, California, USA).
2.2. Refolding and purification
A range of refolding strategies were investigated to maximize the yield of correctly folded CLEC5A protein and to minimize aggregation. Variations in pH, redox-couple component ratios and arginine concentration were tested. Optimal refolding of protein expressed by both constructs, as assessed by size-exclusion gel filtration, was achieved by slow dilution of 52 mg guanidine-solubilized CLEC5A protein into 250 ml 1 M l-arginine, 200 mM Tris pH 8.0, 2 mM EDTA, 5 mM reduced glutathione, 2.5 mM oxidized glutathione, 0.1 M phenylmethylsulfonyl fluoride. The mixture was equilibrated at 277 K for 24 h with slow stirring and was subsequently concentrated under nitrogen to a volume of 5 ml over a 10 kDa exclusion membrane (Millipore, Bedford, Massachusetts, USA). Refolded protein was analyzed and purified by gel-filtration chromatography in 20 mM Tris pH 8.0, 150 mM NaCl with a Superdex 75 26/60 column on an ÄKTA Purifier (GE Healthcare, Uppsala, Sweden). The purity and molecular weight of all samples entered into crystallization trials were assessed by SDS–PAGE and mass spectroscopy, respectively. Liquid-chromatography electrospray ionization mass spectrometry was performed using a reversed-phase C4 column on an Ultima HPLC (Dionex, Sunnyvale, California, USA) connected to a Quadrupole Time-of-Flight Micromass spectrometer (Waters, Milford, Massachusetts, USA).
2.3. Crystallization of CLEC5A
Correctly folded protein expressed from the pOC563 and pOC579 constructs was concentrated to 13.8 or 18.4 mg ml−1, respectively, in 20 mM Tris pH 8.0, 150 mM NaCl for initial crystal trials. Preliminary crystallization screens were performed by sitting-drop vapour diffusion. Solutions promoting crystal growth were dispensed into 96-well Greiner plates (Greiner, Kremsmunster, Austria) using a Robbins Hydra (Robbins Scientific, Solihull, England). Screens sampled included Crystal Screen, Crystal Screen 2, Index, Grid Screen PEG 6000, Grid Screen PEG/LiCl, Grid Screen MPD, Grid Screen Ammonium Sulfate, PEG/Ion (Hampton Research, Aliso Viejo, California, USA) and Wizard I and II (Emerald BioSystems, Bainbridge Island, Washington, USA). A Cartesian dispensing robot and a Cartesian Microsys dispenser (Genomic Solutions, Huntingdon, England) were used to establish crystallization drops with 100 nl protein solution and 100 nl reservoir fluid. Plates were stored in a temperature-controlled TAP Homebase storage vault (The Automation Partnership, Royston, England) at 295 K (Walter et al., 2005 ▶). Crystals of CLEC5A protein expressed from the pOC563 plasmid formed reproducibly within 5 d in a variety of conditions. The largest single crystals formed in 10–30% polyethylene glycol 3350–8000 pH 5.5–9.5 and grew to maximum dimensions of 0.07 mm. Multiple optimization screens were established manually around the best crystal-forming conditions using CLEC5A protein expressed from the pOC579 plasmid. Optimization parameters that were varied included buffer composition and pH, PEG concentration and molecular weight, drop size and the protein solution:reservoir fluid ratio. Maximum crystal dimensions of 0.63 mm were achieved in 100 mM Tris pH 8.0, 30% PEG 6000. Repeat drops of this condition were dispensed onto polypropylene bridges and contained 2.5 µl reservoir fluid and 2.5 µl 18.4 mg ml−1 protein solution in 20 mM Tris pH 8.0, 150 mM NaCl.
2.4. Crystallographic studies
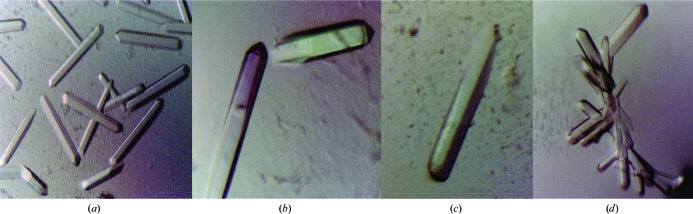
Crystals of CLEC5A protein expressed from the plasmid pOC579 grew in 100 mM Tris pH 8.0, 30% PEG 6000 either as long single crystals of several forms (Figs. 2 ▶ a, 2 ▶ b and 2 ▶ c) or as complex branching needles (Fig. 2 ▶ d). The large single crystals grew to a maximum length of 0.63 mm and were used in all subsequent diffraction studies. Diffraction to 1.56 Å resolution was achieved from a 0.63 mm single crystal grown for 3 d at 295 K. Crystals were frozen at 100 K in a nitrogen cryostream using perfluoropolyether oil as a cryoprotectant. X-ray diffraction data were collected using a MAR 345 detector on beamline 14 at the European Synchrotron Radiation Facility, Grenoble, France (wavelength = 0.976 Å) using 0.55° oscillations. Data collection was implemented using the DNA software package integrated with BM14 at the ESRF (Leslie et al., 2002 ▶). Here, the data-collection rotation range and oscillation angles were determined given the crystal unit cell, orientation, Laue group, spot size and mosaicity. A data set of 111 images was collected from one long three-day-old single crystal mounted in a 0.4–0.7 mm loop. The data were autoindexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
Figure 2.
Crystals of the C-type lectin-like domain of CLEC5A. (a)–(c) show large single crystals, while (d) shows a branching needle crystal form.
3. Results
CLEC5A was initially expressed using the construct pOC563. However, crystals of this protein achieved maximum dimensions of only 0.07 mm. A further truncated form of CLEC5A (pOC579) was therefore generated to increase the likelihood of protein crystallization. Here, the sequence encoded by pOC563 was used as a search template by the SWISS-MODEL comparative protein-modelling program (Schwede et al., 2003 ▶). The subsequent homology model generated by this program represented a construct 117 consecutive residues in length, spanning amino acids 71–187 of CLEC5A, and consisted of two antiparallel β-sheets flanked by a pair of α-helices. The pOC579 construct was designed to match the N- and C-termini of this homology model, with an additional start methionine added to its N-terminus. The protein expressed and purified from the pOC579 construct proved to be a better candidate for crystallography in several respects. The protein yield from plasmid pOC579 was approximately 50 mg per litre of culture and detergent washes of the inclusion bodies generated from this construct resulted in greater than 95% protein purity. Mass spectroscopy revealed that the protein expressed and purified from the pOC579 plasmid had a molecular weight of 13 813.0 Da. Using size-exclusion gel-filtration chomatography, this protein eluted at the size expected for a monomer (Supplementary Fig. 11) and generated long single crystals which produced diffraction data to 1.56 Å resolution. Using SCALEPACK, the space group of these crystals was designated as P31 (Table 1 ▶; Otwinowski & Minor, 1997 ▶). The assignment of this space group was based upon the results of scaling and indexing the data and cannot be formally confirmed until a plausible structural solution has been found. The crystal volume per unit weight (V M) was calculated to be 2.34 Å3 Da−1, with nine molecules per asymmetric unit and a solvent content of 47.38% (Table 1 ▶; Kantardjieff & Rupp, 2003 ▶). CLEC5A plays an important role in the pathophysiology of dengue-virus-induced disease. Diffraction data of this quality provide a good basis for determination of the structure of the virus-binding extracellular domain of CLEC5A.
Table 1. X-ray diffraction data for human CLEC5A.
Values in parentheses are for the last shell.
| Wavelength (Å) | 0.97625 |
| Space group | P31 |
| Unit-cell parameters | |
| a (Å) | 109.096 |
| b (Å) | 109.096 |
| c (Å) | 84.723 |
| Matthews coefficient (Å3 Da−1) | 2.34 |
| Resolution (Å) | 50.00–1.56 (1.62–1.56) |
| Rmerge† (%) | 3.8 (17.3) |
| Completeness (%) | 94.8 (88.9) |
| Average I/σ(I) | 21.2 (3.3) |
| No. of measured reflections | 273689 (23940) |
| No. of unique reflections | 152294 (14271) |
| Redundancy | 1.8 (1.7) |
R
merge = 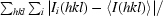
 .
.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309109047915/ll5191sup1.pdf
Acknowledgments
We are grateful to James Brown, Martin Walsh and Benjamin Hall for helpful discussions.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: LL5191).
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res.25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Bakker, A. B., Baker, E., Sutherland, G. R., Phillips, J. H. & Lanier, L. L. (1999). Proc. Natl Acad. Sci. USA, 96, 9792–9796. [DOI] [PMC free article] [PubMed]
- Chen, S. T., Lin, Y. L., Huang, M. T., Wu, M. F., Cheng, S. C., Lei, H. Y., Lee, C. K., Chiou, T. W., Wong, C. H. & Hsieh, S. L. (2008). Nature (London), 453, 672–676. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W., Powell, H. R., Winter, G., Svensson, O., Spruce, D., McSweeney, S., Love, D., Kinder, S., Duke, E. & Nave, C. (2002). Acta Cryst. D58, 1924–1928. [DOI] [PubMed]
- O’Callaghan, C. A., Cerwenka, A., Willcox, B. E., Lanier, L. L. & Bjorkman, P. J. (2001). Immunity, 15, 201–211. [DOI] [PubMed]
- O’Callaghan, C. A., Tormo, J., Willcox, B. E., Blundell, C. D., Jakobsen, B. K., Stuart, D. I., McMichael, A. J., Bell, J. I. & Jones, E. Y. (1998). Protein Sci.7, 1264–1266. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pang, T., Cardosa, M. J. & Guzman, M. G. (2007). Immunol. Cell Biol.85, 43–45. [DOI] [PubMed]
- Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. (2003). Nucleic Acids Res.31, 3381–3385. [DOI] [PMC free article] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]
- Wilder-Smith, A. & Schwartz, E. (2005). N. Engl. J. Med.353, 924–932. [DOI] [PubMed]
- Yim, D., Jie, H. B., Sotiriadis, J., Kim, Y. S. & Kim, Y. B. (2001). Cell. Immunol.209, 42–48. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309109047915/ll5191sup1.pdf
Supplementary material file. DOI: 10.1107/S1744309109047915/ll5191sup1.pdf