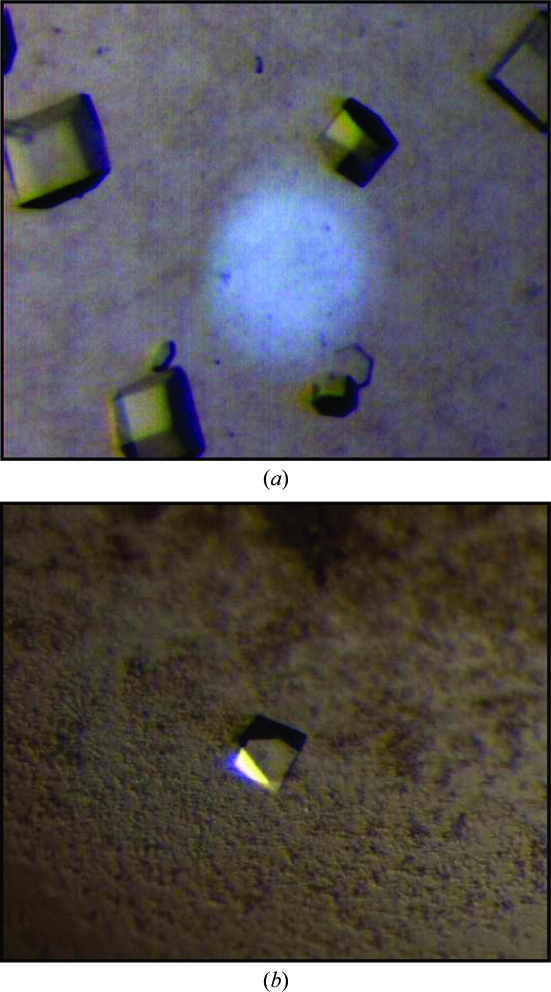
Crystals of S. elodea ATCC 31461 UDP-glucose dehydrogenase (EC 1.1.1.22) were obtained in space groups P622 and P43212 and diffracted to 2.4 and 3.4 Å resolution, respectively.
Keywords: Sphingomonas elodea, gellan gum, exopolysaccharides, UDP-glucose dehydrogenases
Abstract
Gellan gum, a commercial gelling agent produced by Sphingomonas elodea ATCC 31461, is a high-value microbial exopolysaccharide. UDP-glucose dehydrogenase (UGD; EC 1.1.1.22) is responsible for the NAD-dependent twofold oxidation of UDP-glucose to UDP-glucuronic acid, one of the key components for gellan biosynthesis. S. elodea ATCC 31461 UGD, termed UgdG, was cloned, expressed, purified and crystallized in native and SeMet-derivatized forms in hexagonal and tetragonal space groups, respectively; the crystals diffracted X-rays to 2.40 and 3.40 Å resolution, respectively. Experimental phases were obtained for the tetragonal SeMet-derivatized crystal form by a single-wavelength anomalous dispersion experiment. This structure was successfully used as a molecular-replacement probe for the hexagonal crystal form of the native protein.
1. Introduction
Bacterial exopolysaccharides (EPS) are biotechnology products that are of great interest owing to their rheological properties. This is the case for gellan gum, a multifunctional gelling agent that is produced in high yield by the nonpathogenic bacterium Sphingomonas elodea ATCC 31461. It has approval in the US and EU for food use as a gelling, stabilizing and suspending agent, either alone or in combination with other hydrocolloids. In its native form, gellan is a linear high-molecular-weight anionic EPS based on a tetrasaccharide repeat unit composed of two molecules of d-glucose, one of l-rhamnose and one of d-glucuronic acid. The native gellan is partially esterified with acyl substituents (1 mol glycerate and 0.5 mol acetate) per repeat unit (Jay et al., 1998 ▶). Gellan and gellan-like polymers have unique characteristics and have found many applications, particularly in the food, pharmaceutical and biomedical fields (Sá-Correia et al., 2002 ▶; Fialho et al., 2008 ▶).
Owing to the broadness of its commercial applications, the biosynthetic pathway leading to the production of gellan has gained a significant amount of interest. Gellan-gum biosynthesis is a multi-step process that starts with production of the nucleotide-sugar precursors uridine-5′-diphosphoglucose (UDP-glucose or UDP-Glc), UDP-glucuronic acid (UDP-GlcA) and thymine-diphospho-l-rhamnose (dTDP-l-rhamnose or dTDP-LRha), followed by formation of the tetrasaccharide repeating unit, gellan polymerization and export (Aragão et al., 2007 ▶; Sá-Correia et al., 2002 ▶). Three gene clusters that are necessary for gellan production have been described (Harding et al., 2004 ▶), but interestingly the genes encoding the enzymes for UDP-Glc and UDP-GlcA synthesis (pgmG, ugpG and ugdG) are located at different positions of the S. elodea genome, perhaps indicating a broader function of these enzymes (Harding et al., 2004 ▶; Silva et al., 2005 ▶; Aragão et al., 2007 ▶). The identification of key genes such as those mentioned above and the elucidation of the crucial steps in the gellan pathway indicate that possibilities now exist for exerting control of gellan production by acting, for example, on the expression-level activity of these genes/enzymes. Our interest in the S. elodea ATCC 31461 UDP-glucose dehydrogenase, termed UgdG (EC 1.1.1.22), derives from its crucial role in producing UDP-GlcA, which is one of the precursors for gellan biosynthesis. In this work, we report the crystallization and phase-problem determination of UgdG from S. elodea ATCC 31461.
2. Materials and methods
2.1. Cloning and expression
The complete sequence of the ugdG gene from S. elodea ATCC 31461 was amplified by the polymerase chain reaction (PCR) using the cosmid pCG8 as the template DNA (Granja et al., 2007 ▶), Pwo DNA polymerase (Roche Diagnostics, Mannheim, Germany) and the oligonucleotides UGD1 (5′AAAGGATCCGTGCGCATTGCGATG-3′) and UGD2 (5′-AAAAAGCTTTCAGTCGCGGCTCG-3′), which were designed based on the ugdG nucleotide sequence. The PCR product (1320 bp) was digested with BamHI and HindIII (recognition sites are shown in bold in the primers used) and cloned into the vector pWH844 (Schirmer et al., 1997 ▶), generating pUgdG.
This plasmid carries the ugdG gene preceded by a sequence coding for six histidine residues that are present for purification purposes. The DNA insert cloned in pUgdG was sequenced to confirm the fidelity of DNA amplification.
Overexpression of the ugdD gene was carried out by cultivation of Escherichia coli BL21 transformants harbouring the plasmid pUgdG in 600 ml LB medium supplemented with 100 mg ml−1 ampicillin at 310 K until an OD600nm of 0.6 was reached. The cells were then induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 3 h and harvested by centrifugation (10 000g, 30 min, 277 K); the pellets obtained were stored at 253 K.
For the production of SeMet-derivatized UgdG protein (Molecular Dimensions protocol), methionine-auxotrophic strain E. coli B843 (DE3) carrying pUgdG was cultivated in 600 ml LB medium (supplemented with 100 mg ml−1 ampicillin) at 310 K until an OD600nm of 0.6 was reached. The cells were harvested by centrifugation (10 000g, 10 min, 277 K) and gently resuspended in SelenoMet Medium Base (Molecular Dimensions, UK). This process was repeated three times to remove any traces of LB medium containing methionine. Finally, the cells were resuspended in SelenoMet medium and 250× concentrated selenomethionine solution was added (Molecular Dimensions, UK). Induction occurred 3 h after the addition of 0.3 mM IPTG and the cells were harvested by centrifugation (10 000g, 30 min, 277 K). The supernatant was discarded and the pellet was stored at 253 K.
2.2. Purification
The following purification protocol was used for both the native and SeMet-derivative UgdG proteins. Cells were resuspended in 20 mM Na3PO4 pH 7.4, 20 mM imidazole, 1 M NaCl and disrupted in a French press. Crude cell extract was obtained by centrifugation at 27 000g for 40 min at 277 K and the supernatant was applied onto a 5 ml HisTrap column (GE Healthcare) pre-equilibrated with 20 mM Na3PO4 pH 7.4, 20 mM imidazole and 1 M NaCl connected to an ÄKTA Explorer Instrument (GE Healthcare) according to the manufacturer’s recommendations.
The column was washed with buffer A (20 mM Na3PO4 pH 7.4, 20 mM imidazole, 1 M NaCl) to remove any unbound protein and an imidazole gradient (20–500 mM) was applied. Recombinant His6-UgdG protein eluted at approximately 250 mM imidazole in a symmetrical chromatography peak. Imidazole was removed from the eluted fractions of the UgdG protein, replacing the buffer with 25 mM Tris–HCl pH 8.3, 50 mM NaCl, 2.5 mM dithiothreitol (DTT) supplemented with 1 mM oxidized nicotinamide adenine dinucleotide (NAD+) or 0.5 mM UDP-GlcA and 1 mM NAD+ using a PD10 desalting column (GE Healthcare).
In the case of SeMet-derivatized UgdG protein, the eluted fractions were pooled together and buffer-exchanged with 25 mM Tris–HCl pH 8.3, 50 mM NaCl, 2.5 mM DTT and 1 mM NAD+, also using a PD10 desalting column (GE Healthcare).
Both native UgdG protein and SeMet-derivative UgdG protein were concentrated in Vivapore 10/20 concentrators (Vivascience Ltd, UK) to 4.5 and 5.5 mg ml−1, respectively, before storage at 193 K.
The composition and purity of both proteins were confirmed to be homogenous by SDS–PAGE. Protein concentration was determined by the Bradford method (Bradford, 1976 ▶) and by measurement of the absorbance at 280 nm in a NanoDrop ND-1000 spectrophotometer using a extinction coefficient of 18 490 M −1 cm−1.
2.3. Crystallization
An initial crystallization screen for native UgdG in the presence of NAD+ was performed in batch mode with an Oryx6 crystallization robot (Douglas Instruments) using solutions from the Classics Screen (Qiagen Canada Inc., Montreal, Canada). Drops consisting of 0.5 µl protein solution (4.5 mg ml−1 in 25 mM Tris–HCl pH 8.3, 50 mM NaCl, 2.5 mM DTT and 1 mM NAD+) plus 0.5 µl precipitant solution were equilibrated at 293 K under oil. Crystals formed within 24 h using conditions H5 [200 mM Li2SO4, 100 mM Tris–HCl pH 8.5 and 30%(v/v) PEG 4K] and A7 [100 mM sodium citrate pH 5.6, 20% 2-propanol and 20%(v/v) PEG 4K]. The crystals that appeared in condition H5 had dimensions of up to 0.3 × 0.15 × 0.01 mm (thin plates) but could not be reproduced. Other, mostly hexagonal shaped, prismatic crystals appeared in condition A7; these crystals were reproducible and could be further optimized (see Table 1 ▶ and Fig. 1 ▶ a). Crystal cryoprotection was achieved by passing crystals through a solution of their mother liquor including 25%(v/v) glycerol.
Table 1. Optimized crystallization conditions for native and SeMet-derivative UgdG proteins.
| Crystal | Precipitant solution | Drop volume: protein + precipitant (ml) | Time (h) | Temperature (K) | Cryoprotectant | Other notes |
|---|---|---|---|---|---|---|
| Native | 0.1 M Na citrate pH 5.6, 20%(w/v) PEG 4K, 10%(v/v) 2-propanol | 1 + 1 | 24 | 293 | Mother liquor with 25%(v/v) glycerol | Microbatch, additive 0.05 mM NaF |
| SeMet derivative | 0.1 M MES pH 6.5, 40%(w/v) PEG 200 | 1 + 1 | 24–48 | 293 | Mother liquor with 25%(v/v) PEG 400 | Vapour diffusion |
Figure 1.
Crystals of (a) native UgdG in a hexagonal space group (approximate dimensions of 0.04, 0.04, 0.06 mm) and (b) SeMet-derivative UgdG in a tetragonal space group (approximate dimensions of 0.05, 0.05, 0.08 mm).
Crystallization screens for the SeMet-derivatized protein were performed with a ‘Minibee’ Microsys 4000 XL Cartesian Dispensing Systems robot (Genomic Solutions, USA) using the vapour-diffusion method. Drops consisting of 100 nl 5.5 mg ml−1 protein solution in 25 mM Tris–HCl pH 8.3, 50 mM NaCl, 2.5 mM DTT, 1 mM NAD+ and 100 nl precipitant solution were equilibrated against 100 µl of 672 precipitant-solution conditions from the crystallization screens Classics, PEG, MbClass, MbClass II, pHClear, pHClear II and MPD screens from Qiagen Canada Inc. (Montreal, Canada). Crystalline precipitate was found in several conditions from different screens. Of these, solution A7 from the PEG screen [100 mM MES pH 6.5 and 40%(v/v) PEG 200] produced the most promising crystallites. Upon optimization (see Table 1 ▶), octahedral-like crystals appeared within 24–48 h and grew to approximate dimensions of 0.05 × 0.05 × 0.08 mm (Fig. 1 ▶ b) in drops that also contained other precipitated material. Crystals had to be ‘washed’ from the precipitate with mother liquor prior to cryoprotection, which was achieved by quick soaking in mother liquor also including 20%(v/v) PEG 400. Crystals were then flash-cooled and stored in liquid nitrogen for diffraction data collection.
2.4. X-ray diffraction
The diffraction from cryoprotected crystals in a nitrogen stream at 100 K was measured using the European Synchrotron Radiation Facility (ESRF; Grenoble, France) at stations ID23-1 (UgdG) and ID14-3 (SeMet-UgdG). The diffraction images from native protein crystals were integrated with MOSFLM (Leslie, 1992 ▶) and the corresponding intensities were scaled and merged together using SCALA (Evans, 2006 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The diffraction images of the SeMet-derivatized protein were processed with DENZO and the intensities were scaled and merged with SCALEPACK (Otwinowski & Minor, 1997 ▶). The final magnitudes for both crystals were calculated with TRUNCATE (French & Wilson, 1978 ▶).
Crystallographic data and processing statistics are presented in Table 2 ▶.
Table 2. Crystallographic parameters and data-collection statistics.
Values in parentheses are for the outer resolution shell.
| Crystal | UgdG | SeMet-UgdG |
|---|---|---|
| ESRF beamline | ID23-1 | ID14-3 |
| Wavelength (Å) | 1.074 | 0.931 |
| Resolution (Å) | 45.98–2.40 (2.53–2.40) | 48.76–3.40 (3.52–3.40) |
| Space group | P622 | P43212 |
| Unit-cell parameters (Å) | a = b = 167.4, c = 84.4 | a = b = 109.0, c = 175.8 |
| No. of reflections | 335219 (48456) | 197981 (12797) |
| No. of unique reflections | 27740 (3957) | 15086 (1444) |
| Redundancy | 12.1 (12.2) | 13.1 (13.5) |
| I/σ(I) | 5.2 (1.7) | 25.1 (3.78) |
| Rp.i.m.† | 0.029 (0.129) | 0.023 (0.204) |
| Rr.i.m. = Rmeas‡ | 0.102 (0.460) | 0.085 (0.763) |
| Rmerge§ | 0.098 (0.441) | 0.082 (0.729) |
| Ranom¶ | N/A | 0.036 |
| Completeness (%) | 100.0 (100.0) | 99.2 (99.5) |
| Mosaicity (°) | 0.28 | 0.65 |
| VM (Å3 Da−1) | 3.62 | 2.65 |
| Solvent content (%) | 0.66 | 0.53 |
| No. of molecules in ASU | 1 | 2 |
| Wilson B (Å2) | 44.4 | 85.0 |
R
p.i.m. = 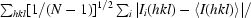
 , where N is the data redundancy, I(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the precision of the final merged and averaged data set (Weiss, 2001 ▶).
, where N is the data redundancy, I(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the precision of the final merged and averaged data set (Weiss, 2001 ▶).
R
r.i.m. = R
meas = 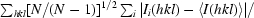
 , where N is the data redundancy, I(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the average spread of the individual measurements (Weiss, 2001 ▶).
, where N is the data redundancy, I(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections. It is an indicator of the average spread of the individual measurements (Weiss, 2001 ▶).
R
merge = 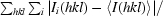
 , where I(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections.
, where I(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections.
R
anom = 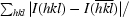
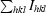 expresses the magnitude of the anomalous signal.
expresses the magnitude of the anomalous signal.
2.5. Phase-problem determination
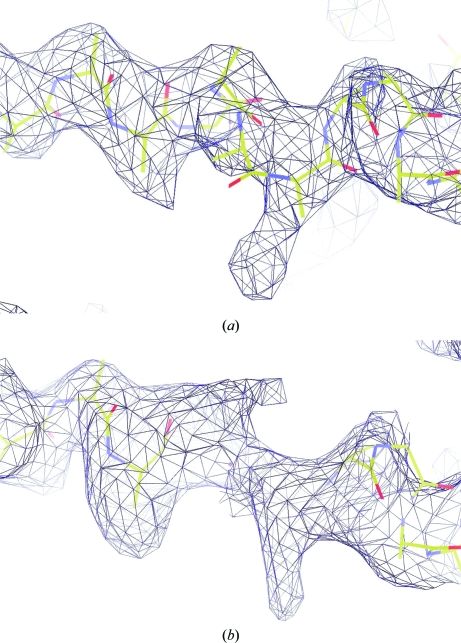
Assuming the presence of one molecule in the asymmetric unit and according to the resolution-dependent distribution of the Matthews coefficient (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶), the hexagonal form of the S. elodea UgdG crystal has a calculated solvent content of 66% and a V M of 3.6 Å3 Da−1 with a probability of 89%. Initial attempts to obtain phases by molecular replacement using UGD from Streptococcus pyogenes (PDB entry 1dlj; Campbell et al., 2000 ▶), which has a sequence identity of 23% to UgdG, as a model were unsuccessful. Single-wavelength anomalous dispersion (SAD) phasing was thus used with the diffraction data from the selenomethionine-derivatized tetragonal crystal form of S. elodea UgdG. This has an estimated solvent content of 53% and a V M of 2.6 Å3 Da−1, assuming the presence of two molecules in the asymmetric unit, with an estimated probability of 89% (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶). The automated protocol in SOLVE (Terwilliger, 2004 ▶) determined 18 selenium sites, as expected for two molecules in the asymmetric unit, and calculated initial experimental phases and electron-density maps. The Se-atom substructure allowed the determination of the NCS operator within the asymmetric unit, which was used by the programs DM (Cowtan & Main, 1998 ▶) and RESOLVE (Terwilliger, 2004 ▶) to improve the experimental phases via NCS averaging of the electron density to a figure of merit of 0.83. RESOLVE (Terwilliger, 2004 ▶) and FFEAR (Cowtan, 1998 ▶) were further used to localize protein-chain fragments, mainly as poly-Ala chains. Their inspection with Xfit (McRee, 1999 ▶) on a stereo-graphics workstation taking the NCS operator into account allowed resolution of their twofold degeneracy and an initial model of the S. elodea UgdG monomer, with 346 out of 438 residues (albeit including 318 poly-Ala residues), was accordingly produced (Fig. 2 ▶). Molecular replacement was attempted once more on the 2.4 Å native data, but this time using this preliminary UgdG monomeric model derived from the selenomethionine data as the search structure. Phaser (McCoy et al., 2007 ▶) successfully located the monomer position in the asymmetric unit of the hexagonal crystal (final Z scores RFZ = 3.5 and TFZ = 9.1). The validity of the solution was corroborated by examination of OMIT maps (Fig. 2 ▶ b). Model completion and structural refinement of the UDP-glucose dehydrogenase from S. elodea ATCC 31461 is in progress.
Figure 2.
Electron-density maps showing a helical motif section (bonds shown as sticks, with carbon in yellow, nitrogen in blue and oxygen in red) of S. elodea UgdG (a) from the experimental SeMet-derivative σA map (Read, 1986 ▶) obtained by RESOLVE (Terwilliger, 2004 ▶) after twofold NCS averaging at 3.4 Å resolution and contoured at 2σ (dark blue mesh) and (b) from the molecular-replacement solution obtained by Phaser (McCoy et al., 2007 ▶): a σA OMIT map (Read, 1986 ▶) at 2.4 Å resolution contoured at 1.5σ (dark blue mesh), where four residues of the helical motif were omitted from the model in the map calculation using REFMAC5 (Murshudov et al., 1997 ▶).
Acknowledgments
The authors gratefully acknowledge the ESRF, Grenoble for provision of synchrotron radiation. JR is the recipient of a PhD fellowship from Fundação para a Ciência e Tecnologia (FCT), Portugal, SFRH/BD/24216/2005. This work was partially supported by FCT grants POCTI/BME/38859/2001, POCI/BIO/58401/2004 and PTDC/QUI/67925/2006.
References
- Aragão, D., Fialho, A. M., Marques, A. R., Mitchell, E. P., Sá-Correia, I. & Frazão, C. (2007). J. Bacteriol.189, 4520–4528. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Campbell, R. E., Mosimann, S. C., van De Rijn, I., Tanner, M. E. & Strynadka, N. C. (2000). Biochemistry, 39, 7012–7023. [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cowtan, K. (1998). Acta Cryst. D54, 750–756. [DOI] [PubMed]
- Cowtan, K. & Main, P. (1998). Acta Cryst. D54, 487–493. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fialho, A. M., Moreira, L. M., Granja, A. T., Popescu, A. O., Hoffmann, K. & Sá-Correia, I. (2008). Appl. Microbiol. Biotechnol.79, 889–900. [DOI] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Granja, A. T., Popescu, A., Marques, A. R., Sá-Correia, I. & Fialho, A. M. (2007). Appl. Microbiol. Biotechnol.76, 1319–1327. [DOI] [PubMed]
- Harding, N. E., Patel, Y. N. & Coleman, R. J. (2004). J. Ind. Microbiol. Biotechnol.31, 70–82. [DOI] [PubMed]
- Jay, A. J., Colquhoun, I. J., Ridout, M. J., Brownsey, G. J., Morris, V. V., Fialho, A. M., Leitao, J. H. & Sá-Correia, I. (1998). Carbohydr. Polym.35, 179–188.
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McRee, D. E. (1999). J. Struct. Biol.125, 156–165. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Read, R. J. (1986). Acta Cryst. A42, 140–149.
- Sá-Correia, I., Fialho, A. M., Videira, P., Moreira, L. M., Marques, A. R. & Albano, H. (2002). J. Ind. Microbiol. Biotechnol.29, 170–176. [DOI] [PubMed]
- Schirmer, F., Ehrt, S. & Hillen, W. (1997). J. Bacteriol.179, 1329–1336. [DOI] [PMC free article] [PubMed]
- Silva, E., Marques, A. R., Fialho, A. M., Granja, A. T. & Sá-Correia, I. (2005). Appl. Environ. Microbiol.71, 4703–4712. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. (2004). J. Synchrotron Rad.11, 49–52. [DOI] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.