This article describes the first successful crystallization of components of eukaryotic ribonucleases P/MRP. Yeast RNase MRP RNA domain P3 was crystallized in a complex with the proteins Pop6 and Pop7; the crystals diffracted to 3.25 Å resolution.
Keywords: ribonuclease MRP, ribonuclease P, ribozyme, Pop6, Pop7, P3 domain, Saccharomyces cerevisiae
Abstract
Eukaryotic ribonucleases P and MRP are closely related RNA-based enzymes which contain a catalytic RNA component and several protein subunits. The roles of the protein subunits in the structure and function of eukaryotic ribonucleases P and MRP are not clear. Crystals of a complex that included a circularly permuted 46-nucleotide-long P3 domain of the RNA component of Saccharomyces cerevisiae ribonuclease MRP and selenomethionine derivatives of the shared ribonuclease P/MRP protein components Pop6 (18.2 kDa) and Pop7 (15.8 kDa) were obtained using the sitting-drop vapour-diffusion method. The crystals belonged to space group P4222 (unit-cell parameters a = b = 127.2, c = 76.8 Å, α = β = γ = 90°) and diffracted to 3.25 Å resolution.
1. Introduction
Ribonuclease (RNase) P is a universal RNA-based enzyme that is found in all kingdoms of life. RNase P is responsible for processing a number of RNA substrates including pre-tRNA and is involved in the regulation of transcription (Guerrier-Takada et al., 1983 ▶; Altman & Kirsebom, 1999 ▶; Reiner et al., 2006 ▶, 2008 ▶; Kazantsev & Pace, 2006 ▶; Coughlin et al., 2008 ▶). A typical RNase P consists of an RNA component which is responsible for catalysis and one or more protein components (Walker & Engelke, 2006 ▶; Kikovska et al., 2007 ▶); all of the protein components are required for the activity of the enzyme in vivo and are essential. The protein moiety of eubacterial RNase P contains one small protein, while archaeal RNase P has at least four proteins and eukaryotic RNase P has even more proteins (nine in Saccharomyces cerevisiae RNase P). The reasons for the increased complexity of the more evolutionarily advanced RNases P are not clear (Walker & Engelke, 2006 ▶; Marvin & Engelke, 2009 ▶).
RNase MRP is a site-specific eukaryotic endoribonuclease (Chang & Clayton, 1987 ▶). RNase MRP has several known functions in the cell. The vast majority of RNase MRP is located in the nucleolus, where it is involved in processing precursor ribosomal RNA (pre-rRNA; Lygerou et al., 1996 ▶; Lindahl et al., 2009 ▶). RNase MRP is also involved in regulating cell-cycle progression by cleaving the 5′-untranslated region (5′-UTR) of CLB2 mRNA, which encodes a B-type cyclin, thus triggering the degradation of this mRNA and aiding cell-cycle progression (Gill et al., 2004 ▶). RNase MRP is a universal eukaryotic enzyme which is required for the survival of the eukaryotic cell. RNase MRP closely resembles eukaryotic RNase P and consists of a presumably catalytic RNA component which shares multiple elements with RNase P as well as a number of protein components, most of which are found in both enzymes.
In S. cerevisiae, RNases MRP and P share eight proteins: Pop1 (100.5 kDa; Lygerou et al., 1994 ▶), Pop3 (22.6 kDa; Dichtl & Tollervey, 1997 ▶), Pop4 (32.9 kDa; Chu et al., 1997 ▶), Pop5 (19.6 kDa), Pop6 (18.2 kDa), Pop7 (15.8 kDa), Pop8 (15.5 kDa; Chamberlain et al., 1998 ▶) and Rpp1 (32.2 kDa; Stolc & Altman, 1997 ▶). RNase MRP has two unique proteins [Snm1 (22.5 kDa; Schmitt & Clayton, 1994 ▶) and Rmp1 (23.6 kDa; Salinas et al., 2005 ▶)], while RNase P has one unique protein, Rpr2 (16.3 kDa; Chamberlain et al., 1998 ▶), a homolog of Snm1. Similar to RNase P, all RNase MRP proteins are essential for the viability of the cell.
The RNA component of S. cerevisiae RNase P is 369 nucleotides in length, while the RNA component of RNase MRP is 340 nucleotides in length. The secondary-structure elements which comprise the putative catalytic center are very similar in RNase P and RNase MRP. The results of a footprinting analysis indicate similar RNA–protein interactions in RNase MRP and eukaryotic RNase P (Esakova et al., 2008 ▶). The similarity between RNase MRP and eukaryotic RNase P strongly suggests that these enzymes have a common ancestor and share a catalytic mechanism but have evolved to have different specificities.
The structural organization of eukaryotic RNases P/MRP is not clear. There is no available structural information on any of their components.
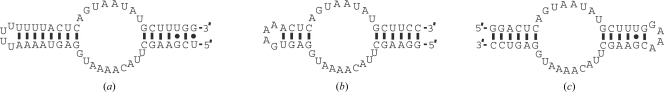
The RNA components of RNases P and MRP have several well defined structural elements (Frank et al., 2000 ▶; Li et al., 2002 ▶; Walker & Avis, 2004 ▶). The P3 domain of the RNA component (Fig. 1 ▶ a) appears to play a unique and crucial role in RNase MRP and eukaryotic RNase P. It is found in practically all eukaryotic RNases P and RNases MRP but not in bacterial or archaeal enzymes. This domain is absolutely essential and is phylogenetically conserved (Lindahl et al., 2000 ▶; Ziehler et al., 2001 ▶). The P3 domain is expected to be a hub for protein binding in the eukaryotic enzymes of the RNase MRP/RNase P family, with several protein components, including Pop1, Pop6 and Pop7, binding to it (Ziehler et al., 2001 ▶; Perederina et al., 2007 ▶).
Figure 1.
(a) The P3 domain of S. cerevisiae RNase MRP RNA; (b) a representative construct from the first set of P3-domain RNAs; (c) the construct from the second set of P3-domain RNAs which crystallized with the Pop6–Pop7 heterodimer.
The crystals reported here will be used to determine the molecular structure of the RNase MRP P3-domain RNA in a complex with RNase P/RNase MRP protein components Pop6 and Pop7 and will provide the first glimpse of the structural organization of the eukaryotic enzymes of the RNase P/RNase MRP family.
2. Materials and methods
2.1. Preparation of Pop6–Pop7–RNA complexes
The production of P3-domain RNA by T7 polymerase-driven run-off transcription has been described previously by Perederina et al. (2007 ▶). Full-length S. cerevisiae Pop6 (YRG030C, NCBI NP_011544) and Pop7 (YBR167C, NCBI NP_009726) proteins were cloned, co-expressed in Escherichia coli and purified as described in Perederina et al. (2007 ▶). The expressed proteins had wild-type sequences without any additions or deletions. Co-expression of Pop6 and Pop7 was necessary for the solubility of Pop7; when co-expressed, Pop6 and Pop7 formed a heterodimer (Perederina et al., 2007 ▶).
A selenomethionine (SeMet) derivative of the Pop6–Pop7 heterodimer was obtained using E. coli strain BL21 and PASM-5052 SeMet-supplemented autoinducing media (Studier, 2005 ▶). The efficiency of SeMet incorporation was checked using mass spectrometry and was found to be better than 95%. The purification of the SeMet-containing Pop6–Pop7 complex did not require any modification of the original protocol (Perederina et al., 2007 ▶). The purified Pop6–Pop7 heterodimer remained stable at 277 K in a buffer containing 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 50 mM KCl, 2 mM DTT, 0.1 mM Na EDTA and 0.1 mM PMSF for several weeks.
Prior to formation of the RNA–protein complex, the RNA was refolded by incubation at 358 K for 2 min in 10 mM Tris–HCl pH 7.4 and cooling to room temperature in a styrofoam rack followed by incubation at 323 K for 10 min in the presence of 5 mM MgCl2 and subsequent cooling to room temperature in a styrofoam rack. KCl was then added to the RNA solution to 200 mM. For the formation of the Pop6–Pop7–RNA complex, 13 µl RNA fragment (5 mg ml−1) in 10 mM Tris–HCl pH 7.4, 5 mM MgCl2, 200 mM KCl was mixed with 7 µl of the Pop6–Pop7 heterodimer (20 mg ml−1) in 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 50 mM KCl, 2 mM DTT, 0.1 mM Na EDTA and 0.1 mM PMSF at room temperature, followed by incubation at 301 K for 15 min. The final RNA–protein complex solution that was used in crystallization contained 3.5 mg ml−1 RNA, 7 mg ml−1 Pop6–Pop7 heterodimer, 10 mM Tris–HCl pH 7.4, 3.3 mM MgCl2, 150 mM KCl, 35 mM NaCl, 0.7 mM DTT, 0.04 mM Na EDTA and 0.04 mM PMSF.
2.2. Crystallization of the Pop6–Pop7 heterodimer–P3-domain RNA complex
Crystallization screening was performed using both commercially available and home-made crystallization screens. Two sets of RNA constructs were used in our crystallization attempts (Figs. 1 ▶ b and 1 ▶ c; see §3.1). The use of a permuted construct from the second set (Fig. 1 ▶ c) resulted in successful crystallization.
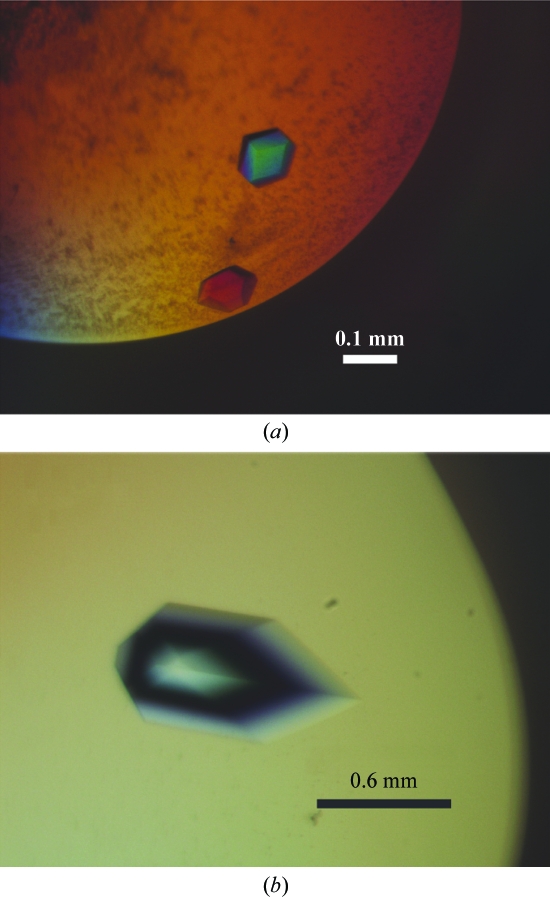
The initial crystallization conditions [sitting-drop method; 2 M ammonium sulfate, 2%(v/v) PEG 400, 100 mM Na HEPES pH 7.5; the equilibration solution in the well was supplemented with 100 mM NaCl and the crystallization temperature was 292 K] yielded crystals which grew to typical dimensions of 0.1 × 0.1 × 0.1 mm (Fig. 2 ▶ a) in 2–5 d and which diffracted to 4.0 Å resolution using an in-house X-ray generator (data not shown).
Figure 2.
Crystals of the P3-domain RNA complexed with Pop6–Pop7. (a) Before optimization of the crystallization conditions; (b) after optimization.
To improve the quality of the crystals, screening for additives and fine-tuning of the crystallization conditions were performed. Upon optimization of the crystallization conditions the crystals grew to typical dimensions of 0.65 × 0.65 × 0.65 mm (Fig. 2 ▶ b). The final crystallization solution was composed of 2 M ammonium sulfate, 2%(v/v) PEG 400, 100 mM HEPES–NaOH pH 7.8, 2 mM ZnCl2 and 5%(w/v) d-trehalose; the equilibration solution in the well was supplemented with 100 mM NaCl.
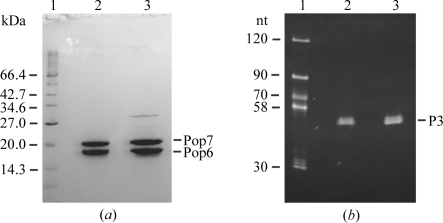
To verify the presence of all the expected components in the crystals, four large crystals were harvested and washed for 1.5 h in 100 µl drops containing mother-liquor solution without RNA and proteins. In the course of this procedure, the crystals were transferred to new mother-liquor drops five times to eliminate RNA and protein carry-over. The washed crystals were dissolved in 20 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM MgCl2, 0.1 mM DTT. The sample was divided into two parts. The first part was analyzed for the presence of the proteins by electrophoresis in a 15% denaturing SDS–polyacrylamide gel. The analysis showed that the crystals did contain the intact Pop6 and Pop7 proteins (Fig. 3 ▶ a; it is interesting to note that crystallization resulted in additional purification of the proteins). The second part of the dissolved crystals sample was used to test for the presence and integrity of the P3-domain RNA. It was run on a 15% denaturing polyacrylamide gel with 8 M urea; upon completion of the electrophoresis the gel was stained with ethidium bromide and photographed in UV light (Fig. 3 ▶ b). This analysis confirmed the presence of the intact P3-domain RNA in the crystals.
Figure 3.
Analysis of the crystal content. (a) Protein analysis. Coomassie-stained 15% denaturing SDS–polyacrylamide gel. Lane 1, markers; lane 2, dissolved crystals; lane 3, purified Pop6–Pop7 heterodimer used in crystallization. Pop7 is known to have anomalously low mobility on SDS gels (Perederina et al., 2007 ▶). (b) RNA analysis. Ethidium bromide-stained 15% denaturing (8 M urea) polyacrylamide gel. Lane 1, markers; lane 2, dissolved crystals; lane 3, initial preparation of the P3-domain RNA used in crystallization.
2.3. Data collection
The crystals were harvested and transferred into a cryoprotectant solution which included all of the components of the crystallization mother liquor supplemented with 40%(w/v) xylitol and were then flash-cooled and stored in liquid nitrogen.
Initial data-set collection and subsequent crystal screening were performed using an in-house rotating-anode generator. The final data-set collection was performed on NSLS beamline X25 (Brookhaven, USA) at 100 K using an ADSC Quantum 315 CCD detector. The crystals proved to be highly sensitive to radiation damage and careful planning of the data-set collection strategy was essential in order to obtain complete data sets. The oscillation range and the number of images required to obtain anomalously complete data sets were optimized using MOSFLM (Collaborative Computational Project, Number 4, 1994 ▶). To optimize the exposure time, one of the crystals was used to collect a test data set with a long (1 min per frame) exposure time; the data set was scaled in real time as the data collection progressed and the maximum allowable total exposure time before the appearance of radiation damage was estimated. The exposure time per frame used for the actual data collection was determined based on the maximal allowable exposure time and the required number of frames.
Since the crystals contained SeMet, three-wavelength data sets were collected (λ = 0.9787, 0.9710 and 0.9792 Å, corresponding to the peak, high remote and inflection point of the Se K edge). In addition, single-wavelength data sets (λ = 0.9787 Å, peak) were collected with longer exposure times to facilitate structure determination.
3. Results and discussion
3.1. Selection of the RNA construct for crystallization
The crystallization of complexes containing a significant fraction of relatively long RNA tends to be challenging and often requires approaches which are not usually applied to the crystallization of protein-only molecules. Instead of screening a very large number of crystallization conditions, it is a common tactic in the field to attempt the crystallization of multiple constructs from different organisms or to introduce modifications in the RNA that should not affect the fold of the region of interest (Spitale & Wedekind, 2009 ▶). Since it is not uncommon to have a reliable prediction for the helical stems in the RNA of interest, the modification of such stems often allows one to screen through a number of similar RNA constructs, thus multiplying the chances of successful crystallization. We used this approach in our crystallization of the Pop6–Pop7 heterodimer–P3-domain RNA complex.
The secondary structure of the P3 domain is well established on the basis of phylogenetic covariation analysis (Lindahl et al., 2000 ▶; Ziehler et al., 2001 ▶; Piccinelli et al., 2005 ▶). The P3 domain contains a single-stranded loop situated between two helical stems (Fig. 1 ▶ a). The results of footprinting studies and mutational analysis indicate that the terminal part of the distal (left in Fig. 1 ▶ a) helical stem is not involved in interactions with proteins and is dispensable (Shadel et al., 2000 ▶; Li et al., 2004 ▶; Esakova et al., 2008 ▶). A biochemical characterization of the P3-domain RNA–Pop6–Pop7 complex also indicates that the terminal part of the proximal (right in Fig. 1 ▶ a) helical stem of the P3 domain is not involved in interactions with Pop6–Pop7 (Perederina et al., 2007 ▶). Combined, these results indicated the possibility of manipulation of the terminal parts of the helical stems in the P3 domain without interfering with Pop6–Pop7 binding.
Two sets of RNA constructs were produced for crystallization trials. The first set included variants of the P3 domain with the terminal UUUU loops replaced by a GAAA tetraloop. The lengths of both helical stems varied from construct to construct. The first two 5′-end nucleotides were replaced by GG to facilitate in vitro transcription with T7 polymerase and matching mutations were introduced into the complementary part of the 3′-terminus of the construct (Fig. 1 ▶ b). The second set of constructs included a circularly permuted P3 domain (Fig. 1 ▶ c). The lengths of the helical stems also varied from construct to construct. As expected, all tested RNA constructs of both types bound the Pop6–Pop7 heterodimer similar to the wild-type P3 domain or whole-length RNase MRP RNA (Perederina et al., 2007 ▶).
The complexes containing the first set of RNA constructs (Fig. 1 ▶ b) failed to produce promising crystals, while the use of one of the constructs from the second set (permuted; Fig. 1 ▶ c) resulted in successful crystallization.
3.2. Preliminary X-ray analysis
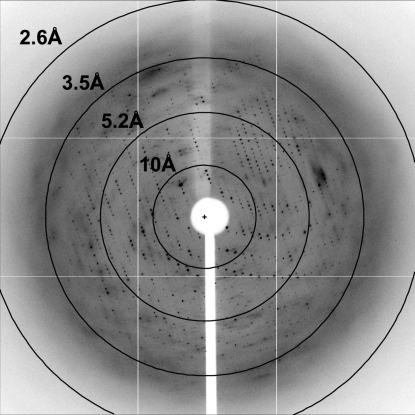
The best crystals diffracted to 3.25 Å resolutin (Table 1 ▶, Fig. 4 ▶). Analysis of the data sets using POINTLESS (Evans, 2006 ▶) allowed us to determine that the crystals belonged to space group P4222, with unit-cell parameters a = b = 127.2, c = 76.8 Å, α = β = γ = 90°.
Table 1. Crystal data and X-ray diffraction data-collection statistics.
Values in parentheses are for the highest resolution bin.
| X-ray source | Beamline X25, NSLS, Brookhaven, USA |
| Detector | ADSC Quantum 315 CCD |
| X-ray wavelength (Å) | 0.9787 |
| Temperature (K) | 100 |
| Space group | P4222 |
| Unit-cell parameters (Å, °) | a = b = 127.2, c = 76.8, α = β = γ = 90 |
| Resolution (Å) | 25–3.25 (3.32–3.25) |
| Total No. of unique reflections | 10060 (620) |
| Redundancy | 4.7 (4.7) |
| Data completeness (%) | 96.8 (98.7) |
| Rmerge (%) | 5.2 (45.0) |
| 〈I/σ(I)〉 | 37.6 (2.0) |
Figure 4.
A representative image (1° oscillation) of the data collected from a crystal of the P3-domain RNA–Pop6–Pop7 complex. The image was collected using an ADSC Quantum 315 CCD detector on beamline X25 at NSLS, Brookhaven, USA.
The data sets were integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The anomalous signal at the peak was used to obtain the initial positions of anomalous scatterers using SOLVE (Brünger et al., 1998 ▶). Both the three-wavelength and the single-wavelength data sets were used in heavy-atom model refinement with SHARP (de La Fortelle & Bricogne, 1997 ▶). The final heavy-atom model contained three zinc ions and three Se atoms. The Se atoms were differentiated from the Zn atoms based on the change in the intensity of the anomalous scattering with the wavelength. The zinc ions are not expected to be of physiological relevance.
The estimated solvent content of the crystals is 65% if there is one molecule of the complex per asymmetric unit (Matthews coefficient of 3.07 Å3 Da−1) and 29% if there are two molecules of the complex per asymmetric unit (Matthews coefficient of 1.54 Å3 Da−1). Considering the estimated solvent content, an absence of apparent noncrystallographic symmetry and the number of observed anomalously scattering Se atoms, we expect one molecule of the complex per asymmetric unit.
Structure determination is now in progress.
Acknowledgments
We thank the staff of beamline X25 (NSLS), N. Yennawar and H. Yennawar (PSU) for their help with data collection. This work was supported by NIH grant GM085149 to ASK. The National Synchrotron Light Source beamline X25 is supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy and by the National Center for Research Resources of NIH. NIH–NCRR shared instrumentation grant S10RR023439 and the X-ray core facility at the Huck Institute of the Life Sciences (Pennsylvania State University) are acknowledged.
References
- Altman, S. & Kirsebom, L. (1999). The RNA World, edited by R. F. Gesteland, T. R. Cech & J. F. Alkins, pp. 351–380. New York: Cold Spring Harbor Laboratory Press.
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chamberlain, J. R., Lee, Y., Lane, W. S. & Engelke, D. R. (1998). Genes Dev.12, 1678–1690. [DOI] [PMC free article] [PubMed]
- Chang, D. D. & Clayton, D. A. (1987). EMBO J.6, 409–417. [DOI] [PMC free article] [PubMed]
- Chu, S., Zengel, J. M. & Lindahl, L. (1997). RNA, 3, 382–391. [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Coughlin, D. J., Pleiss, J. A., Walker, S. C., Whitworth, G. B. & Engelke, D. R. (2008). Proc. Natl Acad. Sci. USA, 105, 12218–12223. [DOI] [PMC free article] [PubMed]
- Dichtl, B. & Tollervey, D. (1997). EMBO J.16, 417–429. [DOI] [PMC free article] [PubMed]
- Esakova, O., Perederina, A., Quan, C., Schmitt, M. E. & Krasilnikov, A. S. (2008). RNA, 14, 1558–1567. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Frank, D. N., Adamidi, C., Ehringer, M. A., Pitulle, C. & Pace, N. R. (2000). RNA, 6, 1895–1904. [DOI] [PMC free article] [PubMed]
- Gill, T., Cai, T., Aulds, J., Wierzbicki, S. & Schmitt, M. E. (2004). Mol. Cell. Biol.24, 945–953. [DOI] [PMC free article] [PubMed]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. (1983). Cell, 35, 849–857. [DOI] [PubMed]
- Kazantsev, A. V. & Pace, N. R. (2006). Nature Rev. Microbiol.4, 729–740. [DOI] [PubMed]
- Kikovska, E., Svard, S. G. & Kirsebom, L. A. (2007). Proc. Natl Acad. Sci. USA, 104, 2062–2067. [DOI] [PMC free article] [PubMed]
- La Fortelle, E. de & Bricogne, G. (1997). Methods Enzymol.276, 472–494. [DOI] [PubMed]
- Li, X., Frank, D. N., Pace, N. R., Zengel, J. M. & Lindahl, L. (2002). RNA, 8, 740–751. [DOI] [PMC free article] [PubMed]
- Li, X., Zaman, S., Langdon, Y., Zengel, J. M. & Lindahl, L. (2004). Nucleic Acids Res. 32, 3703–3711. [DOI] [PMC free article] [PubMed]
- Lindahl, L., Bommankanti, A., Li, X., Hayden, L., Jones, A., Khan, M., Oni, T. & Zengel, J. M. (2009). RNA, 15, 1407–1416. [DOI] [PMC free article] [PubMed]
- Lindahl, L., Fretz, S., Epps, N. & Zengel, J. M. (2000). RNA, 6, 653–658. [DOI] [PMC free article] [PubMed]
- Lygerou, Z., Allmang, C., Tollervey, D. & Seraphin, B. (1996). Science, 272, 268–270. [DOI] [PubMed]
- Lygerou, Z., Mitchell, P., Petfalski, E., Seraphin, B. & Tollervey, D. (1994). Genes Dev.8, 1423–1433. [DOI] [PubMed]
- Marvin, M. C. & Engelke, D. R. (2009). RNA Biol.6, 40–42. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perederina, A., Esakova, O., Koc, H., Schmitt, M. E. & Krasilnikov, A. S. (2007). RNA, 13, 1648–1655. [DOI] [PMC free article] [PubMed]
- Piccinelli, P., Rosenblad, M. A. & Samuelson, T. (2005). Nucleic Acids Res.33, 4485–4495. [DOI] [PMC free article] [PubMed]
- Reiner, R., Ben-Asouli, Y., Krilovetzky, I. & Jarrous, N. (2006). Genes Dev.20, 1621–1635. [DOI] [PMC free article] [PubMed]
- Reiner, R., Krasnov-Yoeli, N., Dehtiar, Y. & Jarrous, N. (2008). PLoS One, 3, e4072. [DOI] [PMC free article] [PubMed]
- Salinas, K., Wierzbicki, S., Zhou, L. & Schmitt, M. E. (2005). J. Biol. Chem.280, 11352–11360. [DOI] [PubMed]
- Schmitt, M. E. & Clayton, D. A. (1994). Genes Dev.8, 2617–2628. [DOI] [PubMed]
- Spitale, R. C. & Wedekind, J. E. (2009). Methods, 49, 87–100. [DOI] [PMC free article] [PubMed]
- Shadel, G. S., Buckenmeyer, G. A., Clayton, D. A. & Schmitt, M. E. (2000). Gene, 245, 175–184. [DOI] [PubMed]
- Stolc, V. & Altman, S. (1997). Genes Dev.11, 2926–2937. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.14, 207–234. [DOI] [PubMed]
- Walker, S. C. & Avis, J. M. (2004). J. Mol. Biol.341, 375–388. [DOI] [PubMed]
- Walker, S. C. & Engelke, D. R. (2006). Crit. Rev. Biochem. Mol. Biol.41, 77–102. [DOI] [PMC free article] [PubMed]
- Ziehler, W. A., Morris, J., Scott, F. H., Millikin, C. & Engelke, D. R. (2001). RNA, 7, 565–575. [DOI] [PMC free article] [PubMed]