Aminoglycoside-2′′-phosphotransferase-IVa [APH(2′′)-IVa] is an enzyme that is responsible for high-level gentamicin resistance in E. casseliflavus isolates. Three different crystals of wild-type substrate-free APH(2′′)-IVa have been prepared and preliminary X-ray diffraction experiments have been undertaken on all three crystal forms.
Keywords: aminoglycoside-2′′-phosphotransferase-IVa, Enterococcus casseliflavus, antibiotic resistance
Abstract
The deactivation of aminoglycoside antibiotics by chemical modification is one of the major sources of bacterial resistance to this family of therapeutic compounds, which includes the clinically relevant drugs streptomycin, kanamycin and gentamicin. The aminoglycoside phosphotransferases (APHs) form one such family of enzymes responsible for this resistance. The gene encoding one of these enzymes, aminoglycoside-2′′-phosphotransferase-IVa [APH(2′′)-IVa] from Enterococcus casseliflavus, has been cloned and the protein (comprising 306 amino-acid residues) has been expressed in Escherichia coli and purified. The enzyme was crystallized in three substrate-free forms. Two of the crystal forms belonged to the orthorhombic space group P212121 with similar unit-cell parameters, although one of the crystal forms had a unit-cell volume that was approximately 13% smaller than the other and a very low solvent content of around 38%. The third crystal form belonged to the monoclinic space group P21 and preliminary X-ray diffraction analysis was consistent with the presence of two molecules in the asymmetric unit. The orthorhombic crystal forms of apo APH(2′′)-IVa both diffracted to 2.2 Å resolution and the monoclinic crystal form diffracted to 2.4 Å resolution; synchrotron diffraction data were collected from these crystals at SSRL (Stanford, California, USA). Structure determination by molecular replacement using the structure of the related enzyme APH(2′′)-IIa is proceeding.
1. Introduction
Bacteria in the environment are engaged in constant chemical warfare in an attempt to gain an advantage over other organisms. Generally, their weapons involve the secretion of specific compounds which kill foreign bacterial cells; because many of these molecules are extremely potent, the bacteria have also co-evolved resistance mechanisms which deactivate their own secreted antibiotics for self-protection (Benveniste & Davies, 1973 ▶). One such example of these potent natural antibiotics are the aminoglycosides, which are broad-spectrum antibiotics that were originally isolated from soil bacteria (Greenwood, 1995 ▶). Streptomycin was the first aminoglycoside antibiotic to be isolated from soil bacteria (Schatz et al., 1944 ▶) and was the first therapeutic found to be effective against Mycobacterium tuberculosis during trials in 1947–1948 (D’Arcy Hart, 1999 ▶). This family of compounds, which now includes the clinically relevant drugs neomycin, kanamycin, gentamicin, tobramycin and amikacin, consist of a central aminocyclitol ring with two or three substituted aminoglycan rings attached at different positions. The aminoglycosides are targeted to the 30S ribosome, binding selectively and irreversibly at the decoding aminoacyl A-site (Carter et al., 2000 ▶; Kotra et al., 2000 ▶; Vicens & Westhof, 2003 ▶), stabilizing the conformation of the tRNA as if it were bound to a cognate mRNA codon (the so-called ‘on’ state) and promoting miscoding owing to the inability of the ribosome to discriminate between cognate and noncognate tRNAs (Wirmer & Westhof, 2006 ▶).
Resistance to these antibiotics is now extremely widespread and several resistance mechanisms have been recognized, including drug efflux, decreased permeability, ribosomal target alteration and drug deactivation by chemical modification (Vakulenko & Mobashery, 2003 ▶), with the latter being the most significant (Wright, 2003 ▶). There are three families of enzymes responsible for the deactivation of the aminoglycosides, the ATP/GTP-dependent phosphotransferases (APH), the ATP-dependent adenylyltransferases (ANT) and the acetyl CoA-dependent acetyltransferases (AAC), which together comprise over 75 different enzymes (Shaw et al., 1993 ▶; Smith & Baker, 2002 ▶; Vakulenko & Mobashery, 2003 ▶). These enzymes have been classified according to the reaction they catalyze, the site they modify on the aminoglycoside and the aminoglycosides with which they interact.
The two most important phosphotransferase subfamilies are the APH(3′) and APH(2′′) enzymes. Three APH structures are currently known: APH(2′′)-IIa (Young et al., 2009 ▶), APH(3′)-IIa (Nurizzo et al., 2003 ▶) and APH(3′)-IIIa (Hon et al., 1997 ▶; Burk et al., 2001 ▶; Fong & Berghuis, 2002 ▶). Four APH(2′′) enzymes have been identified, APH(2′′)-Ia, APH(2′′)-IIa, APH(2′′)-IIIa and APH(2′′)-IVa (Ferretti et al., 1986 ▶; Chow et al., 1997 ▶; Tsai et al., 1998 ▶; Kao et al., 2000 ▶; Toth et al., 2009 ▶), with sequence identities ranging from 21 to 30% (Young et al., 2009 ▶). The APH(2′′)-Ia enzyme is found as one domain of a bifunctional enzyme AAC(6′)-Ie-APH(2′′)-Ia, whereas the other three enzymes are monofunctional enzymes. All four enzymes were initially isolated from enterococci, which are among the most common antibiotic resistant bacteria isolated in nosocomial infections today. APH(2′′)-IVa, initially isolated and named APH(2′′)-Id (Tsai et al., 1998 ▶), has the remarkable ability to use either ATP or GTP as the phosphate substrate, in contrast to APH(2′′)-IIa which uses only ATP. In order to understand the substrate-specificity differences in the APH(2′′) enzymes, we have cloned, purified and crystallized the APH(2′′)-IVa enzyme (306 amino-acid residues, molecular weight 35 400) from Enterococcus casseliflavus.
2. Materials and methods
2.1. Cloning, expression and purification of APH(2′′)-IVa
The aph(2′′)-IVa gene (Genbank accession No. AF016483) was PCR-amplified from the pNC95 construct (Tsai et al., 1998 ▶) by high-fidelity PfuTurbo polymerase (Stratagene) with primers APH-IdNdeD, ATACATATGAGAACTTATACTTTCG (the NdeI site introduced at the start codon of the aph(2′′)-IVa gene is shown in bold), and APH-IdHindR, ATAAAGCTTATTTAATTTTAATGCTTCTG (the HindIII site introduced after the stop codon of the gene is shown in bold). The PCR product was digested by NdeI and HindIII and the gene was recloned into the unique NdeI–HindIII sites of pET22b(+) (Novagen) to produce the pET:APH(2′′)-IVa vector. After transformation into the recipient Escherichia coli BL21 (DE3) strain and selection on LB agar supplemented with 100 µg ml−1 ampicillin, the nucleotide sequence of the cloned aph(2′′)-IVa gene was verified by sequencing of both DNA strands.
APH(2′′)-IVa was expressed and purified as described previously (Toth et al., 2009 ▶). Briefly, bacteria were grown in 350 ml LB broth at 310 K containing 100 µg ml−1 ampicillin until an OD590nm of ∼0.8 was reached. Protein expression was induced by the addition of 0.8 mM IPTG and cells were incubated overnight at 295 K. The bacteria were pelleted by centrifugation (3000g for 15 min), resuspended in buffer A (25 mM HEPES pH 7.5, 0.2 mM DTT) and the cells were disrupted by sonification. The lysate was centrifuged (20 000g for 30 min) and subsequently dialyzed twice against 2 l buffer A. APH(2′′)-IVa was purified by gentamicin–Affigel 15 (Bio-Rad) affinity chromatography. The enzyme was eluted with an NaCl gradient (0–1 M) in 25 mM HEPES pH 7.5 and pooled fractions containing the enzyme were then applied onto a DEAE anion-exchange column and eluted with an NaCl gradient (0–1 M). Fractions containing the APH(2′′)-IVa protein were checked using SDS–PAGE and those fractions containing pure enzyme were pooled, concentrated to 8 mg ml−1, dialyzed against 25 mM HEPES pH 7.5, 1.0 mM DTT and stored at 193 K.
2.2. Crystallization
Initial crystallization screening of substrate-free APH(2′′)-IVa and various complexes was carried out using commercial screens (PEG/Ion Screens I and II, Hampton Research) and a large number of conditions from both screens gave crystals. For screening and subsequent crystal production, crystals were grown at 288 K in Intelli-Plates (Art Robbins Instruments) using a reservoir volume of 75 µl and drops comprising 1 µl of protein complex at approximately 5 mg ml−1 in 25 mM HEPES pH 7.5 and 1 µl reservoir solution. The crystals that were used for X-ray diffraction analysis and data collection were harvested from the drops using cryoloops (Hampton Research) and immersed briefly in cryoprotectant solution composed of reservoir solution with the PEG 3350 concentration raised to 33%(w/v). The crystals were subsequently flash-cooled in liquid nitrogen and stored in a sample cassette designed for use with the Stanford Automated Mounting (SAM) system (Cohen et al., 2002 ▶).
2.3. Data collection and preliminary X-ray analysis
The crystals were transferred to beamline BL9-1 at the Stanford Synchrotron Radiation Laboratory (SSRL) and screened for diffraction quality. X-ray diffraction data were collected using a single crystal maintained at 100 K with an Oxford Cryosystem. Data from diffraction-quality crystals identified during screening were subsequently collected on two different SSRL beamlines: BL9-1, with X-rays from a 16-pole wiggler and an Si(311) side-scattering monochromator, using an ADSC Quantum-315 CCD detector and BL9-2, with X-rays from a 16-pole wiggler and a liquid nitrogen-cooled double Si(111) crystal monochromator, using a Rayonix MX-325 CCD detector. In all cases, diffraction data were collected with an X-ray energy of 12 658 eV (0.9795 Å). The data sets were processed and scaled with the XDS/XSCALE programs (Kabsch, 1993 ▶). Table 1 ▶ gives a summary of the data-collection statistics.
Table 1. Data-collection statistics.
Values in parentheses are for the data in the highest resolution shell (2.3–2.2 Å for forms I and II, and 2.5–2.4 Å for form III).
| Apo form I | Apo form II | Apo form III | |
|---|---|---|---|
| Beamline | BL9-2 | BL9-1 | BL9-1 |
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 |
| Crystal-to-detector distance (mm) | 320 | 350 | 400 |
| No. of images/oscillation angle (°) | 360/0.5 | 156/0.5 | 360/0.5 |
| Space group | P212121 | P212121 | P21 |
| Unit-cell parameters | |||
| a (Å) | 50.06 | 46.38 | 75.94 |
| b (Å) | 63.61 | 62.59 | 65.14 |
| c (Å) | 101.34 | 96.49 | 78.49 |
| β (°) | — | — | 91.7 |
| Resolution range (Å) | 29.8–2.2 | 28.6–2.2 | 28.0–2.4 |
| Observed reflections | 110664 | 45533 | 102252 |
| Unique reflections | 16673 | 16313 | 28901 |
| Rmerge† (%) | 0.060 (0.609) | 0.067 (0.376) | 0.071 (0.512) |
| 〈I/σ(I)〉 | 17.2 (2.3) | 13.8 (2.3) | 15.6 (1.9) |
| Completeness (%) | 97.9 (98.4) | 96.0 (81.0) | 95.6 (72.0) |
R
merge = 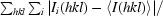
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
3. Results and discussion
3.1. Crystallization
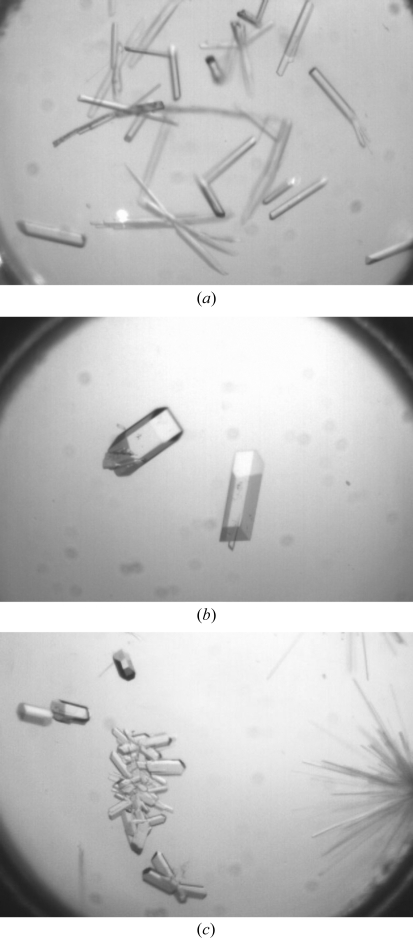
Inspection of the original crystallization screens showed that APH(2′′)-IVa crystallized under a number of conditions scattered across PEG/Ion Screens I and II. In most cases the crystals were rod-like or needle-like in morphology. A wide range of these initial crystals were tested for diffraction and in most cases gave measureable data to better than 3.5 Å resolution. Diffraction-quality crystals (with a resolution better than 2.5 Å) of apo APH(2′′)-IVa were produced in three crystal forms (Fig. 1 ▶). The crystallization conditions were as follows: apo-APH(2′′)-IVa form I, 0.2 M potassium citrate pH 8.0, 20%(w/v) PEG 3350; apo-APH(2′′)-IVa form II, 0.1 M ammonium citrate pH 7.0, 12%(w/v) PEG 3350; apo-APH(2′′)-IVa form III, 4% tacsimate pH 8.0, 12%(w/v) PEG 3350.
Figure 1.
(a) Apo APH(2′′)-IVa form I crystals of approximately 0.25 × 0.05 × 0.05 mm in size. (b) Apo APH(2′′)-IVa form II crystals of approximately 0.4 × 0.1 × 0.05 mm in size. (c) Apo APH(2′′)-IVa form III crystals of approximately 0.15 × 0.06 × 0.05 mm in size.
3.2. Data collection and preliminary X-ray analysis
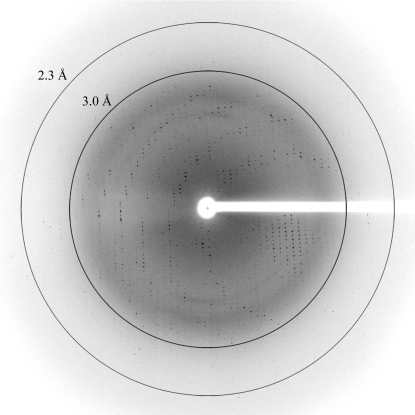
Complete X-ray data sets were collected for these three crystals and the results are summarized in Table 1 ▶. A representative image from one of these crystals is shown in Fig. 2 ▶. The first two apo APH(2′′)-IVa crystal forms both belonged to space group P212121 with slightly different unit-cell parameters. The apo form I crystals (a = 50.06, b = 63.61, c = 101.34 Å) had a unit-cell volume of 322 700 Å3 and a Matthews coefficient (V M; Matthews, 1968 ▶) of 2.28 Å3 Da−1, implying approximately 46% solvent content with one molecule in the asymmetric unit, whilst the apo form II crystals (a = 46.38, b = 62.59, c = 96.49 Å) had a unit-cell volume that was approximately 13% smaller (280 100 Å3) and only 38% solvent content (V M = 1.98 Å3 Da−1). This is significantly lower than the average solvent content for an orthorhombic cell (49% solvent content, V M = 2.51 Å3 Da−1; Chruszcz et al., 2008 ▶). The crystallization conditions for the two apo crystal forms are similar (a citrate salt at pH 8.0 and PEG 3350 as the precipitating agent), with the only difference being the relative concentrations of the salt and the cation (potassium versus ammonium). The significant decrease in unit-cell size and the drop in solvent content is therefore intriguing and could arise from inadvertent dehydration during crystal handling; it will be interesting to analyze the solvation and the crystal packing in these two crystal forms. The third apo APH(2′′)-IVa crystal form belonged to space group P21, with unit-cell parameters a = 75.94, b = 65.14, c = 78.49 Å, β = 91.7° and a V M of 2.74 Å3 Da−1 (55% solvent content) with two molecules in the asymmetric unit.
Figure 2.
Representative diffraction image of apo APH(2′′)-IVa form I. The resolution circles are at approximately 3.0 and 2.3 Å resolution.
Despite the somewhat low sequence identity between the members of the APH(2′′) subfamily (21–31%), structure solution will initially be attempted using the recently determined structure of the APH(2′′)-IIa enzyme (Young et al., 2009 ▶) as a search model for molecular replacement. These structural analyses are currently under way.
Acknowledgments
This work was supported by grant RO1 AI05739 from the NIH. The Stanford Synchrotron Radiation Lightsource is a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy (BES, BER) and by the National Institutes of Health (NCRR, BTP, NIGMS). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
- Benveniste, R. & Davies, J. (1973). Proc. Natl Acad. Sci. USA, 70, 2276–2280. [DOI] [PMC free article] [PubMed]
- Burk, D. L., Hon, W. C., Leung, A. K. & Berghuis, A. M. (2001). Biochemistry, 40, 8756–8764. [DOI] [PubMed]
- Carter, A. P., Clemons, W. M., Broderson, D. E., Morgan-Warren, R. J., Wimberly, B. T. & Ramakrishnan, V. (2000). Nature (London), 407, 340–348. [DOI] [PubMed]
- Chow, J. W., Zervos, M. J., Lerner, S. A., Thal, L. A., Donabedian, S. M., Jaworski, D. D., Tsai, S., Shaw, K. J. & Clewell, D. B. (1997). Antimicrob. Agents Chemother.41, 511–514. [DOI] [PMC free article] [PubMed]
- Chruszcz, M., Potrzebowski, W., Zimmerman, M. D., Grabowski, M., Zheng, H., Lasota, P. & Minor, W. (2008). Protein Sci.17, 623–632. [DOI] [PMC free article] [PubMed]
- Cohen, A. E., Ellis, P. J., Miller, M. D., Deacon, A. M. & Phizackerley, R. P. (2002). J. Appl. Cryst.35, 720–726. [DOI] [PMC free article] [PubMed]
- D’Arcy Hart, P. (1999). BMJ, 319, 572–573. [DOI] [PMC free article] [PubMed]
- Ferretti, J. J., Gilmore, K. S. & Courvalin, P. (1986). J. Bacteriol.167, 631–638. [DOI] [PMC free article] [PubMed]
- Fong, D. & Berghuis, A. M. (2002). EMBO J.21, 2323–2331. [DOI] [PMC free article] [PubMed]
- Greenwood, D. (1995). Editor. Antimicrobial Chemotherapy, pp. 32–48. Oxford University Press.
- Hon, W. C., McKay, G. A., Thompson, P. R., Sweet, R. M., Yang, D. S. C., Wright, G. D. & Berghuis, A. M. (1997). Cell, 89, 887–895. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kao, S. J., You, I., Clewell, D. B., Donabedian, S. M., Zervos, M. J., Petrin, J., Shaw, K. J. & Chow, J. W. (2000). Antimicrob. Agents Chemother.44, 511–514. [DOI] [PMC free article] [PubMed]
- Kotra, L. P., Haddad, J. & Mobashery, S. (2000). Antimicrob. Agents Chemother.44, 3249–3256. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Nurizzo, D., Shewry, S. C., Perlin, M. H., Brown, S. A., Dholakia, J. N., Fuchs, R. L., Deva, T., Baker, E. N. & Smith, C. A. (2003). J. Mol. Biol.327, 491–506. [DOI] [PubMed]
- Schatz, A., Bugie, E. & Waksman, S. A. (1944). Proc. Soc. Exp. Biol. Med.55, 65–69.
- Shaw, K. J., Rather, P. N., Hare, R. S. & Miller, G. H. (1993). Microbiol. Rev.57, 138–163. [DOI] [PMC free article] [PubMed]
- Smith, C. A. & Baker, E. N. (2002). Curr. Drug Targets Infect. Dis.2, 143–160. [DOI] [PubMed]
- Toth, M., Chow, J. W., Mobashery, S. & Vakulenko, S. B. (2009). J. Biol. Chem.284, 6690–6696. [DOI] [PMC free article] [PubMed]
- Tsai, S., Zervos, M. J., Clewell, D. B., Donabedian, S. M., Sahm, D. F. & Chow, J. W. (1998). Antimicrob. Agents Chemother.42, 1229–1232. [DOI] [PMC free article] [PubMed]
- Vakulenko, S. B. & Mobashery, S. (2003). Clin. Microbiol. Rev.16, 430–450. [DOI] [PMC free article] [PubMed]
- Vicens, Q. & Westhof, E. (2003). Biopolymers, 70, 42–57. [DOI] [PubMed]
- Wirmer, J. & Westhof, E. (2006). Methods Enzymol.415, 180–202. [DOI] [PubMed]
- Wright, G. D. (2003). Curr. Opin. Chem. Biol.7, 563–569.
- Young, P. G., Walanj, R., Lakshmi, V., Byrnes, L. J., Metcalf, P., Baker, E. N., Vakulenko, S. & Smith, C. A. (2009). J. Bacteriol.191, 4133–4143. [DOI] [PMC free article] [PubMed]