A complex of the measles virus hemagglutinin and the CD46 receptor representing the initial step of the cell infection has been crystallized.
Keywords: measles virus hemagglutinin, CD46 receptor, virus receptor, virus infection
Abstract
The measles virus (MV) hemagglutinin (MV-H) mediates the attachment of MV particles to cell-surface receptors for entry into host cells. MV uses two receptors for attachment to host cells: the complement-control protein CD46 and the signalling lymphocyte activation molecule (SLAM). The MV-H glycoprotein from an Edmonston MV variant and the MV-binding fragment of the CD46 receptor were overproduced in mammalian cells and used to crystallize an MV-H–CD46 complex. Well diffracting crystals containing two complexes in the asymmetric unit were obtained and the structure of the complex was solved by the molecular-replacement method.
1. Introduction
Measles virus (MV) remains a worldwide cause of morbidity and mortality, especially amongst children, despite the availability of a vaccine (Griffin, 2007 ▶). MV is an enveloped virus that belongs to the Paramyxoviridae family. The virus contains a single segment of negative-stranded RNA encapsulated by a nucleocapsid. The outer surface of the virion is formed by a lipid bilayer that contains two glycoproteins: the measles virus hemagglutinin (MV-H), which mediates attachment to cell-surface receptors, and the fusion protein (MV-F), which is responsible for virus and cell-membrane fusion once the receptor is engaged. MV-H is a disulfide-linked homodimer in the virion envelope. The complement regulatory protein CD46 was first identified as a cellular receptor for the vaccine strain (Edmonston) of MV (Naniche et al., 1993 ▶; Dorig et al., 1993 ▶). SLAM (signalling lymphocyte-activation molecule; CD150), a membrane glycoprotein that is found at the surface of activated T cells, immature thymocytes, memory T cells and certain B cells, was subsequently found to function as a second MV receptor (Tatsuo et al., 2000 ▶). Clinical MV isolates appear to bind specifically to the SLAM receptor, whereas MV variants that bind to CD46 appear during the course of infection (Manchester et al., 2000 ▶). Increase in the virus-binding affinity for CD46 correlated either with an Asn to Tyr mutation at position 481 of the MV-H protein or with a Ser to Gly mutation at position 546 (Lecouturier et al., 1996 ▶; Shibahara et al., 1994 ▶).
The CD46 and SLAM receptor proteins bind to distinct but overlapping sites on the MV-H protein (Santiago et al., 2002 ▶). Extensive mutagenesis studies of MV-H defined several residues that are involved in receptor recognition (Masse et al., 2004 ▶; Vongpunsawad et al., 2004 ▶; Tahara et al., 2007 ▶; Hashiguchi et al., 2007 ▶; Colf et al., 2007 ▶). The virus-binding surfaces were also defined by the crystal structure of the two N-terminal virus-binding short consensus repeats 1 (SCR1) and 2 (SCR2) of the CD46 receptor molecule (Casasnovas et al., 1999 ▶). MV binds to the N-terminal immunoglobulin (Ig) domain of the SLAM protein (Ono et al., 2001 ▶), the structure of which is unknown. Currently, there is no structural information available on MV-H in complex with its receptors. We crystallized a complex of the C-terminal globular region of the MV-H protein and the MV-binding fragment of CD46 comprising the SCR1 and SCR2 domains. Here, we describe the crystallization of the complex and the analysis of the crystals and present preliminary data on structure determination.
2. Preparation of the MV-H and CD46 proteins
The MV-H cDNA comes from a virus variant grown in CV-1 cells (Stern et al., 1995 ▶). The crystallized fragment bears five amino acids (Ile390, Asp416, Ser446, Tyr481 and Gly546) that have been reported to confer high measles virus binding affinity to CD46 (Lecouturier et al., 1996 ▶; Shibahara et al., 1994 ▶; Tahara et al., 2007 ▶). Indeed, the reported CD46-binding affinity of our MV-H protein was about ten times higher than that of the Edmonston B variant (Santiago et al., 2002 ▶; Hashiguchi et al., 2007 ▶), which bears a Ser at position 546. The preparation of recombinant cDNAs coding for a soluble MV-H fragment was carried out as described previously (Santiago et al., 2002 ▶). The recombinant MV-H contained residues 179–617 of the virus protein fused to a HA epitope (YPYDVPDYA) followed by the protein sequence GAQPARSPGIRG and a thrombin-recognition site (LVPRGS) at the N-terminal end. The recombinant MV-H fragment lacked the cysteine residues involved in homodimerization. High-level protein production was carried out in CHO Lec 3.2.8.1 cells (Stanley, 1989 ▶) using the glutamine synthetase system (Casasnovas & Springer, 1995 ▶). The soluble MV-H fragment was purified from supernatants of stable transfected cell clones by affinity chromatography using the MV-H 16CD11 mAb coupled to CNBr-activated Sepharose (GE Healthcare) and was further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) (Santiago et al., 2002 ▶).
A CD46 fragment comprising the two N-terminal SCR1 and SCR2 domains was also prepared in CHO Lec 3.2.8.1 as described previously (Casasnovas et al., 1999 ▶). This region of CD46 is necessary and sufficient to mediate binding to MV (Manchester et al., 1997 ▶).
3. Crystallization
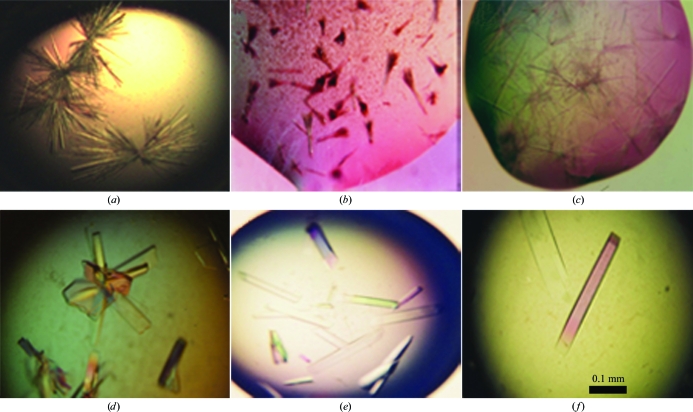
The MV-H and CD46 proteins were both concentrated to about 20 mg ml−1 and the complex was prepared by incubating the proteins at an MV-H:CD46 molar ratio of 1:1.2 for 2 h at room temperature. The final protein concentration was between 10 and 14 mg ml−1. Crystallization was performed using the hanging-drop vapour-diffusion method at 293 K by mixing 1.5 µl protein solution and 1.5 µl reservoir crystallization solution in 24-well plates (Corning Inc.) and equilibrating against 500 µl reservoir solution. Initially, needle-like crystals were obtained for both the MV-H fragment and the MV-H–CD46 complex using a reservoir solution containing 2 M ammonium sulfate, 5% MPD (2-methyl-2,4-pentanediol) and 0.1 M sodium cacodylate pH 6.5 (Figs. 1a–1c). Removal of N-linked glycans by treatment with endoglycosidase H did not improve the quality of the MV-H crystals (Fig. 1 ▶ b). A different rod-like crystal form of the MVH–CD46 complex that grew in clusters was prepared using a reservoir solution containing 12% PEG 8000, 0.2 M ammonium sulfate, 2% PEG 400 and 0.1 M sodium MES pH 6.5 (Fig. 1 ▶ d). The inclusion of 1% 1,2,3-heptanetriol gave isolated single crystals and the subsequent addition of sodium tungstate decreased the number of crystals in the drop and at the same time increased their size (Figs. 1 ▶ e and 1 ▶ f). Protein precipitation without crystals was observed using the PEG 8000 reservoir solution for the MV-H protein alone, suggesting that the crystals contained the complex. Gel electrophoresis and mass spectrometry of redissolved crystals confirmed that they indeed contained both proteins (Fig. 2 ▶). In addition, the identity of both proteins was verified by microsequencing of the crystals, which gave the N-terminal sequences of the recombinant CD46 and MV-H fragments used for crystallization.
Figure 1.
Crystals of the isolated MV-H fragment and the MV-H–CD46 complex. The top row shows crystals prepared with 2 M ammonium sulfate, 5% MPD and 0.1 M sodium cadodylate pH 6.5. The bottom row shows crystals prepared with 12% PEG 8000, 0.2 M ammonium sulfate, 2% PEG 400 and 0.1 M sodium MES pH 6.5. (a) Crystals of the MV-H fragment. (b) Crystals of endoglycosidase H-treated MV-H fragment. (c, d) Crystals of the MV-H–CD46 complex. (e) Crystals of the complex prepared with the addition of 1% 1,2,3-heptanetriol to the PEG 8000 crystallization condition. (f) Crystals of the complex prepared with the addition of 1% 1,2,3-heptanetriol and 1 mM sodium tungstate (Na2WO4).
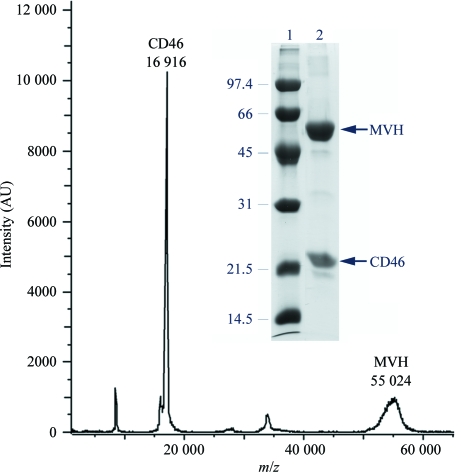
Figure 2.
Identification of the proteins in the MV-H–CD46 crystals. MALDI–TOF analysis of redissolved crystals. The spectrum shows two peaks that correspond to proteins with molecular masses of 55 024 and 16 916 Da, corresponding to MV-H and CD46, respectively. Mass determination was calculated from the single charged ion of the proteins. The inset shows 12% SDS–PAGE analysis under reducing conditions of redissolved crystals (lane 2) and molecular mass markers (lane 1). The mass (kDa) and migration of the marker proteins are indicated.
4. X-ray diffraction data collection
The crystals of the complex were initially harvested directly into the 12% PEG 8000 crystallization solution and dialyzed against crystallization solution containing 20% ethylene glycol as a cryoprotectant in order to flash-cool them in liquid nitrogen. Initial X-ray diffraction data collection revealed high anisotropy and a mosaicity of more than 1.5°. Subsequent adjustment of the harvesting conditions reduced the mosaicity to below 1°. The optimal harvesting buffer was found to be 20% PEG 8000, 0.2 M ammonium sulfate, 2% PEG 400 and 0.1 M sodium MES pH 6.5. Prior to flash-cooling, the crystals were dialyzed against the harvesting solution containing 20% ethylene glycol. Harvesting of crystals grown in 2 M ammonium sulfate was unsuccessful since they dissolved quickly upon harvesting with different solutions.
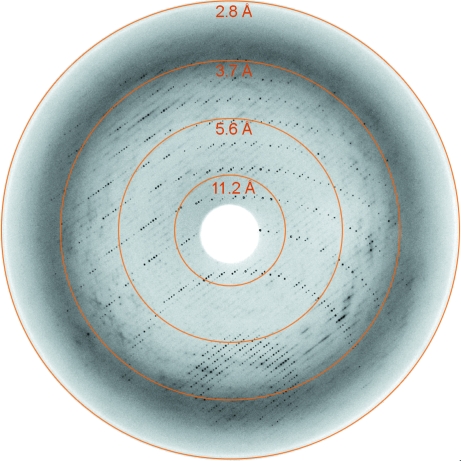
Data collection was carried out at 100 K using crystals mounted in cryoloops on several beamlines at DESY (EMBL Outstation, Hamburg) and on the ID14.4 beamline at ESRF (Table 1 ▶). Although the diffraction pattern was quite anisotropic (Fig. 3 ▶), complete data sets could be collected for several crystals grown in the absence or presence of sodium tungstate, which significantly increased the crystal size (Fig. 1 ▶). Analysis of the crystal diffraction data (Diffraction Anisotropy Server; http://www.doe-mbi.ucla.edu/~sawaya/anisoscale/) showed good-quality data [F/σ(F) > 3] to 3.1 Å resolution for reflections along the b* and c* directions and to about 4 Å resolution for those along a*. The crystals were highly sensitive to radiation damage, particularly at the ID14.4 beamline (data not shown). The best data set was collected using a crystal that was treated with 5% glutaraldehyde (Sigma Ultrapure) for 10 min prior to flash-cooling (Na7 in Table 1 ▶).
Table 1. Data-collection and processing statistics.
Statistics of the X-ray diffraction data collected for the MV-H–CD46 crystals. The Na5 and Na6 crystals were prepared in the absence of sodium tungstate (Fig. 1 ▶). Values in parentheses are for the highest resolution shell.
| Crystal | Na5 | Na6 | Na7 | Na8 |
|---|---|---|---|---|
| Space group | P2221 | P2221 | P2221 | P2221 |
| Unit-cell parameters | ||||
| a (Å) | 81.0 | 80.7 | 81.4 | 79.6 |
| b (Å) | 105.3 | 105.4 | 105.8 | 105.7 |
| c (Å) | 205.9 | 206.3 | 208.7 | 206.1 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Mosaicity (°) | 0.9 | 0.6 | 0.5 | 0.8 |
| Beamline | X13 | X13 | WB7B | ID14.4 |
| Wavelength (Å) | 0.802 | 0.804 | 0.845 | 0.939 |
| Resolution range (Å) | 20–3.1 (3.21–3.1) | 20–3.1 (3.21–3.1) | 20–3.1 (3.21–3.1) | 20–3.1 (3.27–3.1) |
| No. of observed reflections | 781994 | 556154 | 446949 | 138340 |
| No. of unique reflections | 34442 | 36151 | 33285 | 29382 |
| Completeness (%) | 85 (67) | 89 (61) | 96 (96) | 94 (91) |
| Redundancy | 3.8 (2.9) | 3.3 (1.7) | 3.7 (3.2) | 4.3 (3.3) |
| Rmerge | 9.2 (44) | 4.8 (44) | 4.8 (38) | 11.7 (37) |
| I/σ(I) | 11.9 (2.4) | 18.0 (1.5) | 22.7 (2.1) | 3.3 (2.0) |
| B factor (Å2) | 69.3 | 65.8 | 80.3 | 94.0 |
Figure 3.
X-ray diffraction image from a native MV-H–CD46 crystal (Na7 in Table 1 ▶). Rings are shown at the detector edge and at three additional resolution limits.
The MV-H–CD46 crystals belonged to space group P2221, with average unit-cell parameters a = 80.7, b = 105.6, c = 206.8 Å (Table 1 ▶). The crystals contained two macromolecular complexes per asymmetric unit, with a solvent content of about 64%.
5. Structure determination by molecular replacement
The crystal structure was solved using the molecular-replacement method with the program Phaser (Read, 2001 ▶). To locate the MV-H component, the structure of the ligand-free Edmonston B strain protein (PDB code 2zb6; Hashiguchi et al., 2007 ▶) was used as a search model. It was composed of a single MV-H molecule including the β-propeller domain residues 185–607, with only four residues differing from our MV-H fragment (T331A, A392T, G492E and S546G). The search for CD46 was performed with the structures of the same protein containing two N-terminal SCR domains and crystallized either in the absence of ligands (PDB code 1ckl; Casasnovas et al., 1999 ▶) or bound to the adenovirus type 11 knob (PDB code 2o39; Persson et al., 2007 ▶). These two CD46 structures differ in their interdomain orientation. The initial search with the MV-H structure gave a unique solution for the two MV-H molecules in the asymmetric unit, which formed a dimer similar to that described for the structure of a ligand-free disulfide-linked homodimeric MV-H protein (Hashiguchi et al., 2007 ▶). A search with the individual SCR1 and SCR2 domains of CD46 gave two reasonable solutions for the SCR2, in which the domains interacted in the same manner with the two previously identified MV-H molecules. Molecular-replacement solutions obtained with the two-domain fragment structures (SCR1–SCR2; 1ckl and 2o39) were identical to the SCR2 solutions, but the SCR1 of the ligand-free CD46 structure (1ckl) collided with the MV-H molecule (not shown). However, the SCR1–SCR2 structure taken from the complex with the adenovirus type 11 knob gave a solution that did not result in clashes but that had sensible contacts between SCR1 and MV-H. The MV-H β-propeller domain and the CD46 repeats were refined as independent rigid-body entities with CNS (Brünger et al., 1998 ▶), which gave an R free of 43% for 3.1 Å resolution data. Subsequent grouped temperature-factor refinement reduced the R free to 40% (R work of 38.5%). Further structure refinement gave R free and R work values that were below 30% (Santiago et al., 2009 ▶).
6. Conclusions
We crystallized a complex between the MV-H cell-attachment protein and the CD46 receptor using a simple molecular-mixing approach. The crystals of the complex were quite fragile and sensitive to radiation damage, but they could be stabilized by the optimization of both crystallization and harvesting solutions. The described procedure reduced the anisotropy of the diffraction and allowed the collection of complete data sets to 3.1 Å resolution. Structure determination of the MV-H–CD46 complex by the molecular-replacement method gave unique solutions for the two molecular complexes in the asymmetric unit. The MV-H β-propeller domain and the SCR2 domain of CD46 appeared to be well defined in the electron-density maps, whereas the density for the SCR1 repeat of CD46 was somewhat less defined, suggesting higher mobility. The crystal structure of MV-H bound to the CD46 receptor is revealing the MV–receptor binding interface and the MV-H residues critical for recognition of the CD46 receptor protein on the host cell surface (Santiago et al., 2009 ▶).
Acknowledgments
We acknowledge EMBL/DESY (PX-01-168) and ESRF (MX-85, MX-222 and BAG-Madrid) for provision of synchrotron-radiation facilities. This work was sponsored by grants from the Vetenskapsrådet (MFR-12637 and NFR-11994) and the Ministerio de Ciencia e Innovación (BFU2005-05972 and BFU2008-0971). CS was supported by NIH grant AI45716, by the Marie Curie Training Site Programme (Contract No. HPMT-CT-2000-00174) and by the CNB–CSIC. AG-R was the recipient of a Marie Curie fellowship (Contract No. HPMF-CT-2001-01507).
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Casasnovas, J. M., Larvie, M. & Stehle, T. (1999). EMBO J.18, 2911–2922. [DOI] [PMC free article] [PubMed]
- Casasnovas, J. M. & Springer, T. A. (1995). J. Biol. Chem.270, 13216–13224. [DOI] [PubMed]
- Colf, L. A., Juo, Z. S. & Garcia, K. C. (2007). Nature Struct. Mol. Biol.14, 1227–1228. [DOI] [PubMed]
- Dorig, R. E., Marcil, A., Chopra, A. & Richardson, C. D. (1993). Cell, 75, 295–305. [DOI] [PubMed]
- Griffin, D. E. (2007). Fields Virology, Vol. 1, edited by B. N. Fields, D. M. Knipe, P. M. Howley, J. L. Melnick, R. M. Chanock, B. Roizman & T. P. Monath, pp. 1551–1585. Philadelphia: Lippincott, Williams & Wilkins.
- Hashiguchi, T., Kajikawa, M., Maita, N., Takeda, M., Kuroki, K., Sasaki, K., Kohda, D., Yanagi, Y. & Maenaka, K. (2007). Proc. Natl Acad. Sci. USA, 104, 19535–19540. [DOI] [PMC free article] [PubMed]
- Lecouturier, V., Fayolle, J., Caballero, M., Carabana, J., Celma, M., Fernandez-Munoz, R., Wild, T. F. & Buckland, R. (1996). J. Virol.70, 4200–4204. [DOI] [PMC free article] [PubMed]
- Manchester, M., Eto, D. S., Valsamakis, A., Liton, P. B., Fernandez-Muñoz, R., Rota, P. A., Bellini, W. J., Forthal, D. N. & Oldstone, M. B. (2000).J Virol.74, 3967–3974. [DOI] [PMC free article] [PubMed]
- Manchester, M., Gairin, J. E., Patterson, J. B., Alvarez, J., Liszewski, K., Eto, D. S., Atkinson, J. P. & Oldstone, M. B. A. (1997). Virology, 232, 1–8. [DOI] [PubMed]
- Masse, N., Ainouze, M., Neel, B., Wild, T. F., Buckland, R. & Langedijk, J. P. M. (2004). J. Virol.78, 9051–9063. [DOI] [PMC free article] [PubMed]
- Naniche, D., Varior-Krishman, G., Cervoni, F., Wild, T. F., Rossi, B., Rabourdin-Combe, C. & Gerlier, D. (1993). J. Virol.67, 6025–6032. [DOI] [PMC free article] [PubMed]
- Ono, N., Tatsuo, H., Tanaka, K., Minagawa, H. & Yanagi, Y. (2001). J. Virol.75, 1594–1600. [DOI] [PMC free article] [PubMed]
- Persson, B. D., Reiter, D. M., Marttila, M., Mei, Y.-F., Casasnovas, J. M., Arnberg, N. & Stehle, T. (2007). Nature Struct. Mol. Biol.14, 164–166. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Santiago, C., Björling, E., Stehle, T. & Casasnovas, J. M. (2002). J. Biol. Chem.277, 32294–32301. [DOI] [PubMed]
- Santiago, C., Celma, M. L., Stehle, T. & Casasnovas, J. M. (2009). Nature Struct. Mol. Biol. doi:10.1038/nsmb.1726. [DOI] [PubMed]
- Shibahara, K., Hotta, H., Katayama, Y. & Homma, M. (1994). J. Gen. Virol.75, 3511–3516. [DOI] [PubMed]
- Stanley, P. (1989). Mol. Cell. Biol.9, 377–383. [DOI] [PMC free article] [PubMed]
- Stern, L. B., Greenberg, M., Gershoni, J. M. & Rozenblatt, S. (1995). J. Virol.69, 1661–1668. [DOI] [PMC free article] [PubMed]
- Tahara, M., Takeda, M., Seki, F., Hashiguchi, T. & Yanagi, Y. (2007). J. Virol.81, 2564–2572. [DOI] [PMC free article] [PubMed]
- Tatsuo, H., Ono, N., Tanaka, K. & Yanagi, Y. (2000). Nature (London), 406, 893–897. [DOI] [PubMed]
- Vongpunsawad, S., Oezgun, N., Braun, W. & Cattaneo, R. (2004). J. Virol.78, 302–313. [DOI] [PMC free article] [PubMed]