Abstract
Anthocyanidin reductase encoded by the BANYULS (BAN) gene is the core enzyme in proanthocyanidin (PA) biosynthesis. Here, we analyzed the developmental mechanisms that regulate the spatiotemporal expression of BAN in the developing Arabidopsis seed coat. PA-accumulating cells were localized histochemically in the inner integument (seed body and micropyle) and pigment strand (chalaza). BAN promoter activity was detected specifically in these cells. Gain-of-function experiments showed that an 86-bp promoter fragment functioned as an enhancer specific for PA-accumulating cells. Mutations in regulatory genes of PA biosynthesis abolished BAN promoter activity (transparent testa2 [tt2], tt8, and transparent testa glabra1 [ttg1]), modified its spatial pattern (tt1 and tt16), or had no influence (ttg2), thus revealing complex regulatory interactions at several developmental levels. Genetic ablation of PA-accumulating cells targeted by the BAN promoter fused to BARNASE led to the formation of normal plants that produced viable yellow seeds. Importantly, these seeds had no obvious defects in endosperm and embryo development.
INTRODUCTION
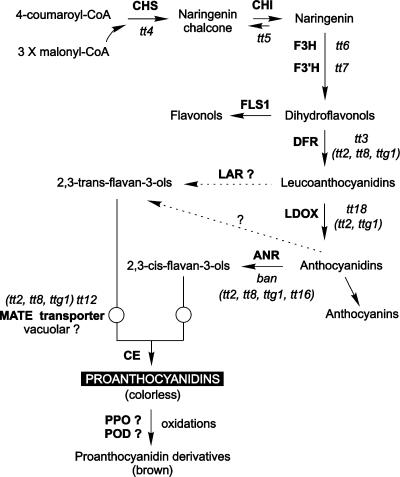
Proanthocyanidins (PAs), also called condensed tannins, are colorless flavonoid polymers that result from the condensation of flavan-3-ol units (Xie et al., 2003) (Figure 1). In Arabidopsis, they are found only in the seed coat (or testa), where they confer a brown color to mature seeds after oxidation (Devic et al., 1999; Debeaujon et al., 2001). Tannin deposition in the testa participates in seed defense against predators and pathogens (Winkel-Shirley, 1998) and in strengthening seed coat–imposed dormancy and longevity (Debeaujon et al., 2000). To date, 22 independent loci have been identified by a mutational approach on the basis of seed coat color deviating from the wild-type brown (Winkel-Shirley, 2002; Shikazono et al., 2003). The corresponding mutants were called transparent testa or tt (tt1 to tt19), transparent testa glabra or ttg (ttg1 and ttg2), and banyuls (ban). Because the seed coat derives from ovular tissue (Schneitz et al., 1995), the phenotypes of the tt, ttg, and ban mutants are determined by the maternal genotype. Sixteen mutants have already led to gene cloning, among which 12 could be placed in the flavonoid pathway (Figure 1). Recently, Abrahams et al. (2002) reported the identification of six complementation groups defective in PA biosynthesis, but whether these groups represent new loci or are allelic to mutants described previously is not known.
Figure 1.
Scheme of the Flavonoid Biosynthetic Pathway.
Enzymes are represented in uppercase boldface letters. Mutants are shown in lowercase italic letters, with regulatory mutants listed in parentheses. Dashed lines represent hypothetical steps. ANR, anthocyanidin reductase; CE, condensing enzyme; CHI, chalcone isomerase; CHS, chalcone synthase; DFR, dihydroflavonol reductase; F3H, flavonol 3-hydroxylase; F3′H, flavonol 3′-hydroxylase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; LDOX, leucocyanidin dioxygenase; POD, peroxidase; PPO, polyphenol oxidase. Adapted from Nesi et al. (2001), Bartel and Matsuda (2003), and Xie et al. (2003).
To elucidate the developmental mechanisms responsible for seed coat pigmentation in Arabidopsis, the functional relationships among genes that affect the type, amount, and spatiotemporal distribution of the pigment synthesized must be analyzed. BAN encodes a dihydroflavonol reductase–like protein belonging to the reductase epimerase dehydrogenase family (Devic et al., 1999). Xie et al. (2003) demonstrated that BAN functions as an anthocyanidin reductase, converting anthocyanidins to their corresponding 2,3-cis-flavan-3-ols (Figure 1). It is a core enzyme of the PA subpathway; therefore, studying the regulation of BAN is of particular importance.
To date in Arabidopsis, six proteins required for the regulation of PA biosynthesis have been described. They are classified according to apparent modalities of action. The first group of regulators involves TT2, TT8, and TTG1. These factors belong to the MYB, MYC/bHLH, and WD40-repeat families, respectively (Walker et al., 1999; Nesi et al., 2000, 2001). They positively regulate BAN expression in the whole seed coat. Indeed, no BAN transcript was detected in tt2, tt8, and ttg1 mutants (Nesi et al., 2000), which is consistent with the fact that tt2, tt8, and ttg1 seeds are completely deprived of PAs and thus are homogeneously yellow (Debeaujon et al., 2001). TT1 and TT16, which were shown to be necessary for PA biosynthesis and normal cell morphology in the seed body but not in the underlying chalaza/micropyle region, constitute a second group of regulators. TT1 is a new zinc finger protein that defines the WIP subfamily of zinc finger transcription factors (Sagasser et al., 2002). TT16 encodes the ARABIDOPSIS BSISTER MADS domain (ABS) transcription factor protein (TT16/ABS) (Nesi et al., 2002). Finally, TTG2 is a zinc finger–like transcription factor of the plant-specific WRKY gene family (Johnson et al., 2002). TTG2 also is required for PA biosynthesis in the whole seed coat, like TT2, TT8, and TTG1. However, because it acts downstream of TTG1 in the PA pathway (Johnson et al., 2002), TTG2 constitutes a third group of regulators.
Here, we focus on an analysis of the endogenous developmental signals that drive the spatiotemporal expression of BAN in the developing Arabidopsis seed coat. We report the in planta functional analysis of the BAN promoter. The distribution of PA pigments strictly matches the expression profile of both ProBAN:uidA and ProBAN:mGFP5-ER chimeric constructs and mRNA localization established by in situ hybridization experiments, suggesting that the cell-specific expression of BAN is regulated largely at the transcriptional level. In parallel, BAN promoter dissection and gain-of-function experiments determined that an 86-bp proximal promoter region was sufficient to activate the BAN promoter in PA-producing cells. Mutants defective in the regulation of PA biosynthesis revealed that a complex network of regulatory factors are involved in the differentiation of PA-accumulating cells for cadastral patterning of the seed coat, ProBAN activation, and tannin biosynthesis. Finally, we conclude from genetic ablation experiments driven by the ProBAN:BARNASE construct that PA-producing cells are not essential for the formation of viable seeds. Some consequences of PA-producing cell ablation on several aspects of seed development and physiology are presented.
RESULTS
Spatiotemporal Accumulation of PAs in the Arabidopsis Seed Coat
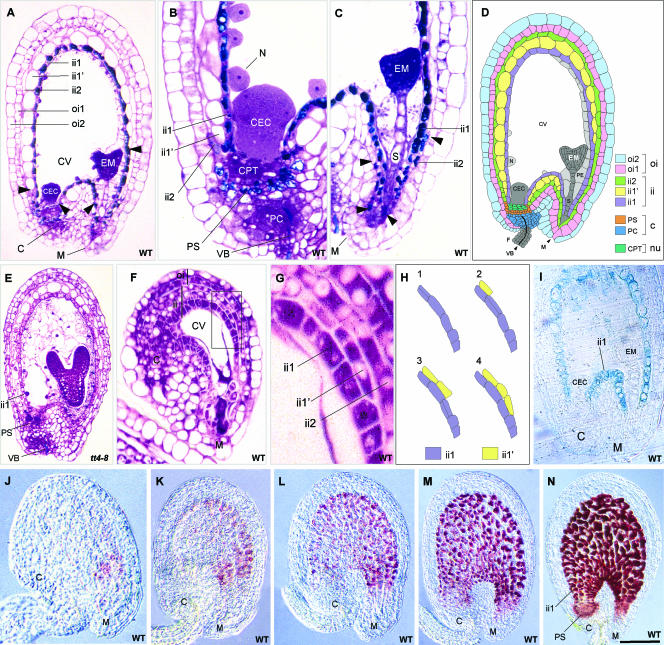
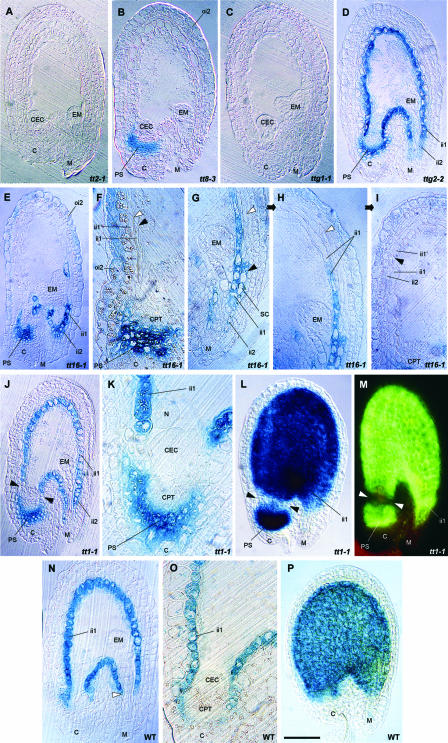
A histochemical analysis of developing Arabidopsis seeds was performed to determine the distribution of PA-accumulating cells. Staining of a transverse section of an immature wild-type seed with toluidine blue O (TBO) revealed the presence of polymeric phenolic compounds, mainly in the innermost layer of the inner integument (endothelium or ii1 layer) from the micropyle to the chalaza (Figure 2A), in cells of the chalazal tissue underlying the endosperm cyst (Figure 2B), in a few cells of the ii2 layer at the micropylar end (Figure 2C), and at the level of the xylem vessels in the vascular bundle of the funiculus and chalaza (Figure 2B). All cells that accumulated polymeric phenolics, except those detected at the xylem level, colocalized with the red products developed by the vanillin assay (Figure 2N), suggesting that they include PAs. The presence of blue staining only at the xylem level on a transverse section of a tt4-8 mutant seed lacking flavonoids but not lignins (Figure 2E) was consistent with this proposition.
Figure 2.
Testa Structure and PA Localization in Developing Seeds.
(A) to (C) Longitudinal sections of wild-type seeds at the heart stage of embryo development stained with TBO.
(A) Whole seed. Arrowheads show the limits of the ii1′ layer.
(B) Chalazal area.
(C) Micropylar area. Arrowheads show the limits of the pigmented ii2 region.
(D) Scheme of Arabidopsis seed anatomy. The integumentary layers are labeled according to Beeckman et al. (2000); the endothelium corresponds to the ii1 layer.
(E) Longitudinal section of a tt4-8 seed lacking flavonoids but not lignins (TBO staining).
(F) and (G) Longitudinal section of a mature wild-type ovule (TBO staining). The micropylar region in which periclinal divisions of endothelial cells took place (boxed area in [F]) is magnified in (G).
(H) Scheme of periclinal division patterning in the boxed region from (F).
(I) Expression pattern of the ProAtML1:uidA fusion construct in a developing seed.
(J) to (N) Accumulation of PAs during seed development (whole-mount vanillin staining). Embryo stages are two cells (J), early globular (K), globular (L), early heart (M), and heart (N).
C, chalaza; CEC, chalazal endosperm cyst; CPT, chalazal proliferating tissue; CV, central vacuole; EM, embryo; F, funiculus; ii, inner integument; M, micropyle; N, nodule; Nu, nucellus; oi, outer integument; PC, placentochalaza; PE, peripheral endosperm; PS, pigment strand; S, suspensor; VB, vascular bundle. Bar in (N) = 65 μm for (A), (D), and (I), 35 μm for (B) and (C), 75 μm for (E) and (M), 50 μm for (K) and (L), 100 μm for (N), 25 μm for (F) and (J), and 10 μm for (G).
Together, these data show that PAs accumulate in three different cell types from the distal to the proximal area of the seed (for schemes, see Figures 2D and 9A): first, a few cells belonging to the ii2 layer and located at the micropylar end (region 1); second, endothelial cells forming the ii1 layer (region 2); and third, chalazal cells lying between the vascular bundle and the nucellar chalazal proliferating tissue (region 3). Region 3 builds a junction between the adaxial and abaxial extremities of the endothelium and forms the pigment strand, analogous to the situation described in wheat by Zee and O'Brien (1970). Here, we consider that the two integuments and the chalaza (placentochalaza and pigment strand) constitute the seed coat (or testa).
A developmental feature of endothelial cells is their capacity to perform one periclinal division that gives rise to the ii1′ cell layer on their outer side in nearly mature ovules (Schneitz et al., 1995). The first periclinal divisions are detected in the chalazal region on both the abaxial and adaxial sides of the endothelium (Figure 2F). They progress toward the micropyle until ∼15 to 20 cells at the abaxial side and 6 to 8 cells at the adaxial side have divided. When the divisions have taken place, ii1′ cells extend and vacuolize until they resemble parenchymatic cells of the above-situated inner integument layers (Figures 2G and 2H). Therefore, daughter cells have completely different fates, one specializing as a tannin-producing cell and the other becoming a parenchymatic cell that participates in the formation of ground tissue. The difference between these cell types is particularly obvious in young seeds (Figure 2A). Additionally, the marker specific to the meristem L1 layer, ProAtML1:uidA (Lu et al., 1996), was expressed in endothelial cells but not in the ii1′ layer (Figure 2I), which is another indication that daughter cell fates are different. Interestingly, no β-glucuronidase (GUS) activity was observed in either region 1 or region 3.
A time-course analysis of PA deposition during seed development was performed by staining PAs and flavan-3-ol precursors with vanillin. It showed that PA accumulation begins in the micropylar region of young seeds approximately at the two-cell stage of embryo development (Figure 2J), progresses in the seed body (Figures 2K to 2M), and ends in the chalazal bulb with the complete filling of cell vacuoles at the heart stage of embryo development (Figure 2N). Unfertilized ovules do not synthesize any PAs (data not shown).
Pattern of BAN Promoter Activity in Wild-Type Developing Seeds
BAN is required for PA biosynthesis; therefore, it has been presented as a marker of early seed coat development (Albert et al., 1997; Devic et al., 1999). Here, we propose that BAN may represent a marker specific for PA-accumulating cells.
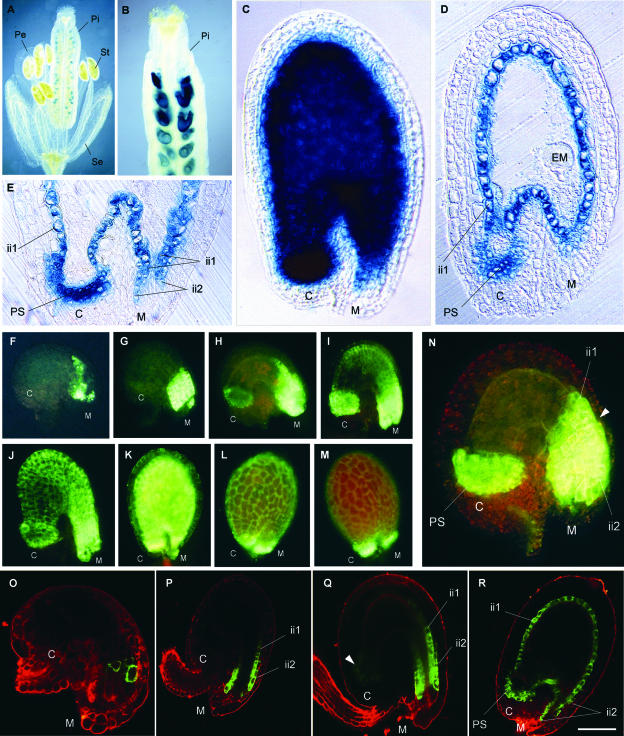
To find support for this proposition, we determined the spatiotemporal activity of the BAN promoter. A 2.3-kb fragment (referred to as ProBAN1) was fused translationally both to the uidA reporter gene, which encodes GUS, and to the mGFP5-ER reporter gene, which encodes the green fluorescent protein (GFP) fused to an endoplasmic reticulum (ER) retention sequence. We located the transcription start site (referred to as position +1) 44 bp upstream of the translation start site (ATG) by a rapid amplification of cDNA ends PCR method designed to amplify cDNAs exclusively from full-length, capped mRNAs. A database comparison of the promoter sequence with PLACE, a collection of promoter elements (http://www.dna.affrc.go.jp/htdocs/PLACE/signalscan.html), revealed a putative TATA box at −28 bp. The chimeric constructs were assayed for the resulting patterns of expression in transgenic Arabidopsis plants (Figure 3). GUS staining was detected essentially in developing seeds (Figures 3A and 3B). At the late globular stage, GUS staining generally was very intense and localized in regions 1, 2, and 3, as described previously (Figures 3C to 3E).
Figure 3.
Tissue-Specific Pattern of BAN Promoter Activity.
(A) to (C) Expression of ProBAN1:uidA. GUS activity is observed on whole mounts, with Nomarski optics.
(A) Flower.
(B) Young silique.
(C) Immature seed at the late-globular stage of embryo development.
(D) and (E) Expression of ProBAN1:uidA. GUS activity is observed on sections of seeds at the globular stage, with Nomarski optics.
(D) Whole seed.
(E) Detailed view of the micropylar/chalazal area.
(F) to (N) Expression of ProBAN1:mGFP5-ER. GFP activity is observed on whole mounts, with a standard fluorescence microscope.
(F) Young unfertilized ovule.
(G) Mature unfertilized ovule.
(H) Young seed at the one-cell stage of embryo development.
(I) Young seed at the two-cell stage.
(J) Early globular stage.
(K) Torpedo stage.
(L) and (M) Cotyledonary stage.
(N) Seven-day-old ovule obtained on a castrated flower showing ProBAN1 activity only in chalazal and micropylar areas. The arrowhead shows the limit of ProBAN activity in the ii2 layer.
(O) to (R) Expression of ProBAN1:mGFP5-ER. GFP activity is observed on confocal sections.
(O) Young unfertilized ovule.
(P) Young seed at the two-cell stage.
(Q) Seed at the early globular stage.
(R) Seed at the late globular stage.
C, chalaza; EM, embryo; ii, inner integument; M, micropyle; Pe, petal; Pi, pistil; PS, pigment strand; Se, sepal; St, stamen. Bar in (R) = 1 mm for (A), 250 μm for (B), 50 μm for (C), (D), (G), and (H), 35 μm for (F) and (O), 60 μm for (I) and (P), 80 μm for (J) and (R), 120 μm for (K), 150 μm for (L) and (M), 25 μm for (N), and 70 μm for (Q) and (E).
The ProBAN1:mGFP5-ER construct was used to monitor BAN promoter activity at various stages of ovule and seed development. GFP signal was detected first at the micropyle level in nearly mature ovules, before floral bud opening, in the absence of fertilization (Figures 3F and 3O). After fertilization, ProBAN1:mGFP5-ER expression progressed in the ii2 layer and in the endothelium, and at the same time it appeared at the chalazal level (Figures 3G to 3I, 3P, and 3Q), until it extended to all of the pigmented tissue (Figures 3J, 3K, and 3R). At the early cotyledonary stage, the GFP signal began to fade (Figure 3L), and in fully developed embryos (Figure 3M), expression was detected only in the micropyle and chalaza. In the absence of fertilization, GFP activity was restricted to the micropylar and chalazal areas (Figure 3N, cf. Figure 3K). The chalazal area corresponded to region 3, but the micropylar area included region 1 and part of the adjacent endothelium (region 2).
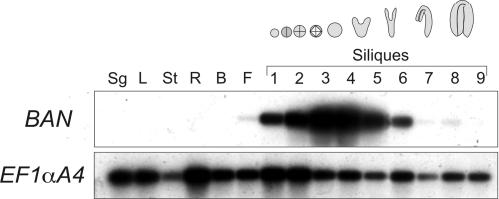
In parallel, an analysis of BAN expression was performed in several organs by semiquantitative reverse transcriptase–mediated (RT) PCR (Figure 4). mRNA was detected exclusively in reproductive tissue, from flowers to siliques at the cotyledonary stage of embryo development, with a peak in siliques at late globular to early heart stage.
Figure 4.
Pattern of BAN mRNA Accumulation.
BAN mRNA was detected in various organs by semiquantitative RT-PCR after 21-cycle PCR amplification and hybridization with a BAN cDNA probe. The elongation factor EF1αA4 was used as a control. The numbers 1 to 9 indicate siliques at various stages of development; the mean embryo stage is indicated by the drawings at top. B, buds; F, flowers; L, rosette leaves; R, roots; Sg, 4-day-old seedlings; St, stems.
Functional Dissection of the BAN Promoter
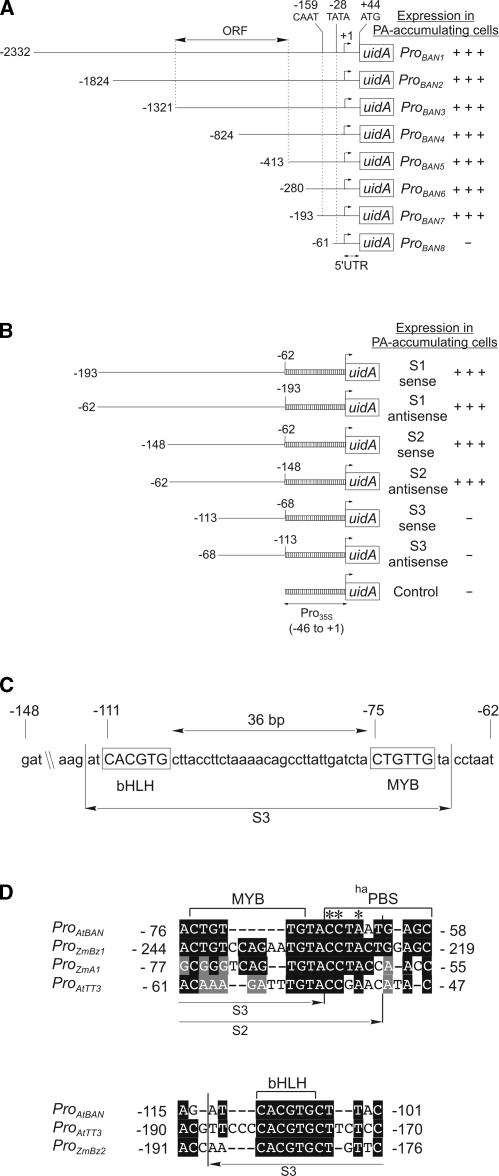
To define the domains within the 2.3-kb BAN promoter that might be important for the transcriptional regulation of tissue specificity, 5′ deletions were generated and in-frame translational fusions to the uidA gene were made (Figure 5A). Deletions to position −193 did not affect the uidA expression pattern and apparent intensity. A further deletion to position −61 resulted in a complete loss of GUS activity. Therefore, the region between −193 and +44 contains the elements necessary for expression in PA-accumulating cells.
Figure 5.
Functional Dissection of the BAN Promoter.
(A) Scheme of the ProBAN:uidA 5′ deletion constructs. The transcription start was used as a reference for numbering (+1). The double-headed arrow indicates the first open reading frame (ORF) upstream of BAN. +++, presence of strong activity; −, absence of activity. UTR, untranslated region.
(B) Scheme of the constructs used for gain-of-function experiments. 5′ and 3′ deletions were fused in both orientations to a minimal 35S promoter.
(C) Putative MYB and bHLH binding sites detected by analysis of the S2 sequence (PA enhancer) with the SignalScan server (PLACE database) are shown. The S3 subfragment, which is inactive in gain-of-function experiments, is indicated.
(D) Sequence similarity between the MYB and bHLH regions of the PA enhancer and promoters of several genes involved in PA and/or anthocyanin biosynthesis. Subfragments S2 and S3, used in gain-of-function experiments, are indicated. Numbering for ProTT3 is from the ATG. Asterisks indicate potentially crucial bases for ProBAN activation. At, Arabidopsis thaliana; haPBS, high-affinity P binding site; Zm, Zea mays.
To determine more precisely the regulatory region of ProBAN7 responsible for expression in pigmented cells, we conducted a gain-of-function analysis with this promoter fragment and subfragments (Figure 5B). The subfragments −193 to −62 (referred to as S1), −148 to −62 (S2), and −114 to −68 (S3) were fused to a minimal (−46 to +1) Cauliflower mosaic virus 35S promoter:uidA reporter construct in the sense and antisense orientations. The pattern and strength of GUS activity displayed by S1- and S2-containing constructs were identical to those conferred by the ProBAN7:uidA construct, regardless of the orientation of the subfragments. By contrast, no GUS activity was detected with the S3-containing construct. We can infer from these data that the region between −148 and −62 is necessary and sufficient for reporter gene expression in pigmented cells. This 86-bp domain is functional in both the sense and antisense orientations and, therefore, behaves as an enhancer element. Hereafter, it will be called the PA enhancer.
Database comparison using PLACE showed the presence of several putative cis-acting regulatory elements in the BAN promoter region corresponding to the PA enhancer. Interestingly, the −111 to −70 region contains a potential MYB binding site (CTGTTG; core consensus sequence CNGTTR) and a basic region helix-loop-helix/Leu zipper (bHLH/ZIP) binding site (core sequence CACGTG; also called the G-box) (Figure 5C). There are different target recognition sites for different groups of MYB proteins (Jin and Martin, 1999). The MYB site CNGTTR is bound by animal c-MYB proteins (Lüscher and Eisenman, 1990). It also was shown previously to be recognized at least by the plant transcription factor MYB.Ph3 from petunia (Solano et al., 1995), which is involved in flavonoid metabolism. The palindromic CACGTG motif belongs to a large family of ACGT-containing cis-acting elements defined in many plant promoters and bound by bZIP and bHLH transcription factors (Weisshaar and Jenkins, 1998).
In addition to BAN, TT2, TT8, and TTG1 also were shown to activate TT3, encoding dihydroflavonol reductase (Nesi et al., 2000), and TT12, encoding a multidrug and toxic compound extrusion transporter (Debeaujon et al., 2001; N. Nesi, I. Debeaujon, and L. Lepiniec, unpublished data). This finding suggests that BAN, TT3, and TT12 expression are coordinated to fulfill PA biosynthesis and compartmentation. To determine whether the corresponding promoters share a common organization with respect to regulation by TT2 and TT8, their 500-bp proximal promoter sequences were aligned with the sequence of the PA enhancer. Comparison with the TT3 promoter revealed significant identity in the MYB and bHLH regions of the PA enhancer (Figure 5D). No sequence homology was encountered with the TT12 promoter. In maize, the promoters of ANTHOCYANINLESS1 (A1), A2, BRONZE1 (BZ1), and BZ2 require both a MYB and a bHLH protein to be activated in a tissue-specific manner (Roth et al., 1991; Tuerck and Fromm, 1994; Bodeau and Walbot, 1996; Lesnick and Chandler, 1998). Alignment of 500-bp promoter sequences of these genes with the PA enhancer sequence revealed significant similarities in the MYB region of the PA enhancer with the BZ1 and A1 promoters and in the bHLH region with the BZ2 promoter (Figure 5D). Interestingly, 6 bp of the conserved MYB region that are absent from the inactive S3 subfragment and present in the active S2 subfragment have strong similarity with part of a “high-affinity” P binding site, CCTACCAACC (haPBS), present in ProZmA1 and also partially present in ProAtTT3 and ProZmBZ1 (Figure 5D). haPBS involves two overlapping MYB P binding sites, CCTACC and CCAACC (Grotewold et al., 1994; Pooma et al., 2002).
Pattern of BAN Promoter Activity in Regulatory Mutant Backgrounds
To gain further insight into the modalities of BAN transcriptional regulation, we studied the GUS expression pattern produced by transferring ProBAN1:uidA in plants defective for genes involved in the regulation of flavonoid metabolism in the seed coat.
Compared with the standard wild-type patterns of GUS and GFP activities described previously (Figure 3), there was a complete absence of GUS staining in the tt2-1 and ttg1-1 backgrounds (Figures 6A and 6C, respectively). In the tt8-3 background, GUS activity was abolished everywhere except in the chalazal pigment strand (Figure 6B). In the ttg2-2 background, no clear difference from the wild-type situation was observed (Figure 6D).
Figure 6.
Effect of Mutations in Flavonoid Regulatory Genes on BAN Promoter Activity.
(A) to (L) Pattern of GUS activity driven by the ProBAN1:uidA construct in regulatory mutant backgrounds.
(A) tt2-1.
(B) tt8-3.
(C) ttg1-1.
(D) ttg2-2.
(E) to (I) tt16-1.
(F) Arrowheads show elongated and vacuolated ii1 (black) and ii1′ (white) cells.
(G) to (I) views of the same seed section showing daughter cells from a supernumerary periclinal division in a uidA-expressing cell (black arrowhead) and supernumerary layers in the inner integument (white arrowhead) (G), supernumerary layers in the inner integument (arrowhead) (H), and atypical localization of the first ii1′ cell in the inner integument (arrowhead) (I).
(J) to (L) tt1-1.
(J) Arrowheads show the absence of GUS activity in a group of endothelial cells.
(K) Magnification of the chalazal region in (J).
(L) Whole mount showing a lack of GUS activity at the base of the endothelium (arrowheads).
(M) Pattern of GFP activity driven by the ProBAN1:mGFP5-ER construct in tt1-1, confirming the lack of ProBAN activity at the base of the endothelium (arrowheads).
(N) to (O) Pattern of GUS activity driven by the ProTT1:uidA construct in the wild type.
(N) Section of a seed at the globular stage.
(O) Detailed view of the micropylar/chalazal area.
(P) Whole mount of a seed at the globular stage observed with Nomarski optics.
C, chalaza; CEC, chalazal endosperm cyst; CPT, chalazal proliferating tissue; EM, embryo; ii, inner integument; M, micropyle; N, nodule; oi, outer integument; PS, pigment strand; SC, supernumerary cell. Bar in (P) = 75 μm for (A) to (E), (J), (L) to (N), and (P), 35 μm for (F) to (I) and (O), and 20 μm for (K).
In the tt16-1 background, ProBAN1:uidA expression was absent from region 2, with the exception of some cells in the micropylar area, but was present in regions 1 and 3 (Figure 6E). Weak staining also was observed systematically in the outer integument (layer oi2). Endothelial cells of tt16-1 were elongated and vacuolized, so that they were not distinguishable from ii1′ cells (Figure 6F, arrowheads). Moreover, the pattern of cell divisions in the inner integument was modified, the outer integument being unaffected. Supernumerary divisions were observed, leading in some parts of the seed to an inner integument with four layers (Figures 6G and 6H). In some cases, the endothelium divided periclinally to give cells expressing the ProBAN1:uidA construct, a situation that was never observed in the wild type (Figure 6G, black arrowhead). In wild-type seeds, the first periclinal division on the chalazal side occurred nearly at the base of the endothelium (Figures 2A and 2D). However, in tt16-1, it occurred nearly at the top of the seed body (Figure 6I, arrowhead). It is remarkable that not all tt16-1 seeds harbor such a disturbed pattern of divisions, which probably is the result of the variable penetrance of the mutation.
In the tt1-1 background, GUS activity was detected in all PA-accumulating cells with the exception of a group of cells localized at the base of the endothelium (Figures 6J to 6L). In tt1-1 plants expressing ProBAN1:mGFP5-ER, this intriguing pattern of activity was even more obvious because of the absence of GFP diffusion (Figure 6M). We noticed no modification of cell shape or abnormal division pattern in the tt1-1 background. As reported previously by Sagasser et al. (2002), GUS activity triggered by ProTT1 in a wild-type background was limited to the endothelium (Figures 6N to 6P). The comparison of both expression patterns led to the suggestion that TT1 is necessary for BAN promoter activity only in the small group of cells at the chalazal boundary of the endothelium (basal part of region 2).
Role of TT2 and TT16 in BAN Regulation and PA Biosynthesis
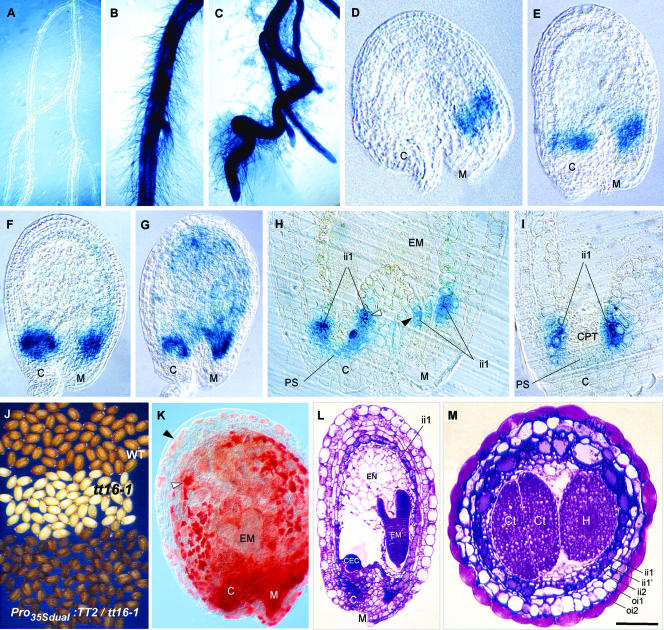
It has been demonstrated that ectopic expression of TT2 in roots was sufficient to trigger BAN mRNA accumulation in this organ (Nesi et al., 2001). We show here that ectopic TT2 expression in roots leads to ProBAN1:uidA activation (Figures 7A to 7C). We deduce from these results that TT2 regulates BAN by activating its promoter. Both approaches emphasize the importance of TT2 in BAN regulation. To understand further the modalities of this regulation, a 2.0-kb promoter from the TT2 gene was fused translationally to the uidA gene, and the in planta pattern of GUS staining was observed (Figures 7D to 7I). Young seeds exhibited strong GUS activity first in the micropylar area (Figure 7D) and then in the chalazal area (Figure 7E). GUS activity in the seed body appeared later and was weak (Figures 7F and 7G). Seed sections revealed that GUS staining was present in cells localized at the base of the endothelium layer on both the chalazal and micropylar sides (Figures 7H and 7I). GUS activity also was detected in vascular strands of leaves and stems, at the boundaries between locules of each theca in anthers, and at the zone of fusion between the outer integument and the funiculus on the adaxial side of seeds (data not shown). Therefore, ProTT2 and ProBAN do not have exactly the same spatial pattern of activity.
Figure 7.
Role of TT2 and TT16 in PA Biosynthesis.
(A) to (C) Roots of 10-day-old transformants expressing ProBAN1:uidA (A), ProBAN1:uidA in a Pro35Sdual:TT2 background (B), and Pro35Sdual:uidA used as a positive control (C).
(D) to (I) Pattern of TT2 promoter activity in developing seeds revealed by the detection of GUS in transformants expressing ProTT2:uidA.
(D) to (G) Whole mounts were observed with Nomarski optics in a young fertilized ovule (D) and during the quadrant stage of embryo development (E), the early globular stage (F), and the globular stage (G).
(H) Seed section showing a detailed view of the micropyle/chalaza area.
(I) Seed section showing a detailed view of GUS activity in endothelial basal cells at the chalaza.
(J) to (M) Ectopic expression of TT2 in the tt16-1 background.
(J) Phenotype of mature dry seeds.
(K) Vanillin assay on a developing seed at the heart stage of embryo development showing the presence of PAs in endothelial cells (white arrowhead) and ectopically in cells of the above-situated integumentary layers (black arrowhead).
(L) Seed longitudinal section.
(M) Seed transverse section.
C, chalaza; CEC, chalazal endosperm cyst; CPT, chalazal proliferating tissue; Ct, cotyledon; EM, embryo; EN, endosperm; H, hypocotyl; ii, inner integument; M, micropyle; PS, pigment strand. Bar in (M) = 35 mm for (A), 25 mm for (B) and (C), 12 mm for (J), 80 μm for (L), 75 μm for (F) and (G), 50 μm for (E) and (M), 30 μm for (D), (H) and (I), and 65 μm for (K).
As shown above, tt16-1 was affected in tannin biosynthesis and cell differentiation in the endothelium and in the pattern of cell divisions in the inner integument. To determine whether these cellular defects are linked to a modification of flavonoid biosynthesis, we analyzed the phenotype of tt16-1 seeds ectopically expressing TT2 under the control of the 35S promoter with dual enhancer. TT2 overexpression led to pigmented seeds with a dark gray-brown color differing from the brown color of wild-type seeds (Figure 7J). It is significant that the vanillin assay detected PAs in the seed body, even if not as much as in wild-type seeds (Figure 7K, cf. Figure 2N). Moreover, ectopic deposition of tannins also was observed in integumentary layers situated above the endothelium. Seed sections showed that endothelial cells do not differ from cells in tt16-1. They confirmed that tannin deposition in endothelial cells was increased (Figure 7L, cf. Figure 2A). These results suggest that cell division and differentiation defects in tt16-1 seeds are not linked to a modification of flavonoid metabolism. Another important conclusion is that tannin biosynthesis does not require TT16 in the presence of TT2, which means that TT2 may act downstream of TT16 in the PA subpathway.
Genetic Ablation of PA-Accumulating Cells
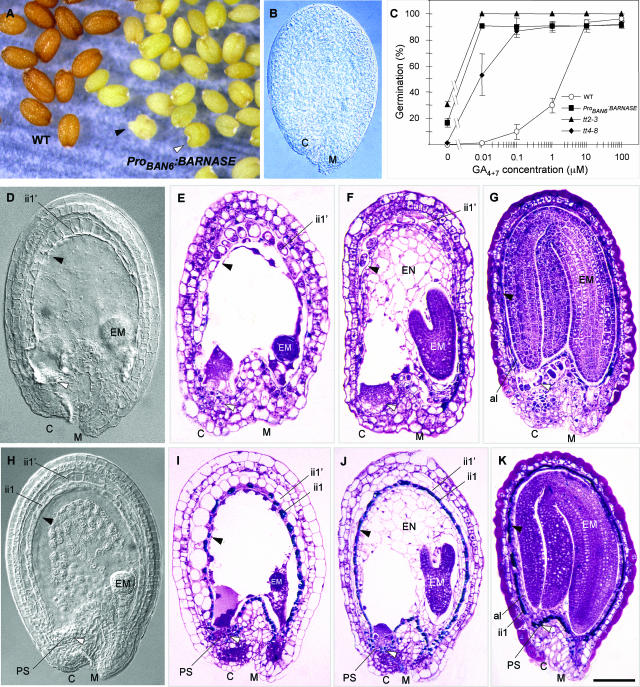
According to the expression data presented above, ProBAN appears to be very specific for the Arabidopsis seed coat pigmented tissue in which PA synthesis occurs. Therefore, it offers the opportunity to investigate the role played by this tissue in seed development and physiology. We made a translational fusion between ProBAN6 and the cytotoxic gene BARNASE. Transgenic plants carrying these constructs were obtained, and at least 20 independent transformants were characterized. Most of them exhibited normal growth and were fully fertile. They set yellow seeds (Figure 8A) as a result of the fact that the seed coat was completely deprived of PAs, as determined by the vanillin assay (Figure 8B). The germination behavior of these seeds was similar to that of tt mutants such as tt2-3 and tt4-8, in the sense that they exhibited reduced seed coat–imposed dormancy. In Figure 8C, this trait is expressed as a reduced requirement for gibberellins to germinate in restrictive conditions (presence of an inhibiting concentration of the gibberellin biosynthesis inhibitor paclobutrazol). A few percent of seeds did not germinate. This result may be attributable to the fact that in an average seed lot, we observed a few individuals with an abnormal heart shape and a tendency to initiate germination in siliques (Figure 8A, arrowheads). These seeds probably do not resist desiccation during dry storage.
Figure 8.
Genetic Ablation of PA-Accumulating Cells.
(A) Phenotype of mature dry seeds. Transgenic seeds are yellow as a result of the complete ablation of PA-accumulating cells. A few seeds exhibit a tendency to vivipary (black arrowhead) or a heart shape (white arrowhead).
(B) The lack of PAs in the seed coat is confirmed by a negative reaction with vanillin.
(C) Germination behavior of seeds from various genotypes. Seeds were sown on 100 μM of the gibberellin biosynthesis inhibitor paclobutrazol for a complete block of wild-type seed germination. Germination was scored at 7 days after sowing.
(D) to (G) Transgenic developing seeds, with arrowheads indicating missing endothelium (black) and chalazal pigment strand (white).
(D) Whole mount observed with Nomarski optics.
(E) to (G) Seed sections stained with TBO at the globular (E), torpedo (F), and cotyledonary (G) stages of embryo development.
(H) to (K) Wild-type developing seeds, with arrowheads showing the presence of endothelium (black) and chalazal pigment strand (white).
(H) Whole mount observed with Nomarski optics.
(I) to (K) Seed sections stained with TBO at the globular (I), torpedo (J), and cotyledonary (K) stages of embryo development.
al, aleurone layer; C, chalaza; CEC, chalazal endosperm cyst; CPT, chalazal proliferating tissue; EM, embryo; EN, endosperm; ii, inner integument; M, micropyle; oi, outer integument; PS, pigment strand. Bar in (K) = 6 mm for (A), 80 μm for (B), (G), and (K), 75 μm for (F) and (J), and 60 μm for (D), (E), (H), and (I).
An analysis of internal seed structures was performed by both whole-mount tissue clearing and sectioning followed by TBO staining. It revealed that the pigmented tissue layer had been removed completely (Figures 8D and 8H). The absence of pigmented cells was confirmed on sections stained with TBO (Figures 8D to 8G, cf. Figures 8I to 8K). In most seeds, the ablation of the pigmented tissue did not appear to disturb embryo development. It did not impede either endosperm formation or normal cellularization (Figures 8F and 8J), and the aleurone layer was similar to that of wild-type seeds (Figures 8G and 8K). The chalazal endosperm was present (Figure 8I).
DISCUSSION
Three Types of Cells Accumulate PAs in Arabidopsis Developing Seeds
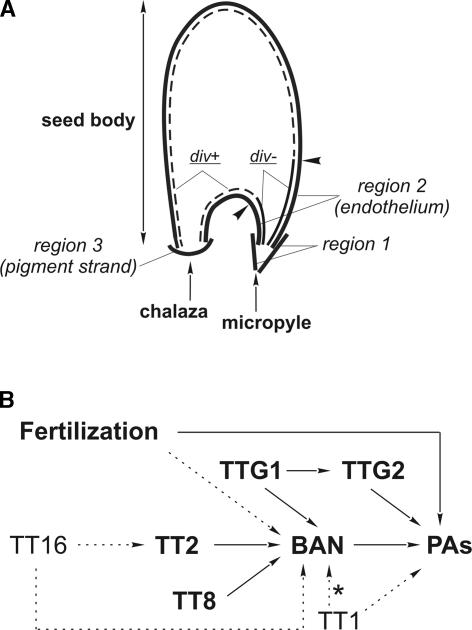
Genetic data suggest the existence of two pigmented regions (seed body and micropyle/chalaza) with different regulatory networks for flavonoid biosynthesis (Debeaujon et al., 2001). Here, we show that the pigment layer is in fact a composite assembly of cells from three different tissues (Figure 9A): micropylar ii2 cells (region 1), endothelial cells (region 2), and chalazal cells (region 3; pigment strand). The three parts are in contact with each other, with ii2 cells spanning several endothelial ii1 cells and region 3 spanning endothelial extremities. Among the mutants described to date, only seeds completely deprived of PAs, or seeds with only region 2 unpigmented, have been recovered (Debeaujon et al., 2001). An exception may be tt16, the seeds of which are pigmented in regions 1 and 3 and the micropylar part of region 2 (Nesi et al., 2002; this work). These data suggest that PA biosynthesis in region 2 occurs only when regions 1 and 3 are themselves pigmented and that pigmentation in regions 1 and 3 may be coregulated. However, we cannot reject the possibility that mutants affected only in regions 1 and 3 may exist but have not been detected because of a mild phenotype.
Figure 9.
Model for the Regulation of PA Biosynthesis in the Seed Coat.
(A) Scheme showing the cadastral organization of PA-accumulating cells in three regions and their subdivisions. Arrowheads indicate the limit between endothelial cells that underwent a periclinal division (div+) and those that did not (div−).
(B) Genetic regulatory pathway. Arrows indicate positive regulations. Gene activities prevailing in both the endothelium and the chalaza/micropyle are shown with boldface letters and solid arrows, and endothelium-specific activities are indicated with lightface letters and dashed arrows. The star indicates that the activity is necessary in a few cells at the endothelium base.
The pattern of PA deposition raises the question of how cells from three different tissues may be recruited to fulfill the same function (i.e., tannin biosynthesis). One hypothesis is that signals common to the three domains may be involved. The chalaza and the integuments share part of their developmental history, because integuments derive ontogenetically from chalazal epidermal cells (Schneitz et al., 1995). Because plant cell identity is determined mainly by positional information (Scheres, 2001), we also may imagine a second hypothesis in which some early signal derived from proximal nucellar tissue in the central chamber plays a role in determining which cells will be competent to respond to downstream regulatory signals for PA biosynthesis. A third hypothesis would be that inducers generated in regions 1 and 3 are transmitted to region 2 by diffusion or signaling.
What is the biological significance of such pigmentation patterning? We may find answers in the fact that PAs have antimicrobial and impermeabilizing properties: by closing the central chamber, region 1 (micropylar ii2 cells) and region 3 (the chalazal pigment strand) may protect the seed from pathogen invasion and also ensure moderate desiccation and imbibition rates during maturation and preparation to germination, respectively. These hypotheses are consistent with the observation that wild-type seeds are pigmented more heavily in the chalaza/micropyle than in the seed body (Debeaujon et al., 2001). Differential control of PA biosynthesis between the chalaza/micropyle and the seed body may guarantee stronger pigmentation in the regions most critical for seed survival. The epistase and the hypostase are described by plant histologists as specialized tissues with tannins that are located at the level of the insertion of the two integuments and in the micropylar parts of the seed, respectively (Boesewinkel and Bouman, 1995). Both structures, which are commonly found in many angiosperm families, are reported to perform a blocking function by closing the chalazal opening and the micropyle in the mature seed. Regions 1 and 3 probably correspond to the epistase and the hypostase, respectively.
The Endothelium Generates Daughter Cells with Different Fates
The ovular endothelium exhibits a cambium-like activity that contributes to the radial growth of the inner integument, from the chalazal region to one-third of the micropylar region (region 2/div+; Figure 9A). Daughter cells enter different differentiation programs, with endothelial cells accumulating PAs and expressing the meristem L1 layer–specific marker ProAtML1:uidA and ii1′ cells forming a ground parenchymatic layer. Endothelial cells at the micropyle (region 2/div−) do not perform such a division. Because tannins also are present in region 2/div−, the absence of periclinal divisions does not affect the competence of the endothelium to perform PA biosynthesis. The ii1′ cells may become vacuolated because of their position inside the inner integument. An example relevant for this discussion is the maize caryopsis, in which periclinal divisions that occur in a cambium-like zone at the periphery of the endosperm give rise to the aleurone layer outside and to the starchy endosperm inside. It has been proposed that positional cues would specify aleurone cell fate (Becraft and Asuncion-Crabb, 2000). A similar mechanism also may occur in Arabidopsis seeds for tannin-producing cell differentiation in the endothelium.
Regulation of BAN Tissue-Specific Expression
In developing Arabidopsis seeds, the pattern of BAN promoter activity matched perfectly BAN mRNA localization and the spatial distribution of PAs. We deduced from these data that ProBAN activity is specific for PA-accumulating cells and thus constitutes an excellent marker for these cells. These data also demonstrate that BAN tissue-specific expression is largely controlled at the transcriptional level, as observed for many flavonoid biosynthetic genes (Mol et al., 1998). In buds, BAN promoter activity but no BAN mRNA was detected. This discrepancy may be attributable to the fact that the amount of mRNA synthesized is too low to be detected by RT-PCR. Another possible explanation is that, in the absence of fertilization, BAN mRNA is less stable than uidA mRNA.
The spatiotemporal pattern of BAN promoter activity reveals the existence of three regions organized along the anteroposterior seed axis. Interestingly, this cadastral organization parallels the three mitotic domains observed in the adjacent endosperm (Boisnard-Lorig et al., 2001; Berger, 2003). BAN promoter activation starts at the micropyle, continues in the chalaza, and ends in the seed body. By contrast, PA accumulation starts at the micropyle, continues in the seed body, and ends at the chalaza. From this observation, we deduce that a mechanism that occurs downstream from BAN transcription may delay tannin accumulation in the chalazal area. Moreover, because the micropyle and chalaza are not adjacent, a signal diffusing from the micropyle to the chalaza through the central chamber may be proposed for ProBAN induction in both compartments.
An 86-bp Region of the BAN Promoter Acts as a PA-Accumulating Cell-Specific Enhancer
The PA enhancer (86 bp) was found to act as a strong positive cis element essential to direct the activity in PA-accumulating cells. A MYB and a bHLH putative binding site are present in the PA enhancer, raising the question of whether they may be binding sites for TT2 and TT8, respectively. Comparison of the PA enhancer with sequences corresponding to the promoters of the coregulated genes TT3 and TT12 revealed homology only with TT3. This finding suggests that binding site sequence similarity is not sufficient to explain a similar regulation. We hypothesize that combinatorial regulation modalities, which may vary according to the regulated gene, involve additional partner(s). Pioneer genetic studies in maize demonstrated that both a MYB and a bHLH transcription factor are necessary to activate the promoters of structural genes involved in anthocyanin biosynthesis (Lesnick and Chandler, 1998). Moreover, they showed that the MYB and bHLH trans-acting factors interact with each other directly to regulate these genes (Goff et al., 1992). Either the MYB C1 and the bHLH R that coactivate the anthocyanin pathway, or the MYB P required alone for phlobaphene biosynthesis, was shown to direct high levels of expression from promoters containing the haPBS (Grotewold et al., 1994; Pooma et al., 2002).
Based on this finding and our results, we hypothesize that a box of 6 bases present in the MYB region of the PA enhancer may be crucial for ProBAN activation. It is possible that TT2 and TT8 factors bind cooperatively to this region of the PA enhancer to activate the BAN promoter. Directed mutagenesis is under way to test this hypothesis. The functional analysis of maize promoters revealed that the region crucial for MYB/bHLH-mediated activation is compact (<200 bp) and located close to the transcription initiation start (Roth et al., 1991; Tuerck and Fromm, 1994; Bodeau and Walbot, 1996; Lesnick and Chandler, 1998). The situation encountered with the Arabidopsis BAN gene resembles the situation in maize, in that a small region located close to the transcription initiation start and containing putative MYB and bHLH binding sites was sufficient for tissue-specific activation of the promoter.
Fertilization Is Required for the Differentiation of PA-Accumulating Cells
In Arabidopsis, fertilization activates seed coat and silique differentiation from ovular and ovary tissues, respectively (Ohad et al., 1999). Fertilization also activates the BAN promoter in region 2/div+, but it is not required for activation in region 1, region 2/div−, and region 3. This finding suggests a correlation between the occurrence of the periclinal division and the need for fertilization. One hypothesis to explain this situation would be that ProBAN activation in region 2/div+ cells requires the presence of an additional activator or the repression of a negative regulator, which would be induced by fertilization. Fertilization also is required to trigger PA biosynthesis in all PA-pigmented cells by inducing ProBAN activity post-transcriptionally or by acting downstream of BAN in the pathway. Possible candidates for the indirect repression of ProBAN activation and PA-accumulating cell differentiation in the absence of fertilization are the FERTILIZATION-INDEPENDENT SEED DEVELOPMENT proteins belonging to the Polycomb group (Luo et al., 1999; Ohad et al., 1999). Tannin deposition progresses from the micropyle to the chalaza via the seed body. Such a pattern of accumulation suggests that a signal triggered in the micropylar area by fertilization may diffuse or be transmitted to the future pigmented tissues to induce PA biosynthesis.
A Complex Regulatory Network Controls BAN Promoter Activity and PA Accumulation
Here, we provide some clues to how the six regulatory factors of PA biosynthesis known to date may activate the BAN promoter and tannin production. Mutations in TT2 and TTG1 block BAN promoter activity in all PA-accumulating cells. By contrast, some activity still is detected in the tt8-3 chalaza at the level of the pigment strand (region 3). It is possible that a bHLH paralog may replace TT8 in the chalaza to activate the BAN promoter. Together, these data strongly suggest that TT2, TT8, and TTG1 act in concert to regulate BAN expression at the transcriptional level. Additional experiments are necessary to determine if they do so by binding directly to ProBAN. It has been demonstrated that TT2 plays a key role in the determination of BAN tissue specificity of expression (Nesi et al., 2001). Here, we show that BAN induction by TT2 works through BAN promoter activation. The spatial pattern of TT2 promoter activity does not exactly match that triggered by the BAN promoter. Thus, positional information for BAN activation also probably is contributed by combinatorial interactions with TTG1, which is known to control epidermal cell fate specification (Berger et al., 1998), and with its potential bHLH partner TT8.
The TT1 zinc finger and TT16 MADS box putative transcription factors belong to another hierarchical level of regulation than the three proteins described previously. Indeed, mutations in corresponding genes abolish PA biosynthesis only in the seed body. This suggests that TT1 and TT16 play a role in the cadastral patterning of pigmentation. According to Sagasser et al. (2002), TT1 is expressed specifically in the endothelium. This finding is in agreement with the fact that, in tt1-1, PAs are present in regions 1 and 3, and it indicates that TT1 is not necessary for PA biosynthesis proper in these areas. We showed that, in the tt1-1 background, ProBAN is active in the entire region 2 (endothelium) with the exception of a group of cells at the boundary with the chalaza (region 3). A hypothesis may be proposed to reconcile these data and explain TT1 function. TT1 may be necessary for tannin biosynthesis by repressing or competing with an endothelium-specific negative regulator that interferes post-transcriptionally with BAN expression (e.g., mRNA stability) or with downstream activities necessary for PA biosynthesis. In support of this hypothesis, a motif resembling a “repression domain” (Ohta et al., 2001) is present in the N-terminal part of TT1. Moreover, TT1 lacks a typical DNA binding domain (Takatsuji, 1999; Sagasser et al., 2002). It is tempting to speculate that TT1 may repress or compete with a factor that would negatively regulate BAN expression at the transcriptional level in the endothelial group of cells located at the boundary with the chalaza. Both proteins would overlap in this expression domain.
As demonstrated previously (Nesi et al., 2002), the absence of TT16 abolishes ProBAN activity and PA biosynthesis in the endothelium, with the exception of a few cells at the micropyle level. Importantly, it also was reported to increase the elongation and degree of vacuolization of endothelial cells. Here, we show that the pattern of periclinal cell divisions in the inner integument also is affected severely in this mutant, in the sense of a modification of their position and an increase in their frequency. Notably, in the tt16-1 background, BAN promoter activity is not affected in region 2/div− or in supernumerary daughter cells issued from their periclinal divisions. Together, these observations suggest that TT16 could act in region 2 by repressing a signal that activates cell division and elongation. Independently, TT16 also may enable cell differentiation toward ProBAN activation and PA biosynthesis in region 2/div+. The ectopic expression of TT16 leads to ectopic PA biosynthesis (Nesi et al., 2002), which confirms that TT16 plays a major role in the differentiation of PA-accumulating cells.
The fact that the ectopic expression of TT2 leads to a complementation of the tt16-1 phenotype for tannin biosynthesis but not for cell shape leads to two conclusions. First, TT16 acts upstream of TT2 in the regulatory hierarchy, possibly by activating it. Second, TT16 performs parallel and independent functions in cell morphogenesis, as proposed above. This situation resembles that reported in petunia (Spelt et al., 2002), in which the bHLH protein AN1 was reported to control pigment synthesis and seed coat development by genetically distinct mechanisms. The differential regulation of PA biosynthesis in the endothelium and in the micropyle/chalaza is puzzling. TT16 and TT1 paralogs may be active in the micropyle/chalaza or may be necessary only to counteract endothelium-specific negative regulators.
The disruption of TTG2 leads to the formation of homogeneously dark yellow seeds and therefore is necessary for PA biosynthesis in all PA-accumulating cells (Johnson et al., 2002). We show here that TTG2 does not perform this function by activating the BAN promoter. Therefore, TTG2 may be involved in either post-transcriptional mechanisms that affect BAN function or in downstream processes that lead to the formation of PA-accumulating cells. This hypothesis is consistent with the proposition of Johnson et al. (2002) that TTG2 acts downstream of TTG1 to enable tannin biosynthesis. TTG2 is needed with, but independent of, the homeodomain-ZIP protein GLABRA2 (GL2) for mucilage production in the outer integument and interacts with GL2 for trichome outgrowth. By analogy with these situations, it is possible that TTG2 acts with a GL2-like homeodomain transcription factor to perform PA biosynthesis. A likely candidate may be an ortholog of ANTHOCYANINLESS2 (Kubo et al., 1999) in seeds.
On the basis of the original information presented here and data from the literature, we propose a model for gene interactions involved in PA-accumulating cell differentiation and PA biosynthesis (Figure 9).
The Role of PA-Accumulating Cells in Seed Development
Targeted ablation of PA-accumulating cells leads to the formation of viable yellow seeds with normal embryo and endosperm that behave like tt seeds in germination experiments. In addition, plants that express ProBAN6:BARNASE have a wild-type phenotype. These data confirm that ProBAN is specific for PA-accumulating cells. Moreover, they clearly demonstrate that PA-accumulating cells are not crucial for seed development. Finally, they reinforce the previous assertion that testa flavonoids are major determinants of seed coat–imposed dormancy.
The endothelium has been described by some authors as the “integumentary tapetum” (Kapil and Tiwari, 1978), because it may have a nutritive role for embryo and endosperm. However, in Arabidopsis, endosperm development and embryo nutrition apparently occur correctly, even in the absence of a functional endothelium. Therefore, we cannot designate the endothelium as a real integumentary tapetum on the model of the anther tapetum. This situation differs from that observed in petunia seeds cosuppressed for the Floral Binding Protein 7 (FBP7) and FBP11 genes, in which endothelium abortion leads to degeneration of the adjacent endosperm (Colombo et al., 1997). This difference may result from the fact that FBP7 and FBP11 act earlier than BAN during seed development. Their precocious absence may lead to more drastic developmental defects than if they were absent only from the mature ovule stage onward.
According to Nguyen et al. (2000), the chalazal endosperm cyst, with its basal haustorial portion, also plays an important role in the loading of maternal resources into developing Arabidopsis seeds. Therefore, region 3 (pigment strand) may have a strategic location, spanning both endothelial extremities and therefore disrupting nutrient loading. Indeed, it is known that PAs have impermeabilizing properties (Debeaujon et al., 2000). Consistent with this hypothesis, a major function of the developing pigment strand in cereals is to provide an efficient means of controlling, and eventually blocking, grain development (Felker et al., 1984). In Arabidopsis, PA biosynthesis in region 3 ends at approximately the late heart stage of embryo development, which corresponds approximately to the end of endosperm cellularization. At that time, we may consider the seed to be nearly nutrient autonomous, with the embryo translocating endosperm reserves in its cotyledons (maturation stage). However, our data suggest that the suppression of the pigment strand also has no obvious impact on embryo nutrition and development.
Thus, we cannot exclude the possibility that the amount and quality of endosperm and embryo reserve composition may be modified. TBO staining showed that the parenchymatic integument layers are overloaded with substances that may be nutrients diffusing from the chalazal area. Because wild-type seeds do not exhibit such a pattern of accumulation, this may mean that the endothelium would channelize and possibly metabolize nutrients. Beeckman et al. (2000) showed the presence of a cuticle on the inner cell wall of endothelial cells facing the embryo sac. The lipidic nature of this cuticle may lead to an impermeabilization of the endothelium, and it is possible that the nutrients that enter the seed through the funiculus may be guided directly into the embryo sac via the chalazal tissue without first diffusing into the testa. Therefore, transgenic seeds need to be characterized thoroughly at the biochemical level by quantitative and qualitative analyses of reserves to ascertain that PA-accumulating cell ablation does not affect reserve deposition.
Our data demonstrate that the BAN promoter is a powerful tool to investigate not only the regulation of PA-accumulating cell differentiation but also seed coat development. Future work will aim to analyze further how the six regulatory factors interact to regulate BAN expression and PA biosynthesis from promoter activation to protein synthesis.
METHODS
Arabidopsis Strains and Growth Conditions
The Arabidopsis thaliana tt1-1, tt2-1, tt4-1, ttg1-1, and ttg2-1 alleles (ecotype Landsberg erecta) were described previously (Debeaujon et al., 2000; Johnson et al., 2002). The tt2-3 (DRO55), tt4-8 (DFW34), tt8-3 (DEB122), tt16-1 (DXT32), and ttg2-2 (CTA18) alleles (ecotype Wassilewskija) were identified in the Versailles T-DNA collection (Devic et al., 1999; Nesi et al., 2000, 2001, 2002; I. Debeaujon and L. Lepiniec, unpublished data). The Pro35Sdual:TT2 line was described by Nesi et al. (2001). The ProTT1:uidA and ProAtML1:uidA lines were described by Sagasser et al. (2002) and Sessions et al. (1999), respectively. Plant growth conditions were as described by Nesi et al. (2000). The germination assay was performed according to Debeaujon and Koornneef (2000).
Genetic Crosses
To determine allelism, the DFW34 and CTA18 lines were crossed with tt4-1 and ttg2-1, respectively. In both cases, F2 seeds exhibited a mutant phenotype, indicating that the mutations are allelic. The new alleles were called tt4-8 and ttg2-2, taking into account that previous alleles were described previously by Saslowsky et al. (2000) and Johnson et al. (2002), respectively.
To construct the tt1-1, tt2-1, tt8-3, tt16-1, ttg1-1, and ttg2-2 lines containing ProBAN1:uidA and the tt1-1 line containing ProBAN1:mGFP5-ER, each mutant was crossed with a transformant expressing the corresponding fusion in a strong and representative manner. Hygromycin-resistant F2 plants producing tt seeds were considered for GUS and GFP detection. For ectopic activation experiments, the ProBAN1:uidA transformant also was crossed to a line carrying a Pro35Sdual:TT2 fusion. GUS was observed on hygromycin-resistant F2 plantlets containing both constructs, as ascertained by PCR genotyping.
Molecular Techniques
To generate pBS-GUS, the uidA:t35S cassette was released from plasmid pRTL2-GUS (Carrington and Freed, 1990) with XhoI-HindIII and cloned into XhoI-HindIII–digested pBluescript SK+ (pBS; Stratagene, La Jolla, CA). The 5′ deletion subclones of a 2.3-kb BAN promoter fragment (Figure 5A) were amplified with a proofreading DNA polymerase Pfu (PfuTurbo; Stratagene) using BAC clone T13M11 as the template. Primers used for amplification were modified by the addition of a SalI restriction site preceded by 3 random bases at the 5′ end and by the creation of a NcoI site at the first codon (ATG). Their sequences are as follows: ProBAN-NcoI (5′-GAGTCTGGTCCATGGTTGTACTT-3′), ProBAN1-SalI (5′-GTAGTCGACATTTAGGCCAAGATTTTAGGAG-3′), ProBAN2-SalI (5′-GTAGTCGACTTCTCTTCTGCTTGAAGGATTC-3′), ProBAN3-SalI (5′-GTAGTCGACAGGAGGTTTTCAAAGACTATGG-3′), ProBAN4-SalI (5′-GTAGTCGACGTATTATGTGAAGCAGCAATGG-3′), ProBAN5-SalI (5′-GTAGTCGACTGACTCGTGTAATTCGTGAAAC-3′), ProBAN6-SalI (5′-GTAGTCGACTAACAGAACCTTACTGTAACAC-3′), ProBAN7-SalI (5′-GTAGTCGACTTGGTAGATGATAACAAATCAG-3′), and ProBAN8-SalI (5′-GTAGTCGACGAGCAAGGACTATTTGCAAATC-3′).
PCR products were digested with SalI-NcoI and cloned into XhoI-NcoI–digested pBS-GUS. The ProBAN:uidA:t35S cassettes were cloned as KpnI-SmaI fragments into the binary vector pBIB-HYG (Becker, 1990). To build ProBAN1:mGFP5-ER, the ProBAN1 subclone was amplified by PCR as described above using the primers ProBAN1-5′ (5′-ATTTAGGCCAAGATTTTAGGAG-3′) and ProBAN-3′ (5′-GATTGTACTTTTGAAATTACAGAG-3′) involving the full 5′ untranslated region. The PCR product was ligated directly into the pBIN-mGFP5-ER plasmid (http://www.plantsci.cam.ac.uk/Haseloff/techniques/indexFrame.html) digested previously with HindIII-XbaI and blunted with T4 DNA polymerase (Invitrogen, Carlsbad, CA). To construct Pro35Sdual:uidA, the Pro35Sdual:uidA:t35S cassette from pBS-GUS was cloned into pBIB-HYG as a HindIII fragment. For gain-of-function experiments (Figure 5B), three subdomains of the BAN promoter (S1, S2, and S3) were amplified by PCR and cloned in the sense and antisense orientations in front of a minimum 35S promoter (−46 to +1).
The minimum 35S promoter was amplified from the pRTL2-GUS plasmid with the primers Pro35S-XhoI (5′-GTACTCGAGCAAGACCCTTCCTCTATATAAG-3′) and Pro35S-NcoI (5′-GTACCATGGTCCTCTCCAAATGAAATGAAC-3′). The PCR product was cloned into pBS-GUS as a XhoI-NcoI fragment to generate the Pro35S(-46)-GUS plasmid. BAN subdomains were obtained by PCR amplification on BAC clone T13M11 with the following primers: S1-5′ (5′-TTGGTAGATGATAACAAATCAG-3′), S1-3′ (5′-ATTAGGTACAACAGTAGATCAATAAG-3′), S2-5′ (5′-GATTCTTAGGTGAAGACAAGTTGG-3′), S2-3′ (same as S1-3′), S3-5′ (5′-ATCACGTGCTTACCTTCTAAAAC-3′), and S3-3′ (5′-TACAACAGTAGATCAATAAGGC-3′). The PCR products were cloned directly at the blunted XhoI site of Pro35S(-46)-GUS. Sx:35S:uidA cassettes were cloned as KpnI-SmaI fragments into pBIB-HYG. A 2.0-kb TT2 promoter was amplified by PCR from the MOK9 BAC as described above.
Primers used for amplification were modified by the addition of a XhoI restriction site preceded by 3 random bases at the 5′ end and by the creation of a NcoI site at the ATG. Their sequences are as follows: ProTT2-XhoI (5′-GTACTCGAGGTCAAAGTAAATTCAAACTACACG-3′) and ProTT2-NcoI (5′-CTCTTTCCCATGGTCACTTTTCTCTCTCT-3′). The amplified product was cloned as a XhoI-NcoI fragment into pBS-GUS. The ProTT2:uidA cassette was cloned into pBIB-HYG as a KpnI-SmaI fragment. For the genetic ablation experiment, The ProBAN6 promoter (see above) was cloned as a SalI-NcoI fragment into the plasmid pWP146 containing the cassette ProA6:BARNASE:BARSTAR:t35S (P. Perez, unpublished data) after ProA6 removal. The ProBAN6:BARNASE:BARSTAR translational fusion was digested with XhoI and cloned into the SalI site of pBIB-HYG. All constructs were checked by sequencing.
Binary plasmids were electroporated into Agrobacterium tumefaciens strain C58C1 GV3101 (pMP90) and used to transform Arabidopsis Wassilewskija plants as described by Nesi et al. (2000). Primary transformants were selected by plating seed progeny from infiltrated plants on B5 medium containing hygromycin (50 mg/L). For each construct, at least 10 independent transgenic lines were considered. Although some variations in the intensity of reporter gene expression were observed among lines carrying a given construct, the patterns of expression were mostly similar.
Semiquantitative reverse transcriptase–mediated PCR analysis was performed as described previously (Nesi et al., 2000). The transcription initiation site for the BAN gene was determined with the RNA Ligase-Mediated Rapid Amplification of cDNA Ends kit (FirstChoice Ambion, Austin, TX). The gene-specific primer sequences were BAN-5′outer (5′-GCACGAACGAGATCGAGCCAG-3′) and BAN-5′inner (5′-CGCGAATTCGCCAAGTTCTTGAAGTTGCC-3′). The PCR product was cloned in pBS at the SmaI site and sequenced.
Microscopy
Developing seeds for resin embedding were fixed in 2% (v/v) glutaraldehyde and 1% (v/v) paraformaldehyde in 0.1 M phosphate buffer, pH 7.2. After incubation at 4°C for 24 h, samples were washed in phosphate buffer, dehydrated using a series of graded ethanol solutions, and embedded in Technovit 7100 resin (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. Sections (4 μm) were made on a rotary Jung RM 2055 microtome (Leica Microsystems, Heidelberg, Germany) equipped with metallic blades (Heraeus Kulzer) and stained for 10 s in 0.5% (w/v) toluidine blue O (TBO; Research Organics, Cleveland, OH) in 0.1 M phosphate buffer, pH 7.2, according to Feder and O'Brien (1968) after 10 s of prestaining oxidation with pure sodium hypochlorite (9.6% active chlorine) according to Gutmann (1995), with modification. Observations were performed with an Axioplan 2 microscope (Zeiss, Jena, Germany) equipped with bright-field optics. The metachromatic stain TBO develops a greenish-blue color when associated with polyphenolic compounds such as PAs and lignins. It also stains pectic substances pink and nucleic acids and proteins purple. The whole-mount vanillin assay for PA detection was handled as described previously (Debeaujon et al., 2000). Vanillin (vanilaldehyde) can condense specifically to PAs and flavan-3-ol precursors to give a bright-red product in acidic conditions. Clearing of young seeds was obtained after overnight incubation in chloral hydrate:distilled water:glycerol (8:2:1, w/v/v). Microscopic observations were performed with Nomarski differential interference contrast optics.
For histochemical detection of GUS activity, tissues were prefixed in 90% (v/v) ice-cold acetone:water for 30 min at 4°C. After three rinses in 0.1 M phosphate buffer, pH 7.2 (PB), they were transferred in a PB solution containing 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Duchefa, Haarlem, The Netherlands), 0.1% (v/v) Triton X-100:water, 0.5 to 10 mM each of potassium ferrocyanide and potassium ferricyanide, depending on the construct, and 10 mM Na2-EDTA. Vacuum was applied for 1 h before incubating for 12 h at 37°C in the dark. Chlorophyll was removed by room temperature incubation in 70% (v/v) ethanol:water. Afterward, stained tissues were either cleared with a chloral hydrate solution (whole mount) or embedded in resin after fixation as described above.
Imaging of living tissues expressing the mGFP5-ER reporter gene was performed with a standard Axioplan 2 fluorescence microscope equipped with epifluorescence optics after mounting in water. Observations also were made with an inverted Leica TCS-NT confocal laser scanning microscope equipped with an argon/krypton laser (Omnichrome, Chino, CA) and an acousto-optical tunable filter for excitation after mounting in the lipophilic dye FM 4-64 (Molecular Probes, Eugene, OR). Photographs were digitized using an ARCUS 1200 scanner (Agfa-Gevaert, Mortsel, Belgium). The images were converted to TIFF file format and processed using Adobe Photoshop 5.0 LE (Mountain View, CA).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Loïc Lepiniec, lepiniec@versailles.inra.fr.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: AC005882 (Arabidopsis BAC clone T13M11); AB015477 (Arabidopsis BAC clone MOK9); At1g61720 (Arabidopsis BAN); At5g42800 (Arabidopsis TT3); X05068 (maize A1); X07937 (maize BZ1); and U14599 (maize BZ2).
Acknowledgments
We thank Maarten Koornneef and Mark Tepfer for critical reading of the manuscript. We also thank Jim Haseloff and James Carrington for the pBIN-mGFP5-ER and the pRTL2-GUS plasmids, respectively; Bernd Weisshaar and Allen Sessions for transgenic seeds containing the ProTT1:uidA and ProAtML1:uidA constructs, respectively; David Smyth for ttg2-1 seeds; Marcel Salanoubat for BAC clone T13M11; and Bruno Letarnec and Krystyna Gofron for supplying excellent greenhouse services. We thank Jean-Noël Plagès for his constant support. This work was supported by postdoctoral fellowships from Limagrain and Biogemma to I.D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014043.
References
- Abrahams, S., Tanner, G.J., Larkin, P.J., and Ashton, A.R. (2002). Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol. 130, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, S., Delseny, M., and Devic, M. (1997). BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 11, 289–299. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Matsuda, S.P.T. (2003). Seeing red. Science 299, 352–353. [DOI] [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft, P.W., and Asuncion-Crabb, Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127, 4039–4048. [DOI] [PubMed] [Google Scholar]
- Beeckman, T., De Rycke, R., Viane, R., and Inzé, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113, 139–148. [Google Scholar]
- Berger, F. (2003). Endosperm, the crossroad of seed development. Curr. Opin. Plant Biol. 6, 42–50. [DOI] [PubMed] [Google Scholar]
- Berger, F., Hung, C.-Y., Dolan, L., and Schiefelbein, J. (1998). Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194, 235–245. [DOI] [PubMed] [Google Scholar]
- Bodeau, J.P., and Walbot, V. (1996). Structure and regulation of the maize Bronze2 promoter. Plant Mol. Biol. 32, 599–609. [DOI] [PubMed] [Google Scholar]
- Boesewinkel, F.D., and Bouman, F. (1995). The seed: Structure and function. In Seed Development and Germination, J. Kigel and G. Galili, eds (New York: Marcel Dekker), pp. 1–24.
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Freed, D.D. (1990). Cap-independent enhancement of translation by a plant potyvirus 5′ non-translated region. J. Virol. 64, 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, L., Franken, J., Van der Krol, A.R., Wittich, P.E., Dons, H.J., and Angenent, G.C. (1997). Downregulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development. Plant Cell 9, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Léon-Kloosterziel, K.M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiol. 122, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Léon-Kloosterziel, K.M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter–like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic, M., Guilleminot, J., Debeaujon, I., Bechtold, N., Bensaude, E., Koornneef, M., Pelletier, G., and Delseny, M. (1999). The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 19, 387–398. [DOI] [PubMed] [Google Scholar]
- Feder, N., and O'Brien, T.P. (1968). Plant microtechnique: Some principles and new methods. Am. J. Bot. 55, 123–142. [Google Scholar]
- Felker, F.C., Peterson, D.M., and Nelson, O.E. (1984). Development of tannin vacuoles in chalaza and seed coat of barley in relation to early chalazal necrosis in the seg1 mutant. Planta 161, 540–549. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. [DOI] [PubMed] [Google Scholar]
- Gutmann, M. (1995). Improved staining procedures for photographic documentation of phenolic deposits in semi-thin sections of plant tissue. J. Microsc. 179, 277–281. [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Johnson, C.S., Kolevski, B., and Smyth, D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapil, R.N., and Tiwari, S.C. (1978). The integumentary tapetum. Bot. Rev. 44, 457–490. [Google Scholar]
- Kubo, H., Peeters, A.J., Aarts, M.G., Pereira, A., and Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick, M.L., and Chandler, V.L. (1998). Activation of the maize anthocyanin gene a2 is mediated by an element conserved in many anthocyanin promoters. Plant Physiol. 117, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P., Porat, R., Nadeau, J.A., and O'Neill, S.D. (1996). Identification of a meristem L1 layer–specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher, B., and Eisenman, R.N. (1990). New light on Myc and Myb. Part II. Myb. Genes Dev. 4, 2235–2241. [DOI] [PubMed] [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Stewart, A.J., Jenkins, G.I., Caboche, M., and Lepiniec, L. (2002). The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14, 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H., Brown, R.C., and Lemmon, B.E. (2000). The specialized chalazal endosperm in Arabidopsis thaliana and Lepidium virginicum (Brassicaceae). Protoplasma 212, 99–110. [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooma, W., Gersos, C., and Grotewold, E. (2002). Transposon insertions in the promoter of Zea mays a1 gene differentially affect transcription by the Myb factors P and C1. Genetics 161, 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, B.A., Goff, S.A., Klein, T.M., and Fromm, M.E. (1991). C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell 3, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasser, M., Lu, G.H., Hahlbrock, K., and Weisshaar, B. (2002). A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 16, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky, D.E., Dana, C.D., and Winkel-Shirley, B. (2000). An allelic series for the chalcone synthase locus in Arabidopsis. Gene 255, 127–138. [DOI] [PubMed] [Google Scholar]
- Scheres, B. (2001). Plant cell identity: The role of position and lineage. Plant Physiol. 125, 112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hülskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana, a light microscope study of cleared whole-mount tissue. Plant J. 7, 731–749. [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263. [DOI] [PubMed] [Google Scholar]
- Shikazono, N., Yokota, Y., Kitamura, S., Suzuki, C., Watanabe, H., Tano, S., and Tanaka, A. (2003). Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon ions. Genetics 163, 1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Nieto, C., Avila, J., Canas, L., Diaz, I., and Paz-Ares, J. (1995). Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14, 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J., and Koes, R. (2002). ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14, 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji, H. (1999). Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078. [DOI] [PubMed] [Google Scholar]
- Tuerck, J.A., and Fromm, M.E. (1994). Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell 6, 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40-repeat protein. Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar, B., and Jenkins, G.I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1, 251–257. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (1998). Flavonoids in seeds and grains: Physiological function, agronomic importance and the genetics of biosynthesis. Seed Sci. Res. 8, 415–422. [Google Scholar]
- Winkel-Shirley, B. (2002). A mutational approach to dissection of flavonoid biosynthesis in Arabidopsis. In Recent Advances in Phytochemistry: Proceedings of the Annual Meeting of the Phytochemical Society of North America, Vol. 36, J.T. Romeo, ed (New York: Elsevier), pp. 95–110.
- Xie, D.-Y., Sharma, S.B., Paiva, N.L., Ferreira, D., and Dixon, R.A. (2003). Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399. [DOI] [PubMed] [Google Scholar]
- Zee, S.-Y., and O'Brien, T.P. (1970). Studies on the ontogeny of the pigment strand in the caryopsis of wheat. Aust. J. Biol. Sci. 23, 1153–1171. [Google Scholar]