Abstract
Cytokinins are hormones that regulate cell division and development. As a result of a lack of specific mutants and biochemical tools, it has not been possible to study the consequences of cytokinin deficiency. Cytokinin-deficient plants are expected to yield information about processes in which cytokinins are limiting and that, therefore, they might regulate. We have engineered transgenic Arabidopsis plants that overexpress individually six different members of the cytokinin oxidase/dehydrogenase (AtCKX) gene family and have undertaken a detailed phenotypic analysis. Transgenic plants had increased cytokinin breakdown (30 to 45% of wild-type cytokinin content) and reduced expression of the cytokinin reporter gene ARR5:GUS (β-glucuronidase). Cytokinin deficiency resulted in diminished activity of the vegetative and floral shoot apical meristems and leaf primordia, indicating an absolute requirement for the hormone. By contrast, cytokinins are negative regulators of root growth and lateral root formation. We show that the increased growth of the primary root is linked to an enhanced meristematic cell number, suggesting that cytokinins control the exit of cells from the root meristem. Different AtCKX-green fluorescent protein fusion proteins were localized to the vacuoles or the endoplasmic reticulum and possibly to the extracellular space, indicating that subcellular compartmentation plays an important role in cytokinin biology. Analyses of promoter:GUS fusion genes showed differential expression of AtCKX genes during plant development, the activity being confined predominantly to zones of active growth. Our results are consistent with the hypothesis that cytokinins have central, but opposite, regulatory functions in root and shoot meristems and indicate that a fine-tuned control of catabolism plays an important role in ensuring the proper regulation of cytokinin functions.
INTRODUCTION
Cytokinins are a class of plant hormones that play roles in many aspects of plant growth and development, including apical dominance, the formation and activity of shoot meristems, leaf senescence, nutrient mobilization, seed germination, and pathogen responses. They also appear to mediate a number of light-regulated processes, such as deetiolation and chloroplast differentiation (Mok, 1994). Some cytokinin functions are executed primarily through the control of cell cycle activity. The most prevalent naturally occurring cytokinins are N6-substituted adenine derivatives with an unsaturated isoprenoid side chain (e.g., zeatin [Z] and isopentenyladenine [iP]). Their homeostasis is regulated by the rate of de novo synthesis, the import rate, the formation and breakdown of cytokinin conjugates (which are mainly glycosides), and the rate of export and catabolism (Mok and Mok, 2001).
Much of our knowledge about the biological activities of cytokinins is based on experiments that studied the consequences of the exogenous addition of cytokinin or the endogenous enhancement of cytokinin content (Smart et al., 1991; Faiss et al., 1997; Rupp et al., 1999). Although these gain-of-function experiments have yielded significant information about processes that can be influenced by cytokinins, they might not always reflect true in vivo functions. It is possible that the additional cytokinins trigger processes that are not normally under cytokinin control. Plants with reduced cytokinin content are expected to be more informative, because the lack of cytokinin might cause a loss-of-function phenotype for physiological and developmental traits in which cytokinins are limiting. In the past, because of the lack of specific mutants and biochemical tools, it was not possible to analyze cytokinin-deficient plants (Faure and Howell, 1999). Experimentally enhanced cytokinin catabolism in genetically engineered transgenic plants is a novel tool with which to study the consequences of cytokinin deficiency. Cytokinin oxidase/dehydrogenase (CKX; EC 1.5.99.12) catalyzes the irreversible degradation of cytokinins and in many plant species is responsible for the majority of metabolic cytokinin inactivation (Mok and Mok, 2001). The enzyme is a flavin adenine dinucleotide–containing oxidoreductase that selectively cleaves unsaturated N6 side chains from Z, iP, and their corresponding ribosides (Armstrong, 1994; Jones and Schreiber, 1997). The reaction products of iP catabolism are adenine and the unsaturated aldehyde 3-methyl-2-butenal (McGaw and Horgan, 1983). By contrast, cytokinin nucleotides, O-glucosides, and cytokinins with saturated side chains are not CKX substrates (Armstrong, 1994).
Genes that encode enzymes that catalyze cytokinin breakdown have been isolated from maize (Houba-Hérin et al., 1999; Morris et al., 1999), Arabidopsis (Bilyeu et al., 2001; Werner et al., 2001), and orchids (Yang et al., 2003). The Arabidopsis AtCKX gene family has seven members (AtCKX1 to AtCKX7) (for review, see Schmülling et al., 2003). The predicted proteins are similar in size (∼60 kD) and have a conserved binding site for the cofactor flavin adenine dinucleotide and small highly conserved domains that may play roles in substrate recognition and electron transport. Individual N-terminal signal peptides indicate different subcellular localizations of the AtCKX proteins (Schmülling et al., 2003).
In a previous study, we showed that overexpression of AtCKX genes in transgenic tobacco plants reduces the endogenous cytokinin content and profoundly influences root and shoot development (Werner et al., 2001). Here, we report the functional analysis of six gene family members in the model plant Arabidopsis. We show that all six predicted genes form functional products and that their individual overexpression in transgenic plants causes similar, although quantitatively different, cytokinin deficiency traits. We demonstrate that an important function of cytokinins is the control of cell proliferation in meristems, with opposite roles in root and shoot meristems. We show that individual members of the gene family differ in biochemical characteristics, subcellular compartmentation, and the regulation of their expression. These results indicate a pivotal role of cytokinin metabolism control in coordinating the multiple cytokinin functions in growth and development.
RESULTS
AtCKX Transgenic Arabidopsis Shows Enhanced Cytokinin Breakdown
To explore the consequences of decreased cytokinin content, we transformed Arabidopsis with individual AtCKX genes (AtCKX1 to AtCKX6) under the control of the constitutive 35S promoter of Cauliflower mosaic virus. At least 10 independent transgenic lines that showed phenotypes similar to those described below were obtained for each construct. The phenotypic changes were qualitatively similar for all genes, but overexpression of AtCKX1, AtCKX3, and AtCKX5 resulted in stronger changes than overexpression of AtCKX2, AtCKX4, and AtCKX6. Most quantitative phenotypic analyses were performed with homozygotic progeny of AtCKX1 (35S:AtCKX1-11 and 35S:AtCKX1-15), AtCKX2 (35S:AtCKX2-2 and 35S:AtCKX2-9), AtCKX3 (35S:AtCKX3-9 and 35S:AtCKX3-12), and AtCKX4 (35S:AtCKX4-37 and 35S:AtCKX4-41). All lines showed strong alterations typical of the two phenotypic classes. Detailed analyses were performed with plants overexpressing AtCKX1 and/or AtCKX2 as representatives of their respective classes. Fewer quantitative data are available for 35S:AtCKX5 and 35S:AtCKX6 transgenic plants. However, they all showed symptoms typical of cytokinin deficiency, indicating that all six analyzed genes encode functional products.
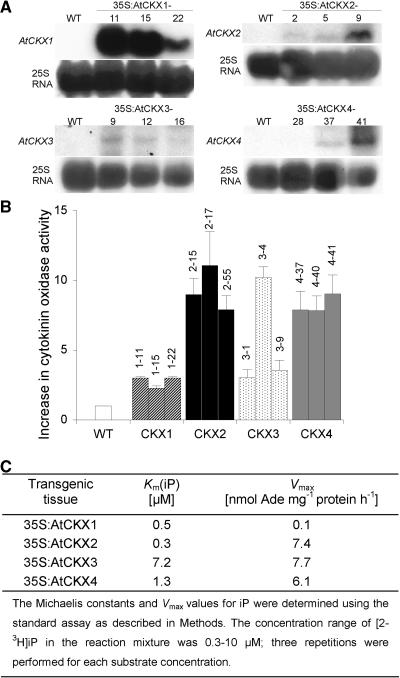
RNA gel blot analyses showed that 35S-driven gene expression enhanced the endogenous steady state mRNA level of each AtCKX gene (Figure 1A; data not shown for 35S:AtCKX5 and 35S:AtCKX6 transgenic lines). Callus derived from transgenic and wild-type root tissue was analyzed for the levels of CKX activity using 2-3H-iP as a substrate. All transgenic lines showed a 3- to 10-fold increase in CKX activity compared with the wild type. The greatest increases were found in calli overexpressing AtCKX2, AtCKX4, and AtCKX6 genes, whereas AtCKX1, AtCKX3, and AtCKX5 overexpressers had only up to threefold the wild-type activity (Figure 1B). Similarly, a threefold to fivefold enhancement of CKX activity was found in leaves of AtCKX1-overexpressing plants compared with wild-type plants (data not shown). The apparent Km(iP) values for AtCKX1, AtCKX2, and AtCKX4 enzymes extracted from transgenic callus tissue were between 0.3 and 1.3 μM, whereas the apparent Km(iP) of AtCKX3 was significantly greater (Figure 1C). Interestingly, AtCKX1 showed a considerably lower maximum velocity than the other AtCKX proteins, indicating that the enzyme has a lower turnover capacity, at least with iP as a substrate.
Figure 1.
AtCKX Gene Expression and Enzyme Activity in Transgenic Arabidopsis Plants.
(A) RNA gel blots (50 μg of total RNA) of individual transformants were probed with gene-specific probes that covered the whole genomic sequences. Only clones with enhanced AtCKX transcripts showed a phenotype. A control hybridization was performed with 25S rRNA. WT, wild type.
(B) Increase in CKX enzymatic activity in Arabidopsis callus overexpressing single AtCKX genes compared with wild-type callus. The specific activity of extracts of wild-type callus was 32.1 ± 6.4 pmol adenine·mg−1 protein·h−1. Error bars represent se; n = 3.
(C) Apparent Km(iP) and maximum velocity (Vmax) values of CKX extracts of Arabidopsis callus overexpressing single AtCKX genes.
Ectopic AtCKX Expression Decreases the Cytokinin Content and ARR5:β-Glucuronidase Reporter Gene Expression
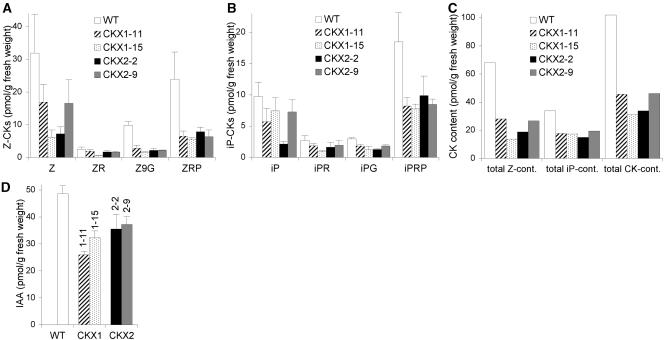
The endogenous cytokinin content was determined for two independent 35S:AtCKX1 and 35S:AtCKX2 transgenic clones. In shoots harvested 14 days after germination (DAG), most of the eight different cytokinin metabolites analyzed were reduced significantly compared with wild-type tissue (Figure 2). Z-derived metabolites are the predominant cytokinins of Arabidopsis, their concentration being approximately twofold greater than that of iP derivatives. The more abundant Z cytokinins were more strongly reduced (20 to 41% of wild type) than were iP cytokinins (44 to 58% of wild type). The total content of the measured iP and Z metabolites in individual transgenic clones ranged between 30 and 45% of wild-type concentrations. Interestingly, the overall changes in cytokinin content, as well as the metabolic spectra, were similar for both AtCKX1 and AtCKX2 overexpressers (Figure 2C), although there are apparent differences in the expressivity of phenotypic traits between these transgenic lines (see below). The concentration of dihydrozeatin-type cytokinins was either very low or below the detection limit in both wild-type and transgenic seedlings (data not shown).
Figure 2.
35S:AtCKX Transgenic Seedlings Have Lower Concentrations of Cytokinins and IAA.
(A) Endogenous concentrations of Z-type cytokinin metabolites.
(B) Endogenous concentrations of iP-type cytokinin metabolites.
(C) Total content of all measured Z-type cytokinin metabolites and iP-type cytokinin metabolites, and the sum of all measured Z- and iP-type cytokinin metabolites.
(D) Endogenous concentration of IAA.
Seedlings for hormone analysis were grown on soil under long-day conditions. Aerial tissue was collected at the same developmental stage of the plant (i.e., when six leaves were formed). Wild-type and 35S:AtCKX2 plants reached the six-leaf stage at 13 DAG, and 35S:AtCKX1 transgenic plants reached the six-leaf stage at 15 DAG. Three independently pooled samples of ∼150 mg were analyzed for each clone. Error bars represent se; n = 3. CK, cytokinin; IAA, indole-3-acetic acid; iP, N6-(Δ2isopentenyl)adenine; iPG, N6-(Δ2isopentenyl)adenine glucoside; iPR, N6-(Δ2isopentenyl)adenosine; iPRP, N6-(Δ2isopentenyl)adenosine 5′-monophosphate; WT, wild type; Z, zeatin; Z9G, zeatin 9-glucoside; ZR, zeatin riboside; ZRP, zeatin riboside 5′-monophosphate.
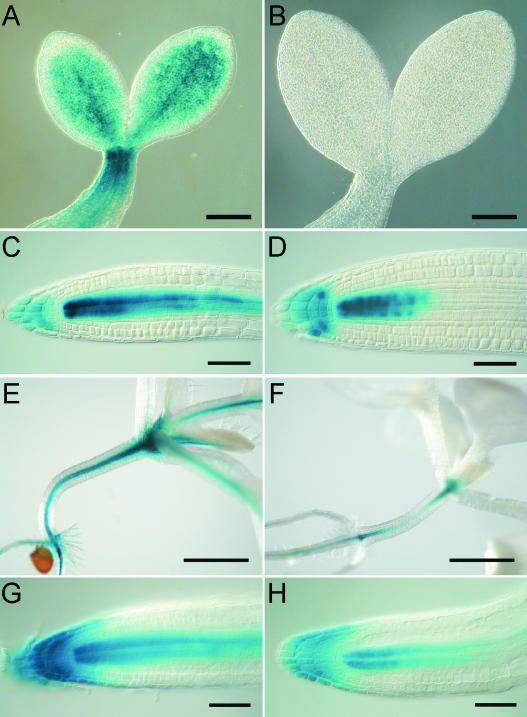
The activity of hormone reporter genes reflects both hormone content and hormone sensitivity. ARR5 is a response regulator gene of the two-component system that is transcriptionally upregulated by cytokinin (D'Agostino et al., 2000). To investigate whether the reduction in the endogenous cytokinin content was reflected by changes in the expression of a cytokinin response gene, we analyzed ARR5:GUS (β-glucuronidase) expression in the transgenic background. ARR5:GUS activity was reduced strongly during various developmental stages of 35S:AtCKX1 plants (Figure 3). The reduction was more severe in the shoot than in the root. At 2 DAG, ARR5:GUS activity was completely absent in the shoot apical meristem (SAM) of 35S:AtCKX1 transgenic plants, whereas in the root meristem, the signal was weaker but still detectable (Figures 3B and 3D). A similar but somewhat weaker reduction was observed at later stages of development (Figures 3E to 3H show plants at 9 DAG). The reporter gene assay confirms the results of the hormonal measurements and indicates that the reduction in cytokinin content was not, or not fully, compensated for by an increase in sensitivity, but that the plant reacted with a sustained reduction of the cytokinin response system. Moreover, the reduction in ARR5:GUS expression suggests that altered or reduced signaling through the two-component system may be involved causally in the phenotypic changes in the transgenic plants.
Figure 3.
Reduced ARR5:GUS Expression in the 35S:AtCKX1 Transgenic Background.
(A) to (D) ARR5:GUS expression in wild-type plants ([A] and [C]) and 35S:AtCKX1 transgenic plants ([B] and [D]) at 2 DAG.
(E) to (H) ARR5:GUS expression in wild-type plants ([E] and [G]) and 35S:AtCKX1 transgenic plants ([F] and [H]) at 9 DAG.
The duration of staining was 14 h for (A) to (F) and 80 min for (G) and (H). Bars = 200 μm for (A) and (B), 50 μm for (C), (D), (G), and (H), and 1 mm for (E) and (F).
Cytokinins frequently act in conjunction with other hormonal signals. Interactions between cytokinin and auxin have been described as occurring at multiple levels, including mutual regulation at the level of active hormone (Coenen and Lomax, 1997). Cytokinin-overproducing tobacco plants have been shown to contain lower levels of free indole-3-acetic acid (IAA) and reduced rates of IAA synthesis and turnover (Eklöf et al., 1997, 2000). It was suggested that cytokinins act as downregulators of IAA and vice versa. In contrast to this hypothesis, the levels of IAA were reduced significantly to 53 to 66% and 73 to 76% of the wild-type content in plants expressing 35S:AtCKX1 and 35S:AtCKX2, respectively (Figure 2D). This finding does not necessarily indicate a direct regulation of auxin metabolism by cytokinins. For example, it is possible that the different tissue composition in transgenic plants leads to a lower proportion of auxin-producing tissue.
Shoot Development of 35S:AtCKX Transgenic Plants Is Retarded
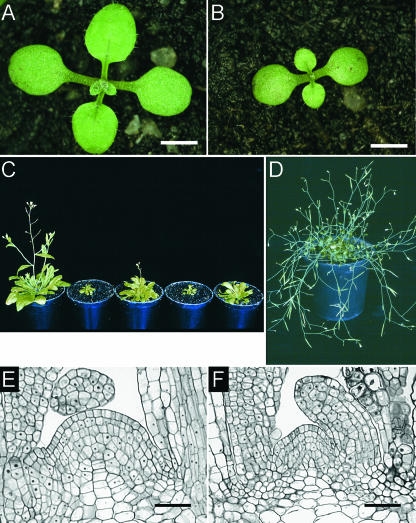
Cytokinin deficiency caused pleiotropic developmental changes during all phases of the growth cycle. In shoots, a retardation of development was noticeable soon after germination (Figure 4). The formation of new rosette leaves was delayed throughout vegetative growth in 35S:AtCKX1 and 35S:AtCKX3 transgenic plants but not in 35S:AtCKX2 and 35S:AtCKX4 transgenic plants (Figures 4A to 4C, Table 1). In ∼30% of the seedlings of homozygotic strong expressers of 35S:AtCKX1 and 35S:AtCKX3, shoot growth stopped completely at the two- to four-leaf stage (data not shown).
Figure 4.
Shoots of 35S:AtCKX-Expressing Arabidopsis Plants Show Retarded Development.
(A) and (B) Phenotypes of a wild-type seedling (A) and a homozygote 35S:AtCKX3 transgenic seedling (B) at 10 DAG.
(C) Six-week-old plants. From left to right: wild-type, 35S:AtCKX1, 35S:AtCKX2, 35S:AtCKX3, and 35S:AtCKX4 transgenic plants.
(D) A 35S:AtCKX1-expressing Arabidopsis plant grown for 4 months under long-day conditions in the greenhouse. Pot diameter is 6 cm.
(E) and (F) Median longitudinal sections of the SAMs of wild-type (E) and 35S:AtCKX1-expressing (F) plants.
Bars = 2 mm for (A) and (B) and 25 μm for (E) and (F).
Table 1.
Leaf Formation and Flowering in 35S:AtCKX Transgenic Plants
| Line | Rosette Leaf No. at 17 DAG |
Rosette Leaf No. at 27 DAG |
Bolting Time a | Leaf No. at the Time of Bolting |
|---|---|---|---|---|
| Wild type | 6.1 ± 0.1 | 12.1 ± 0.2 | 32.6 ± 0.4 | 15.5 ± 0.4 |
| 35S:AtCKX1-11 | 5.1 ± 0.1 | 8.9 ± 0.3 | 54.4 ± 3.8 | 13.1 ± 0.7 |
| 35S:AtCKX1-15 | 5.0 ± 0.2 | 8.7 ± 0.3 | 57.2 ± 2.7 | 14.6 ± 0.6 |
| 35S:AtCKX2-2 | 6.5 ± 0.1 | 11.6 ± 0.2 | 30.9 ± 0.3 | 14.8 ± 0.3 |
| 35S:AtCKX2-9 | 6.4 ± 0.1 | 11.1 ± 0.3 | 32.3 ± 0.2 | 14.3 ± 0.4 |
| 35S:AtCKX3-9 | 4.1 ± 0.1 | 6.4 ± 0.2 | 71.2 ± 2.6 | 15.8 ± 0.8 |
| 35S:AtCKX3-12 | 4.3 ± 0.1 | 8.1 ± 0.3 | 60.1 ± 3.6 | 15.8 ± 1.1 |
| 35S:AtCKX4-37 | 6.4 ± 0.2 | 11.8 ± 0.3 | 33.5 ± 0.4 | 15.9 ± 0.2 |
| 35S:AtCKX4-41 | 7.1 ± 0.1 | 12.3 ± 0.2 | 31.1 ± 0.3 | 15.9 ± 0.3 |
For each transgenic clone, two independent homozygous lines were analyzed. Data shown are mean values ±se; n ≥ 10.
Time when an inflorescence of ∼0.5 cm was apparent.
In tandem with these phenotypical alterations were distinct histological differences of AtCKX transgenic shoot tissues. The morphology of the vegetative SAM was examined in longitudinal sections of wild-type and 35S:AtCKX1 transgenic plants at 7 DAG (Figures 4E and 4F). AtCKX1 overexpression resulted in a strong reduction of the diameter and height of the meristem, which was attributable mainly to fewer meristematic cells (11.1 ± 0.8 cells across the epidermal layer of the 35S:AtCKX1 SAM versus 17.7 ± 1.1 cells for the wild-type SAM; n = 4) but also to the significantly decreased size of the meristematic cells (Figures 4E and 4F). Unlike the quantitative parameters, the typical regular structure and organization of the SAM were not altered in 35S:AtCKX1 transgenic plants.
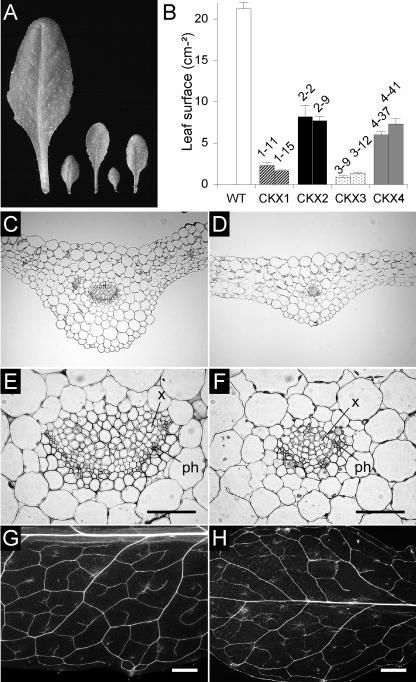
In addition to the slower formation of primordia, leaf expansion was reduced strongly in transgenic plants (Figure 5A). Rosette leaves expanded more slowly than wild-type leaves until they reached their final size, and unlike wild-type leaves, the leaves of 35S:AtCKX1 and 35S:AtCKX3 transgenic plants continued to expand significantly after the transition to flowering. Figure 5B shows that at the end of the vegetative growth phase, the surface area of leaves from the main rosette was 8 to 12% of the wild-type area for 35S:AtCKX1 and 35S:AtCKX3 transgenic plants and 30 to 38% of the wild-type area for 35S:AtCKX2 and 35S:AtCKX4 transgenic plants. To examine whether the differences in leaf size were caused by a reduced cell number or by changes in cell expansion, we determined the cell density of wild-type and 35S:AtCKX1 transgenic leaves. The number of epidermal cells per square unit was counted on the abaxial surface of the seventh fully expanded rosette leaf, in the center between the midvein and the leaf margin. The cells of 35S:AtCKX1 leaves were slightly enlarged (cell density 82% of wild type), showing that the reduced leaf size was primarily the result of a decreased cell number. Leaves of 35S:AtCKX transgenic plants showed no earlier onset of senescence, either in intact plants or in detached leaves that were kept in the dark (data not shown).
Figure 5.
Leaf Development of 35S:AtCKX-Expressing Arabidopsis Plants.
(A) Leaf size comparison. The seventh rosette leaves detached from 6-week-old plants are shown. From left to right: leaves from wild-type, 35S:AtCKX1, 35S:AtCKX2, 35S:AtCKX3, and 35S:AtCKX4 transgenic plants.
(B) Leaf surface area of the wild type (WT) and two independent homozygous lines of plants overexpressing AtCKX1, AtCKX2, AtCKX3, and AtCKX4. The surface area of leaves from the main rosette was determined at the time of bolting.
(C) and (D) Transverse sections through the central part of a fully developed wild-type leaf (C) and a leaf from a 35S:AtCKX1-expressing plant (D).
(E) and (F) Magnifications of (C) and (D) showing details of the leaf vascular systems of wild-type (E) and 35S:AtCKX1-expressing (F) plants.
(G) and (H) Vascular pattern in the fully expanded seventh rosette leaves of wild-type (G) and 35S:AtCKX1 transgenic (H) plants.
ph, phloem; x, xylem. Bars = 50 μm for (E) and (F) and 1 mm for (G) and (H).
Transverse sections through the central part of the fully developed seventh leaf showed that fewer cells were formed in the dorsoventral direction and that the extent of intercellular air spaces between blade mesophyll cells was greater than that in wild-type leaves (Figures 5C and 5D). In addition, a lower number of xylem and phloem cells resulted in a decrease in the diameter of vascular bundles in 35S:AtCKX1 transgenic plants (Figures 5E and 5F). A comparison of the vascular pattern, as visualized in whole-mount preparations of cleared leaves, showed that the leaf vasculature in 35S:AtCKX1 leaves was reduced greatly compared with that in wild-type leaves. The number and spatial density of veins of higher order (tertiary and quaternary veins and freely ending veinlets) was decreased (Figures 5G and 5H).
The reduced size of the SAM together with the retarded leaf formation and reduced cell production in the leaves provides evidence that cytokinins are required as a positive regulator of cell division activity in the Arabidopsis shoot.
The Reproductive Development of Cytokinin-Deficient Plants Is Altered
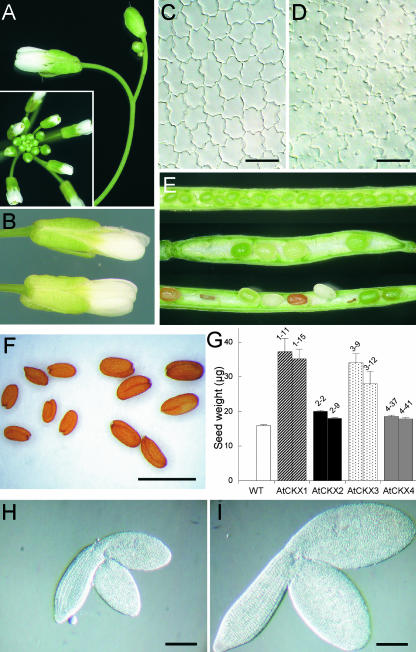
Under long-day conditions, the onset of flowering in 35S:AtCKX1 and 35S:AtCKX3 transgenic plants was delayed by up to 5 weeks compared with that in the wild type, but it was unaffected in AtCKX2 and AtCKX4 overexpressers (Table 1). However, the developmental timing of the floral transition was affected minimally, or not at all, because both wild-type and transgenic plants initiated flowering after producing approximately the same number of rosette leaves (Table 1). Occasionally, 35S:AtCKX1 and 35S:AtCKX3 transgenic plants did not flower but either remained in the vegetative stage or eventually died. During the prolonged reproductive phase and after the initiation of flowering, AtCKX1- and AtCKX3-overexpressing plants formed more axillary branches than did wild-type plants, indicating decreased apical dominance. This is the opposite effect of what was expected from decreased cytokinin content. This phenotypic trait may be the result of the decreased activity of the shoot apex, which leads to decreased auxin production. The axillary rosettes acquired the size of the main rosette, giving the plant a somewhat bushy appearance. The plants formed thin inflorescence stems that bent downward (Figure 4D). AtCKX1 and AtCKX3 transgenic plants formed very few flowers on each single inflorescence stem, indicating that the capability of apical inflorescence meristems to form new flower primordia was reduced (Figure 6A).
Figure 6.
Changes in Reproductive Development of Plants Overexpressing AtCKX Genes.
(A) Apical part of an inflorescence of a 35S:AtCKX1 transgenic plant. A wild-type inflorescence is shown in the inset.
(B) Comparison of fully developed flowers from wild-type (top) and 35S:AtCKX1 (bottom) plants.
(C) and (D) Epidermal cells from the abaxial, distal portion of fully mature wild-type (C) and 35S:AtCKX1 (D) petals.
(E) Comparison of silique development in wild type (top) and 35S:AtCKX1 (middle and bottom) plants. 35S:AtCKX1 siliques of two different developmental stages are shown. Both young fruits (middle) and older fruits (bottom) show nonsynchronous ripening and occasionally fail to develop.
(F) Comparison of mature seeds of wild-type (left) and AtCKX1-overexpressing (right) plants.
(G) Increased biomass of seeds from plants expressing AtCKX genes. The weight of one seed was calculated from the weight of pools of 200 seeds. Error bars represent se; n = 10. WT, wild type.
(H) and (I) Whole-mount preparations of the mature embryos of wild-type (H) and AtCKX1-overproducing (I) plants.
Bars = 20 μm for (C) and (D), 1 mm for (F), and 200 μm for (H) and (I).
The structure, morphology, and size of the flowers were similar to those of the wild type (Figure 6B). We examined the cell size and cell number in the distal portion of the petal epidermis. These cells have been shown to be diploid and uniform in size and shape (Mizukami and Ma, 1992). We found that fully mature petals of 35S:AtCKX1 had fewer cells per square unit than wild-type petals and that the cells were 80% larger than normal (Figures 6C and 6D). This finding suggests that fewer cells were produced per organ. However, the transgenic flowers reached the wild-type size, because the larger 35S:AtCKX1 cells compensated for the decrease in cell numbers. The fertility of flowers was diminished strongly. The first flowers did not form any pollen. Flowers formed later produced a small amount of pollen and were able to self-fertilize. Approximately 8 to 20 viable seeds were in the siliques of 35S:AtCKX1 and 35S:CKX3 transgenic plants, whereas wild-type siliques harbor up to 60 seeds. 35S:AtCKX1 and 35S:AtCKX3 siliques were not filled completely: seeds in one silique matured unequally, and some aborted during development (Figure 6E). Interestingly, the mature viable seeds of 35S:AtCKX1 and 35S:AtCKX3 transgenic clones were enlarged. Their weight was approximately two times the wild-type weight (Figures 6F and 6G). A similar enlargement was observed for transgenic embryos, which was attributable to increases in both cell number and cell size (Figures 6H and 6I and data not shown). It is not clear whether this finding suggests a role for cytokinins during embryogenesis or whether other factors, such as the reduced seed set, are involved in producing the increased seed biomass. To summarize, the reproductive development of cytokinin-deficient plants indicates that cytokinins regulate meristem activity and also limit cell formation in developing organs during the reproductive phase of development.
Root Formation and Growth of 35S:AtCKX Transgenic Plants Are Enhanced
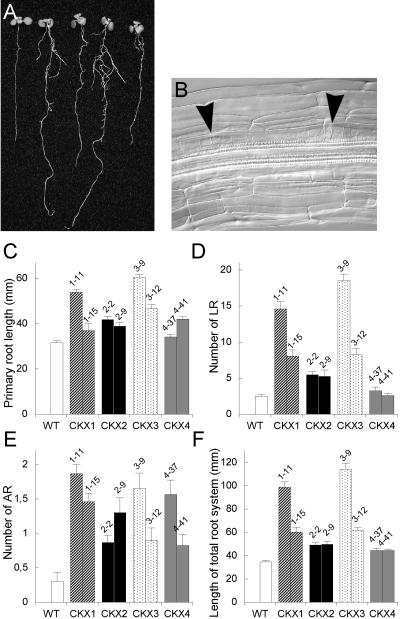
In contrast to the retarded shoot development, the root growth of 35S:AtCKX transgenic Arabidopsis plants was enhanced (Figure 7A). 35S:AtCKX1- and 35S:AtCKX3-expressing seedlings grown for 8 days under in vitro conditions had primary roots whose rates of elongation were 20 to 70% and 50 to 90% greater than that of wild-type seedlings, respectively (Figure 7C). The increase in primary root length for AtCKX2- and AtCKX4-overexpressing seedlings was less pronounced (i.e., 10 to 30% increase compared with that in the wild type). Additionally, the formation of lateral and adventitious roots was enhanced strongly (Figures 7D and 7E). The number of initiated lateral and adventitious roots in 35S:AtCKX1- and 35S:AtCKX3-expressing plants was up to seven times greater than that of the wild type. Moreover, the lateral root primordia in these transgenic clones often were initiated in much closer proximity than those in wild-type plants (Figure 7B). A significant increase of adventitious root formation, but not of lateral roots, was observed in 35S:AtCKX2 and 35S:AtCKX4 transgenic seedlings (Figures 7D and 7E). As a result, the length of the total root system increased up to three times in 35S:AtCKX1 and 35S:AtCKX3 transgenic plants and ∼1.4 times in plants expressing 35S:AtCKX2 or 35S:AtCKX4 (Figure 7F). These results suggest that cytokinins are involved in controlling both root growth rate and the generation of new root meristems.
Figure 7.
Root Phenotypes of AtCKX-Expressing Transgenic Arabidopsis Plants.
(A) Seedlings grown in vitro for 8 days. From left to right: wild-type, 35S:AtCKX1, 35S:AtCKX2, 35S:AtCKX3, and 35S:AtCKX4 transgenic seedlings.
(B) Lateral root primordia initiated in close proximity frequently were observed in 35S:AtCKX1 and 35S:AtCKX3 roots but never in wild-type roots.
(C) to (F) Morphometric analysis of root growth and development at 8 DAG. Error bars represent se; n ≥ 18. WT, wild type.
(C) Length of the primary root.
(D) Number of emerged lateral roots (LR).
(E) Number of adventitious roots (AR).
(F) Length of the entire root system, summarizing the length of the primary root, lateral roots, and adventitious roots.
Wild-type lateral root growth showed an acropetal developmental gradient. Longer lateral roots were positioned closer to the root-hypocotyl junction, leading to a cone-like structure of the root system. This developmental pattern was altered in 35S:AtCKX1- and 35S:AtCKX3-expressing plants, in which lateral roots of different lengths were distributed randomly (data not shown). This finding suggests that in the transgenic plants, the lateral root primordia were not initiated sequentially in the acropetal direction or that the growth rate of individual lateral roots differed from one to the other.
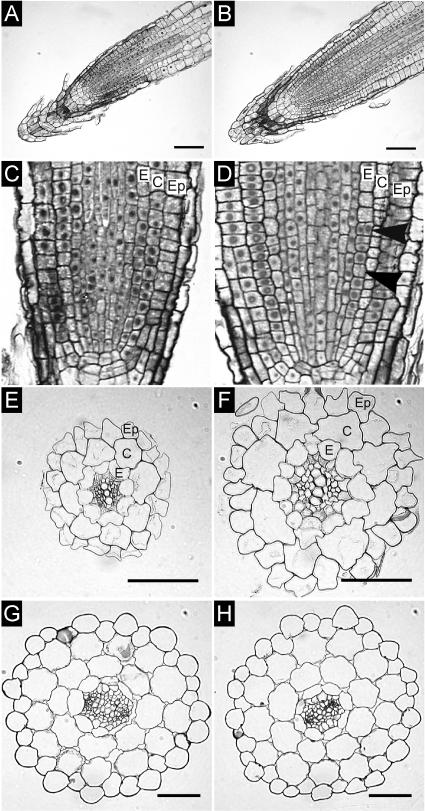
To address the question of whether the altered root growth was associated with changes in morphology and/or tissue pattern, we analyzed root sections of 35S:AtCKX1 transgenic plants and compared them with those of wild-type plants. Longitudinal sections revealed that the 35S:AtCKX1 primary root apical meristem (RAM) was enlarged in both the longitudinal and lateral directions and that the number of cells in the RAM was increased. However, the overall regular structure of the root meristem was maintained in 35S:AtCKX1 transgenic plants (Figures 8A to 8D). Transverse sections through the mature root revealed an increased number or size of cells in some cell files (Figures 8E and 8F). Whereas the wild-type epidermis was composed of 19 to 21 cell files, the 35S:AtCKX1 transgenic root contained between 24 and 27 epidermal cell files. Similarly, the 35S:AtCKX1 root contained 10 to 13 endodermal files, unlike the wild-type endodermis, which was composed invariably of 8 cell files. By contrast, the number of cortex cells was identical in wild-type and transgenic roots. Because both cortex and endodermis cell files originate from the same initials, this finding suggests that the extra endodermal cell files were formed after the basic radial pattern was established. Indeed, extra divisions of endodermal cells were visible within the meristematic region of transgenic roots (Figure 8D), supporting the notion that additional cell files did not originate from an increased number of the corresponding initial cells. Moreover, an increased radial expansion of all cell types, including xylem and phloem cells, resulted in an increased root diameter in 35S:AtCKX1 plants. In contrast to the changes in the radial root organization, cell numbers and cell sizes were not altered in most outer tissues of the hypocotyl (Figures 8G and 8H). It is noteworthy that the lower number of vascular cells in 35S:AtCKX1 transgenic hypocotyls is reminiscent of the reduced vasculature in the shoot part.
Figure 8.
Tissue Organization in Roots and Hypocotyls of 35S:AtCKX-Expressing Arabidopsis Plants.
(A) and (B) Longitudinal sections through the root meristems of a wild-type plant (A) and a 35S:AtCKX1 transgenic plant (B).
(C) and (D) Periclinal cell divisions in the endodermal tissue of a 35S:AtCKX1 root meristem ([D], arrowheads) were not observed in the wild type (C).
(E) and (F) Cross-sections of roots of wild-type (E) and 35S:AtCKX1 transgenic (F) plants.
(G) and (H) Cross-sections of hypocotyls of wild-type (G) and 35S:AtCKX1 transgenic (H) plants. Note the decreased diameter of the vascular cylinder.
C, cortex; E, endodermis; Ep, epidermis. Bars = 50 μm.
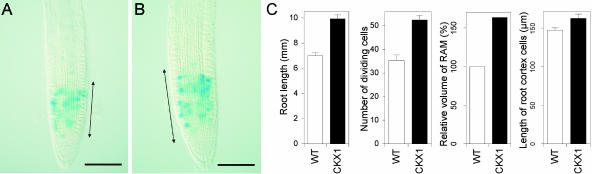
To investigate the mechanism by which decreased cytokinin content increases the root elongation rate, we examined the roots of 35S:AtCKX1 transgenic plants in detail. To examine the pattern of mitotic activity in the root meristems, we analyzed the activity of a CycB1:GUS fusion gene in the transgenic background. Owing to a mitotic degradation signal in the protein, reporter gene activity marks only actively dividing cells (Colón-Carmona et al., 1999) (Figures 9A and 9B). At 4 DAG, the primary root length of 35S:AtCKX1 seedlings was 44% greater than that of the wild type (Figure 9C). The number of dividing meristematic cells, as visualized by CycB1:GUS expression, was increased from 33.4 ± 8.6 in wild-type roots to 49.7 ± 7.7 in 35S:AtCKX1 transgenic roots (Figure 8C). The length of the cell division zone, defined here by the most distally stained mitotic cells, was increased from 174.4 ± 8.6 μm (n = 16) in the wild type to 215.9 ± 7.7 μm in 35S:AtCKX1 (n = 16). The volume of the RAM and the number of mitotic cells was estimated to be proportionally larger (Figure 8C). Thus, the number of dividing cells per RAM volume unit was not changed significantly in 35S:AtCKX1 plants. The length of mature cortical cells was increased slightly in 35S:AtCKX1 plants (Figure 8C). However, this increase was small compared with the increase in root elongation rate. Therefore, the enhanced root growth in 35S:AtCKX1 plants was attributed largely to the increased number of dividing cells in the RAM. This result suggests that cytokinins regulate the root growth rate by determining the number of dividing cells in the RAM by controlling the exit of cells from the RAM.
Figure 9.
Increased Root Meristem Activity in 35S:AtCKX1 Transgenic Plants.
(A) and (B) CycB1:GUS expression in mitotic cells of the wild-type root meristem (A) and in the 35S:AtCKX1 transgenic background (B) at 4 DAG. The arrows indicate the zone of dividing cells, defined by the stained meristematic cells. Bars = 100 μm.
(C) Statistical evaluation of root length, number of dividing cells per root meristem, relative volume of the RAM (wild-type value of 1.13 × 106 ± 0.06 × 106 μm3 was set as 100%), and average final length of root cortical cells. Error bars represent se; n ≥ 15. WT, wild type.
AtCKX Proteins Have Different Subcellular Localizations
The precise sites of cytokinin metabolism are unknown at the cellular and subcellular levels. It has been shown that specific spectra of cytokinin metabolites occur extracellularly as well as in subcellular compartments such as chloroplasts (Faiss et al., 1997; Benková et al., 1999). Sequence analyses of AtCKX proteins predict different subcellular localizations (see below), and the analyses described above of AtCKX overexpressers indicated that the site of enhanced cytokinin degradation might be relevant to the expression of the cytokinin deficiency syndrome. To contribute to a better understanding of the subcellular compartmentation of cytokinin catabolism, we attempted to analyze the subcellular localizations of several AtCKX proteins.
Although different AtCKX proteins share low sequence identity in their N-terminal regions, hydropathy plots revealed a highly hydrophobic N-terminal domain as a common feature of all AtCKX proteins, suggesting the presence of N-terminal target sequences (data not shown). The only exception is AtCKX7, which was not analyzed in this work. For most of the AtCKX proteins (AtCKX2, AtCKX4, AtCKX5, and AtCKX6), the cellular localization programs TargetP (Emanuelsson et al., 2000) and iPSORT (Bannai et al., 2002) predicted a short N-terminal signal peptide for targeting to the endoplasmic reticulum (ER) and subsequent transport through the secretory pathway. The results for AtCKX4, AtCKX5, and AtCKX6 were classified as highly reliable, whereas the AtCKX2 signal peptide prediction belonged to the lowest reliability class (RC5). However, the AtCKX2 protein has been excreted in heterologous yeast expression systems (Bilyeu et al., 2001; Werner et al., 2001).
TargetP predicted that both AtCKX1 and AtCKX3 proteins contain mitochondria-targeting peptides with RC2 and RC5, respectively. Consistently, iPSORT predicted AtCKX1 to be a mitochondrial protein, but AtCKX3 was classified as an excreted protein. Moreover, cleavage of the AtCKX3 N-terminal targeting signal as predicted by TargetP would partially delete the conserved flavin adenine dinucleotide cofactor binding site of AtCKX3, which makes the TargetP prediction unlikely. These partially ambiguous predictions and the fact that computer algorithms occasionally fail to predict subcellular localizations correctly (Emanuelsson and von Heijne, 2001; Schwacke et al., 2003) emphasized the need for in vivo targeting experiments.
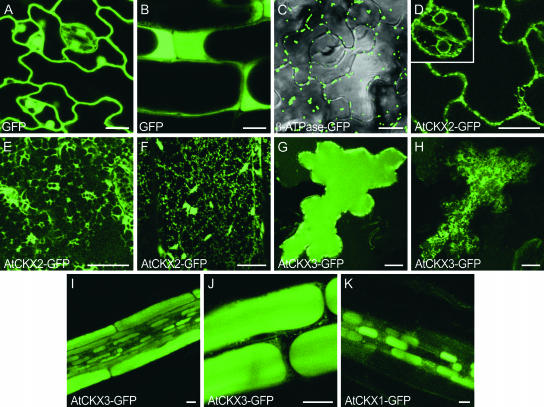
We fused the green fluorescent protein (GFP) to the C termini of AtCKX1, AtCKX2, and AtCKX3 and expressed these fusion proteins under the control of the 35S promoter in stably transformed Arabidopsis plants. Transgenic lines expressing individual AtCKX-GFP fusion genes phenocopied the corresponding AtCKX overexpressing lines, indicating that the fusion proteins were functional. As controls, we used plants expressing either GFP alone or GFP with an N-terminal mitochondrial target sequence (β-ATPase-GFP) (Logan and Leaver, 2000). The GFP distribution was monitored in abaxial epidermal leaf cells and in root cells using confocal microscopy. In control plants expressing untargeted GFP, the fluorescence signal showed the typical distribution in nuclei, in cytoplasm displaced to the cell periphery by the large central vacuole, and in transvacuolar cytoplasmic strands in young leaf epidermal cells (Figures 10A and 10B). The GFP protein targeted to mitochondria accumulated in discrete fluorescent particles distributed in the cytoplasm (Figure 10C).
Figure 10.
AtCKX1-GFP, AtCKX2-GFP, and AtCKX3-GFP Fusion Proteins Show Different Subcellular Localizations in Stably Transformed Arabidopsis Plants.
(A) and (B) Control plants, expressing 35S:GFP, typically had nuclear GFP fluorescence along with cytoplasmic signals. Optical sections through the centers of leaf epidermal cells (A) and root cells (B) are shown.
(C) Mitochondrial localization of β-ATPase-GFP in the lower leaf epidermis. Two optical sections through the middle of cells and the cell cortex were merged with the transmission image.
(D) to (F) AtCKX2-GFP is associated with the ER in lower epidermal cells of the leaf blade ([D] and [E]) and the petiole (F). The same cell is presented in different confocal sections through the center of the cell (D) and at the level of the cortical ER (E). The inset in (D) shows perinuclear fluorescence in stomatal guard cells.
(G) and (H) AtCKX3-GFP is localized in the central vacuole (G) (optical section through the center of the cell) and to ER-like structures in the cortical cytoplasm (H) (section close to the surface of the same cell as in [G]).
(I) and (J) Localization of AtCKX3-GFP in vacuoles of different root cell types (I) and higher magnification of epidermal root cells (J) (cf. control shown in [B]).
(K) AtCKX1-GFP accumulated in vacuoles of root cells in the central cylinder.
Cells were visualized with a confocal laser scanning microscope. Images in (E), (F), and (H) represent projections of three 1-μm sections close to the cell surface. Bars = 10 μm.
In comparison, optical sections through the center of cells revealed AtCKX2-GFP fluorescence to be distributed as a thin, more or less discontinuous line at the cell periphery and around nuclei (Figure 10D). In an optical section at the level of the cortex cytoplasm, the fluorescence appeared as a reticulate polygonal network often linked with punctuated structures (Figure 10E). Based on published data, this distinct pattern can be identified as indicating localization to the ER (Boevink et al., 1996, 1998; Batoko et al., 2000). The ER localization of AtCKX2-GFP was supported further by the visualization of motile, spindle-shaped fluorescent bodies in the epidermis of petioles (Figure 10F). These structures were shown previously to reside within the lumen of the ER and are typically seen in Arabidopsis plants expressing both ER-retained and secreted forms of GFP (Hawes et al., 2001). Considering that AtCKX2 does not contain a consensus H/KDEL ER-retrieval motif, it is conceivable that the protein was finally secreted. However, we have no experimental proof for this assumption, besides the accumulation of extracellular AtCKX2 enzymatic activity in yeast (Werner et al., 2001).
In contrast to the AtCKX2-GFP staining pattern, epidermal leaf cells of plants expressing high levels of AtCKX3-GFP accumulated fluorescence in the large central vacuole (Figure 10G). The uniformly stained vacuole was surrounded at the periphery by small, highly fluorescent particles that may represent intermediate transport compartments. Additionally, the ER-like fluorescent network was observed in peripheral confocal sections (Figure 10H), indicating that AtCKX3-GFP entered the ER lumen and was directed subsequently from the default secretory pathway to the vacuole. The ER-associated fluorescence was detected in most of the leaf epidermal cells, but strong vacuolar staining was confined to only a small number of cells. This might be the result of the large volume of vacuoles requiring high levels of AtCKX3-GFP expression for visualization or of the fusion protein being rapidly turned over by vacuolar proteases. Vacuolar targeting of AtCKX3-GFP was confirmed in root tissue, in which fluorescence was localized to central vacuoles, ER, and small fluorescence particles in different cell types (Figures 10I and 10J). Like AtCKX3-GFP, the AtCKX1-GFP fusion marked the ER network in leaf epidermal cells, but it was not detected in vacuoles. However, AtCKX1-GFP fluorescence was detected in vacuoles of mainly smaller root cells, such as those of the vascular cylinder (Figure 10K). This finding suggests that vacuolar targeting of AtCKX1-GFP might be specific to certain cell types, or, more likely, that the AtCKX1-GFP expression level was lower and thus stained only smaller vacuoles. Together, these data support the conclusion that the final destination of AtCKX1 and AtCKX3 is the vacuole.
Individual Members of the AtCKX Gene Family Are Expressed Differentially
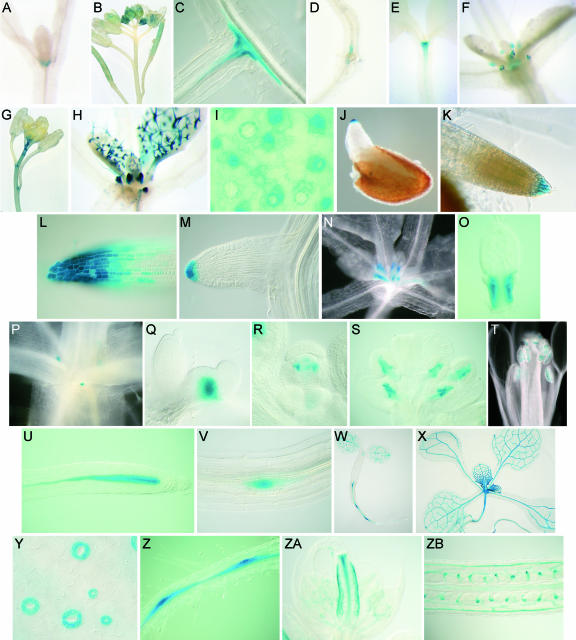
To gain further information about the individual roles of the AtCKX genes during plant development, we studied their expression patterns using promoter:GUS fusion genes (Figure 11). The AtCKX1 promoter drove gene expression in the shoot apex (Figure 11A), lateral shoot meristems, and growing tissues of young flowers (Figure 11B). In roots, AtCKX1:GUS expression was localized in close proximity to the vascular cylinder at the site of growing lateral roots (Figure 11C). GUS activity gradually increased with the increasing elongation of lateral roots. Weak AtCKX1:GUS expression was detected at the root-hypocotyl junction (Figure 11D). GUS activity disappeared in roots when the AtCKX1 promoter fragment was shortened to 1300 bp, indicating the presence of the regulatory cis element between positions −1300 and −1800 (data not shown). Expression of the AtCKX2 promoter was detected in the shoot apex (Figure 11E), in stipules (Figure 11F), and occasionally in the most apical part of inflorescence stems (Figure 11G). No GUS activity was observed in roots. Of the 30 analyzed T1 AtCKX3:GUS transformants, in which GUS expression was driven by a 1.46-kb promoter fragment, we occasionally detected very weak expression in the young shoot tissues ∼2 weeks after germination (data not shown).
Figure 11.
Expression Analysis of AtCKX Gene Promoters.
(A) to (D) Localization of AtCKX1:GUS activity. AtCKX1:GUS is expressed in the shoot apex (A) and in young floral tissues (B). In roots, AtCKX1:GUS expression is detected in the pericycle around the root branching points (C) and at the root-hypocotyl junction (D).
(E) to (G) Localization of AtCKX2:GUS activity. AtCKX2:GUS expression is observed in the shoot apex (E), stipules (F), and in the apical stem of flowering plants (G).
(I) to (M) Localization of AtCKX4:GUS activity. AtCKX:GUS4 is expressed in stipules and young trichomes (H). In leaves, the strongest expression is observed in stomatal meristemoids and clonally related cells (I). In roots, the earliest expression is detected in the root caps of germinating seedlings (J). Expression remains high in the central and lateral columnella at 5 DAG (10-min staining [K] and 80-min staining [L]). Side roots express AtCKX4:GUS at the very tip after their complete emergence (M).
(N) to (V) Localization of AtCKX5:GUS activity. AtCKX5:GUS expression is localized at the very base of the youngest emerging leaves, marking the developing leaf petiole ([N] and [O]), and after bolting in the rib zone of the axillary meristems ([P] and [Q]). In flowers, expression is detected in developing stamen ([R] and [S]) and in ripening pollen grains (T). In roots, expression is confined to the vascular cylinder within the apical meristem (U). Expression is strongest in the vascular initials directly adjacent to the quiescent center. The earliest expression in lateral root primordia is detected at the start of vascular proliferation (V).
(W) to (ZB) Localization of AtCKX6:GUS activity. Expression in the shoot is confined mainly to the vascular system of young tissues (e.g., cotyledons [W] and expanding leaves [X]) and developing stomatal guard cells of young leaves (Y). In postembryonic roots, expression is first detected at 2 DAG in the vasculature, with expression maxima around lateral root primordia ([W] and [Z]). During later root development, AtCKX6:GUS is expressed equally throughout the whole root vascular cylinder, not reaching the root meristem ([X] and data not shown). In flowers, expression is detected in the gynoecium (ZA) and the funiculus (ZB).
High AtCKX4:GUS activity was localized to trichomes, which are polyploid (Traas et al., 1998) (Figure 11H). According to the basipetal developmental pattern of trichomes in the growing leaves (Hülskamp et al., 1994), the strongest GUS activity was associated with developing trichomes in the basal part of expanding leaves, and it gradually decreased in maturing trichomes toward the distal end of the leaves (Figure 11H). A similar basipetal staining gradient was observed in developing stomata of young growing leaves. The greatest GUS activity was present in stomatal meristemoids and their clonally related cells (von Groll and Altmann, 2001) and ceased during the differentiation of guard cells (Figure 11I). Faint GUS staining was present in epidermal pavement cells. Very strong AtCKX4:GUS expression was detected in stipules, organs shown to consist of endoreduplicating cells (Jacqmard et al., 1999) (Figure 11H). Hence, the activity of the AtCKX4 promoter is linked to the mitotic and endoreduplicating cells of the vegetative Arabidopsis shoot. In roots, AtCKX4:GUS was expressed highly and specifically in the root cap. It was first detectable in the very tip of the radicle after its emergence (Figure 11J). GUS activity extended progressively in the basal direction as the lateral root columella expanded. At this developmental stage, GUS activity was very high, being detectable after a few minutes of staining in the outermost layer of the root cap (Figure 11K). Even after staining for 15 h, the root meristem proper was not stained and the stained area remained restricted to the root cap (Figure 11L). A similar staining pattern was observed during lateral root development. The onset of gene expression occurred after the complete emergence of the root primordia (Figure 11M).
In the shoot, AtCKX5:GUS expression was confined to the edges at the most basal part of the youngest leaves, specifically marking the developing leaf petiole (Figures 11N and 11O). Furthermore, it was detected in the rib zone of the axillary shoot meristems (Figures 11P and 11Q). This expression was associated with the growing axillary shoots rather than with the dormant buds, because the GUS activity was first detected soon after bolting. The number of stained axillary meristems increased when apical dominance was reduced by removing the main apex (data not shown). During flower development, AtCKX5:GUS expression was detected in young developing stamen primordia (Figure 11R) and later became confined to the central part of growing anthers (Figure 11S). Before and during pollination, expression was restricted to the maturing pollen grains (Figure 11T). In the primary root, the AtCKX5 promoter was active in the vascular cylinder within the RAM (Figure 11U). From the center of the meristem, including the initial cells proximal to the quiescent center, GUS activity decreased gradually approaching the elongation zone, and it was only faintly detectable in the differentiated root. In lateral roots, expression followed the same pattern. GUS activity was detectable in the center of lateral root primordia from developmental stage V onward (Malamy and Benfey, 1997) (Figure 11V).
AtCKX6:GUS expression was localized primarily to the vascular system of developing cotyledons, leaves, and roots. GUS activity was strongest in the vasculature of young growing leaves. It gradually decreased basipetally in older leaves and was undetectable in fully expanded leaves (Figures 11W and 11X). Likewise, expression was detected only in the vascular bundles in the most apical portion of the growing stem. In addition, the AtCKX6 promoter was active during the maturation phase of stomatal guard cells but not in fully mature stomata in older leaves (Figure 11Y), suggesting that AtCKX6 expression plays a role in stomatal development rather than in stomatal function. In the root vasculature, GUS activity was detectable soon after germination, with localized maxima forming around the lateral root primordia (Figure 11Z). Later in development, the expression expanded equally throughout the whole root vascular cylinder but did not reach the RAM. In flowers, AtCKX6:GUS expression was confined to the vascular bundles and transmitting tissue of developing gynoeciae (Figure 11ZA). When the ovules developed, GUS activity became visible in funiculi (Figure 11ZB).
DISCUSSION
We undertook a detailed analysis of the AtCKX gene family of Arabidopsis and have shown functionality for six of the seven gene family members. One important aim of this investigation was to explore the consequences of reduced cytokinin content on the development of Arabidopsis plants. This study revealed that cytokinins play crucial and opposite roles in shoots and roots.
Cytokinin Is a Positive Regulator of Shoot Meristem Activity
Soon after the discovery of cytokinins, work by Skoog and Miller (1957) showed that the hormone is able to induce the formation of shoots in unorganized growing callus tissue. However, it was unknown to what extent cytokinins are required during further development of the shoot, in particular in the shoot apex. A crucial function for cytokinins in cell proliferation in the SAM clearly is indicated by the fact that the SAM of cytokinin-deficient plants contains significantly fewer cells than that of wild-type plants and even stops its activity completely in strong AtCKX expressers. As a consequence of the decreased activity of the SAM, we observed slower leaf and flower primordia formation in 35S:AtCKX1 and 35S:AtCKX3 transgenic plants. A role for cytokinins in the SAM also was indicated by the expression of AtCKX1 and AtCKX2, which are members of the two different AtCKX classes, in the shoot apex (Figures 11A and 11E). A possibly mutual interaction of cytokinins and homeobox genes of the SAM has been reported (Sinha et al., 1993; Ori et al., 1999; Rupp et al., 1999; Frugis et al., 2001). These could present elements of a regulatory pathway that quantitatively control meristem activity. The reduced size of the SAM of cytokinin-deficient plants suggests that cytokinins not only control the cell proliferation rate but also are involved in regulating the transition from undifferentiated stem cells to differentiated primordia. If cytokinins do have a role in the establishment of specific local developmental fields, which determine the developmental fate of cells, a function of CKX could be to participate in the maintenance of such fields. In this context, it will be interesting to determine the consequences of cytokinin deficiency on meristem size in the background of known mutants affected in the meristematic transition of cell differentiation, such as clv (Clark et al., 1996), amp1 (Nogué et al., 2000), and mgo (Laufs et al., 1998).
Cytokinin Controls the Frequency of Leaf Initiation and Leaf Growth
During leaf formation, a phase of cell division is followed by cell expansion and functional differentiation. The maintenance of the meristematic competence of leaf cells defines the total cell number and therefore the size of the organs. The ability of individual cells to undergo the cell cycle, maintain the competence to proliferate, cease the cell cycle activity, and expand and differentiate depends on various external and endogenous signals and developmental cues. In our study, overexpression of different AtCKX genes led to a significant reduction of the leaf area, caused primarily by decreased cell division during leaf development, which was compensated for only partially by increased cell size. Apparently, cytokinin deficiency reduces the cell division rate and/or causes an earlier termination of leaf cell differentiation. A similar leaf phenotype has been described for Arabidopsis plants overproducing the cyclin-dependent kinase inhibitor KRP2. In these plants, cell cycle progression was limited and the uncoupling of cell growth from cell division resulted in an increase of the final cell size (De Veylder et al., 2001). By contrast, constitutive overexpression of the D-type cyclin CYCD3 (Dewitte et al., 2003) and the cell cycle–regulating transcription factors E2Fa and DPa (De Veylder et al., 2002) was shown to cause the hyperproliferation of smaller and incompletely differentiated leaf cells. These comparisons indicate that the altered leaf phenotype of AtCKX transgenic plants could be achieved through the regulation of cell cycle genes. In accordance with such a function, the expression of several AtCKX genes was associated, in leaves, with dividing cells (Figure 11). Another factor controlling cell proliferation during leaf organogenesis is AINTEGUMENTA (ANT), an APETALA2-like transcription factor (Mizukami and Fischer, 2000). Loss- and gain-of-function analyses revealed that its regulatory function is in the control of plant organ cell number and organ size. Thus, the observed reduced cell division in AtCKX plants could involve ANT.
What could be the cause of the premature arrest of leaf cell division? Based on experimental data from Drosophila, Day and Lawrence (2000) suggested a model in which individual cells monitor a concentration gradient of morphogen established along the axis of the organ. According to this model, cells grow and divide as long as the morphogen gradient is sufficiently steep. In plants, Ljung et al. (2001) showed that the IAA concentration in developing tobacco leaves exhibits a clear basipetal gradient, which is correlated with the spatial distribution of cell mitotic activity. Based on the observed reduction of cell division in cytokinin-deficient plants, it is tempting to speculate that a similar gradient of cytokinin could coordinate and limit cell proliferation during leaf growth.
Cytokinin-Deficient Leaves Do Not Exhibit an Earlier Onset of Senescence
Cytokinin has been reported to delay leaf senescence (Gan and Amasino, 1995), and it was hypothesized that decreasing the cytokinin concentration below a certain threshold level could trigger the onset of leaf senescence. In contrast to this expectation, leaf greening was not compromised in AtCKX transgenic leaves, and leaf senescence, determined visually, did not occur earlier than in the wild type. By contrast, leaves stayed green even longer in 35S:AtCKX transgenic plants, and detached leaves did not senesce more rapidly. This finding argues against the hypothesis that cytokinins act as triggers of leaf senescence if their concentration is decreased beyond a threshold level. As a note of caution, altered sink-source relations in the transgenic plants might interfere with the normal mechanism of senescence.
Cytokinin Deficiency Alters Vascular Development
Cytokinins are thought to be essential to support the continuous division of vascular cambium cells, which provides precursor cells for xylem and phloem (Aloni, 1995). Cytokinins stimulate early stages of vascular differentiation. At optimal concentrations in the presence of auxin, cytokinins favor phloem formation (Aloni, 1995). In shoots of cytokinin-deficient plants, the number of both xylem and phloem cells was reduced, indicating that the changes are the result of an overall reduction of cambial activity without affecting cell differentiation specifically. This finding supports a role for cytokinins in cambial cell division and argues against a role in xylem or phloem cell specification. Moreover, as with the apical meristems of shoots and roots, the cambial activity responds to cytokinin deficiency in an organ-specific manner. In roots, both xylem and phloem cells were slightly more abundant in cytokinin-deficient plants than in control plants.
A Role for Cytokinins during Reproductive Development
The promoter activity of AtCKX genes in various parts of the developing flowers and developmental changes shown by cytokinin-deficient plants (reduced number of flowers and increased size of embryos) indicate a role for the hormone during reproductive development. Enlarged embryos suggest that cytokinins may play a role in controlling the cell division rate in maturing embryos. Similarly, a specific role for cytokinins in controlling cell divisions during the establishment of the embryonic vasculature was demonstrated recently in a mutant of the cytokinin receptor CRE1/WOL/AHK4 (Mähönen et al., 2000). Early consequences of cytokinin deficiency might have been missed in the AtCKX-overexpressing plants, because the 35S promoter driving AtCKX expression becomes active late during embryogenesis (Custers et al., 1999). More indirect reasons for embryo enlargement, such as differences in nutrient mobilization, cannot be excluded at present.
Of particular interest is the expression of AtCKX6 in the funiculi (Figure 11ZB). The funiculus is the feeding tract for the growing embryo. A role for CKX in this tissue could be the control of sink strength. This control could be exerted locally through the regulation of invertase and hexose transporters, which are important during assimilate import and have been shown to be regulated by cytokinins (Ehness and Roitsch, 1997). Alternatively, cytokinin import into embryonic sink tissue might be controlled in the funiculus, thus controlling sink strength in a more indirect manner.
A Role for CKX Enzymes in the Cell Cycle?
Another important function of CKX proteins could be the degradation of cell cycle–derived cytokinins. The level of cytokinins increases dramatically and transiently during short periods of the cell cycle (Redig et al., 1996b). It is not known how the rapid readjustment of the original level is achieved, but CKX proteins certainly are relevant. In cell division zones, CKX may simply be relevant for the retrieval of the purine moiety of the hormone or serve as a protectant from cytokinins derived from neighboring cells, thus preserving the cytokinin autonomy of each single cell. The latter view implies that, during cell division, each cell makes its own cytokinin for its own exclusive use. Furthermore, CKX may degrade cytokinins to reset the cytokinin-sensing system to a basal level. The expression of AtCKX genes in zones of active cell division is in agreement with the proposed functions.
Cytokinins Are a Negative Regulator of Root Meristem Activity
In contrast to its promotional role in shoot organs, the root phenotype of cytokinin-deficient plants indicates that cytokinins have a negative regulatory function in root growth. Transgenic plants displayed an overall enhanced root system as a result of the more rapid elongation of primary and lateral roots and because of the increased formation of lateral and adventitious roots. We found that the enhanced growth of primary roots correlated with an increased number of dividing cells in the root meristem. The density of dividing cells was comparable to that of the wild type, suggesting that the cell division rate was not altered. We conclude that cytokinins regulate the number of divisions that cells undergo before they leave the meristem and thus are involved in the control of the exit from the proliferative state at the basal border of the meristem. This conclusion is in good agreement with the work of Beemster and Baskin (2000), who showed that exogenous cytokinin inhibits the elongation of wild-type roots primarily by reducing the number of dividing cells and the size of the meristem rather than by reducing the cell division rate.
What could be the molecular link between cytokinin and root meristem activity? One of the possible components of the downstream signaling pathway that controls meristem activity is STUNTED PLANT1 (STP1) (Baskin et al., 1995). As a result of slower cell production, stp1 mutant roots had a slower elongation rate and, more specifically, a reduced response toward cytokinin. This finding indicates that STP1 is required to mediate the cytokinin effect upon root expansion. Additionally, cytokinin-treated wild-type roots phenocopied the stp1 mutation (Beemster and Baskin, 2000). It was hypothesized that STP1 is an elongation-promoting factor that is downregulated by cytokinins. In this model, the reduced cytokinin content in AtCKX-expressing plants would lead to more STP1 protein, which in turn would allow for faster root elongation. Another possible regulatory target for cytokinins is CycB1, whose overexpression causes increased root elongation (Doerner et al., 1996).
Cytokinins Are a Regulatory Factor in Root Branching
Lateral root formation is initiated in the root differentiation zone by anticlinal divisions in pericycle cells adjacent to the protoxylem poles (Malamy and Benfey, 1997; Casimiro et al., 2001). A subsequent series of highly ordered transverse cell divisions leads to the emergence of the newly formed lateral root (Malamy and Benfey, 1997). Numerous studies have shown that auxin, and in particular its polar transport, is necessary for the initiation and the later development of lateral roots (Ruegger et al., 1997; Reed et al., 1998; Casimiro et al., 2001). The initiation phase of lateral root development is dependent on basipetal polar auxin transport and can be blocked by the auxin transport inhibitor N-1-naphthylphthalamic acid (Casimiro et al., 2001). Subsequently, shoot-derived auxin plays a role early in lateral root emergence (Casimiro et al., 2001; Bhalerao et al., 2002). Consistently, the stm1 mutant, which lacks the auxin source in the shoot, is able to initiate a number of lateral root primordia similar to those of the wild-type plant, which elongate more slowly and irregularly (Casimiro et al., 2001). Given that AtCKX transgenic plants have less auxin-producing shoot tissue than wild-type plants and have a lower auxin content, it is surprising that they initiated more lateral root primordia that elongated faster than those in the wild type. This finding suggests that cytokinin functions as an auxin antagonist during lateral root formation and must be considered an important regulatory factor in primordia initiation.
Differential Subcellular Targeting of AtCKX Proteins
We analyzed the in planta subcellular targeting of three AtCKX-GFP fusion proteins. Surprisingly, AtCKX1-GFP and AtCKX3-GFP were targeted to the plant vacuoles, although computer algorithms predicted mitochondrial localization. The enzymatic activity of AtCKX1, but not of AtCKX2, was significantly greater under acidic conditions in vitro, which is in agreement with a vacuolar localization for the former (P. Galuszka and T. Werner, unpublished data). Vacuolar soluble proteins require a sorting signal to be separated from the proteins destined for secretion. In plants, three classes of vacuolar sorting determinants have been identified (reviewed by Matsuoka and Neuhaus, 1999). Sequence-specific signals in the N-terminal propeptides containing the conserved consensus sequence NPIR or NPIXL target proteins to lytic vacuoles. The second group contains C-terminal signals with no obvious consensus sequence. They are found in proteins destined for protein storage vacuoles. The third type was defined as an integral structural domain within the mature protein. None of these motifs was found in AtCKX proteins, with the exception of NPSDIR starting at position 51 of AtCKX1, which closely resembles the consensual NPIR targeting signal. Overexpression of AtCKX1 and AtCKX3 caused a stronger cytokinin deficiency syndrome than did the overexpression of proteins with a predicted extracellular localization. It is possible that vacuolar localization creates a stronger intracellular sink for cytokinins or that a more relevant fraction of the hormone is degraded. In any case, the results indicate an important role for subcellular compartmentation in cytokinin function.
Fluorescence accumulated in AtCKX2-GFP transgenic plants primarily in the reticulate structures typical of the ER. This finding, and the fact that AtCKX2 does not contain an ER retention motif, is in agreement with the assumption that AtCKX2 is an extracellular protein. This was indicated previously by its excretion from yeast cells (Werner et al., 2001). Our inability to detect the GFP signal in the apoplast could be explained by the weak fluorescence of secreted GFP (Batoko et al., 2000). Interestingly, an increase in the extracellular CKX fraction was triggered in tobacco cell cultures by cytokinin and was correlated with enhanced CKX glycosylation (Motyka et al., 2003). Based on the facts that AtCKX proteins contain consensus N-glycosylation sites (Schmülling et al., 2003) and that protein glycosylation occurs during transport through the secretory pathway, we hypothesize that ER-located AtCKX enzymes are released to the apoplast and that the release is at least partly dependent on a triggering signal, which could be cytokinin. However, additional experiments are required to prove an extracellular localization and to demonstrate a regulatory function of AtCKX protein glycosylation in subcellular targeting and/or activity. It was shown recently that the activity of another cytokinin-metabolizing enzyme, zeatin O-xylosyltransferase, is regulated by auxin-dependent post-translational processing (Martin et al., 1997). Post-translational modification of enzymes may be a common but largely unexplored aspect of hormone metabolism.
Conclusions
We have demonstrated that cytokinin degradation is regulated in Arabidopsis by members of a small gene family. Distinct, almost nonoverlapping expression domains of the AtCKX gene members and different subcellular localizations suggest that specific developmental and physiological functions are fulfilled by each gene and that the tissue-specific regulation of the endogenous cytokinin content is an important prerequisite in the regulation of cytokinin functions in planta. The results of this study and similar phenotypic alterations described for cytokinin-deficient tobacco plants (Werner et al., 2001) suggest that conclusions drawn about the role of cytokinins in the control of root and shoot growth may be generalized.
METHODS
Generation of AtCKX-Overexpressing Plants and Transgenic Plants Harboring AtCKX Promoter:GUS Fusions
Binary plasmids harboring the AtCKX1, AtCKX2, AtCKX3, and AtCKX4 genes under the control of a 35S promoter have been described (Werner et al., 2001). Numbering of AtCKX genes was according to their appearance in the database, as described by Schmülling et al. (2003). The genomic sequences of the AtCKX5 (MIPS gene code At1g75450; http://mips.gsf.de/proj/thal/) and AtCKX6 (At3g63440) genes were amplified from DNA of Arabidopsis thaliana Columbia (Col-0) using the following primers: 5′-ggggtaccTTGATGAATCGTGAAATGAC-3′ (forward) and 5′-ggggtaccCTTTCCTCTTGGTTTTGTCCTGT-3′ (reverse) for AtCKX5 and 5′-cccgggTCAGGAAAAGAACCATGCTTATAG-3′ (forward) and 5′-cccgggTCATGAGTATGAGACTGCCTTTTG-3′ (reverse) for AtCKX6 (nonhomologous sequences used for cloning are shown in lowercase letters). The resulting gene fragments were cloned into pBINHygTx downstream of the 35S promoter of Cauliflower mosaic virus (Gatz et al., 1992).
To obtain promoter:GUS fusion genes, the promoter sequences of the AtCKX1, AtCKX2, AtCKX3, AtCKX4, AtCKX5, and AtCKX6 genes were amplified by PCR from DNA of Arabidopsis Col-0. Primers were as follows: AtCKX1 promoter, 5′-gctctagaAAATGCTCTGTTATCAATGTGTC-3′ (forward) and 5′-gctcccgggCTACTTTGTTGAGAGAAATTGCA-3′ (reverse); AtCKX2 promoter, 5′-gcgtcgacCATTCCTCAACCAATATCTGCAAC-3′ (forward) and 5′-gctctagaTATGTTTCTCTCTCTCTCTGATTTG-3′ (reverse); AtCKX3 promoter, 5′-gcgtcgacCAAAGTTTGGCCTACGATTGTT-3′ (forward) and 5′-gctctagaGCTTGATTCTTATCAATGAAGAGTAG-3′ (reverse); AtCKX4 promoter, 5′-cccaagcttCGGTTATTTTATGGGCCAGTT-3′ (forward) and 5′-cccaagcttAACAAACGGGTAGGTTAATGG-3′ (reverse); AtCKX5 promoter, 5′-aagcttATCGACAAAGAGCAAATTATGAA-3′ (forward) and 5′-tctagaGAAACAAGAATCAAGATTGAGGA-3′ (reverse); and AtCKX6 promoter, 5′-aagcttCTTCAAGTGGACCGTTTATCTCT-3′ (forward) and 5′- cccgggATAAGGCCTCTTGATTTCTGAGA-3′ (reverse). The lengths of the amplified sequences were 1867 bp (AtCKX1), 1603 bp (AtCKX2), 1452 bp (AtCKX3), 1926 bp (AtCKX4), 1817 bp (AtCKX5), and 1828 bp (AtCKX6). All promoter sequences were inserted into vector pGPTV-BAR (Becker et al., 1992) upstream of the GUS reading frame. Dideoxy sequencing of the amplified DNA fragments was performed with an ABI PRISM BigDye Terminator cycle sequencing reaction kit (Perkin-Elmer Applied Biosystems Division) to exclude PCR and cloning errors.
Generation of AtCKX1-GFP, AtCKX2-GFP, and AtCKX3-GFP Translational Fusion Genes
Soluble modified GFP (smGFP) was amplified with primers 5′-ggggtacccgggtctagactagtctcgagtATGAGTAAAGGAGAAGAACTTTTCACTGGA-3′ (forward) and 5′-gcgtcgacttaagagctcgggcccTTATTTGTATAGTTCATCCATGCCATGT-3′ (reverse) from pSMGFP (Davis and Vierstra, 1998), creating KpnI, SmaI, XbaI, SpeI, and XhoI linker sequences at the 5′ end and ApaI, SacI, and SalI linker sequences at the 3′ end. The amplified fragment was inserted downstream of the 35S promoter in the unique KpnI and SalI of pBINHygTx (Gatz et al., 1992), yielding the vector pBinSMGFP. To generate the p35S:AtCKX1-GFP reporter plasmid, the AtCKX1 stop codon was mutated in the initial pUC19-AtCKX1 construct by exchanging a 41-nucleotide KpnI fragment at the 3′ end. The mutated full-length AtCKX1 genomic fragment was fused via the XhoI site to the N-terminal end of the GFP coding region in the vector pBinSMGFP. A C-terminally truncated, 2386-bp genomic fragment of the AtCKX2 gene was subcloned from vector pUC19-AtCKX2 and added to vector pBinSMGFP as an N-terminal fusion to GFP via the KpnI and SmaI sites, generating the plasmid p35S:AtCKX2-GFP. A XhoI site 6 bp upstream of the AtCKX3 stop codon was used to excise a KpnI-XhoI AtCKX3 genomic fragment from plasmid pBS-AtCKX3 and to generate an N-terminal fusion with GFP in the plasmid pBinSMGFP, yielding the vector p35S:AtCKX3-GFP. Seeds of Arabidopsis plants expressing the mitochondrially localized β-ATPase-GFP fusion protein were a kind gift of D.C. Logan (University of St. Andrews, St. Andrews, UK).
The protein sequence analyses were performed using TargetP version 1.0 (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2000) and iPSORT (http://www.hypothesiscreator.net/iPSORT/) (Bannai et al., 2002).
Plant Transformation and Culture Conditions
Arabidopsis Col-0 plants were transformed according to the flower-dip method (Bechtold et al., 1993). Transgenic AtCKX-containing progeny were selected after surface sterilization of seeds on MS medium (Murashige and Skoog, 1962) containing 15 mg/L hygromycin. Arabidopsis plants containing promoter:GUS fusions were selected on soil, 5 days after sowing, by spraying once with 0.1% BASTA. Plants were cultured in vitro on MS medium under 16-h-light/8-h-dark cycles at 20°C. Growth conditions in a glasshouse were 24 to 20°C and 16-h-light/8-h-dark cycles. All phenotypic characterizations were performed with homozygous progeny obtained by self-fertilizing. Callus material for CKX enzymatic measurements was induced from root explants and maintained on MS medium containing 1 mg/L naphthylacetic acid and 1 mg/L kinetin. Transgenic plants that were used as a source for callus material looked similar to the transformants shown in Figures 4C and 7A.
RNA Analysis
Total RNA was extracted from leaf tissue according to Verwoerd et al. (1989). RNA gel blot analysis with 50 μg of total RNA from transgenic plants (T1) was performed essentially as described (Faiss et al., 1997). The lowest stringency wash after hybridization with gene-specific probes covering the full gene length was performed in 0.1× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 60°C. The filters were rehybridized to a 25S rDNA-specific probe as a loading control.
Hormone Analyses
Frozen plant samples were ground in liquid nitrogen, transferred into Bieleski solution (Bieleski, 1964), and extracted overnight at −20°C. For IAA analysis, 50 pmol of 13C6-IAA (phenyl-13C6-indole-3-acetic acid; Cambridge Isotope Laboratories, Andover, MA) was added to the sample before centrifugation (20,000 rpm for 15 min at 4°C). For each cytokinin compound determined, 10 pmol of 2H5-dihydrozeatin, 2H5-dihydrozeatin riboside, 2H5-dihydrozeatin 9-glucoside, 2H5-dihydrozeatin riboside 9-glucoside, 2H6-N6-(Δ2isopentenyl)adenine, 2H6-N6-(Δ2isopentenyl)adenosine, 2H6-N6-(Δ2isopentenyl)adenine glucoside, and 2H6-N6-(Δ2isopentenyl)adenosine 5′-monophosphate (APEX International, Honiton, Devon, UK) was added. The pellet was resuspended for 1 h at 4°C in 80% methanol and centrifuged once more. The supernatants of both fractions were pooled and dried. After dissolving in water, cytokinins were purified by a combination of solid-phase and immunoaffinity chromatography as described by Redig et al. (1996a). After the immunoaffinity treatment, IAA was concentrated on a C18 cartridge and methylated before analyses (Prinsen et al., 2000). Hormones were quantified by micro-liquid chromatography-positive electrospray-tandem mass spectrometry in multiple reactant monitoring mode (Prinsen et al., 1998). The chromatograms obtained were processed by means of Masslynx software (Micromass, Manchester, UK). Concentrations were calculated according to the principles of isotope dilution and expressed in picomoles per gram fresh weight.
CKX Measurement
The assay of CKX activity was based on the conversion of 2-3H-iP to adenine as described elsewhere (Motyka et al., 1996). The reaction mixture contained 100 mM 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid–NaOH buffer, pH 8.5, 2 μM 2-3H-iP (7.4 Bq/mol), and enzyme preparations equivalent to 2.5 to 500 mg of fresh tissue in a total volume of 50 μL. After incubation at 37°C, the reaction was stopped by the addition of 120 μL of 0.75 mM adenine and iP in 95% (v/v) cold ethanol and 10 μL of 200 mM Na4EDTA. Precipitated protein was removed by centrifugation, and adenine and iP were separated by thin layer chromatography on microcrystalline cellulose plates (Aldrich) developed with the upper phase of a 4:1:2 (v/v/v) mixture of ethyl acetate:n-propanol:water. The radioactivity of adenine- and iP-containing zones was determined by liquid scintillation using the Packard TRI-CARB 2500 TR scintillation counter. The apparent Km(iP) and maximum velocity values of CKX preparations were determined in the same assay mixture described above with 2-3H-iP as the substrate used in the concentration range of 0.3 to 10 μM. The apparent Km(iP) and maximum velocity values were calculated on the basis of a Lineweaver-Burk double reciprocal plot of CKX activity as a function of iP concentration.
GUS Staining and Light Microscopy
Histochemical analysis of the GUS reporter enzyme was performed essentially according to Jefferson et al. (1987) as modified by Hemerly et al. (1993). Sample tissues were fixed in 90% ice-cold acetone for 1 h and incubated for 1 to 12 h in reaction buffer. After staining, seedlings were cleared and mounted according to Malamy and Benfey (1997). Endogenous pigments were destained with 70% ethanol, and the GUS staining pattern was recorded with a stereomicroscope (SZX12; Olympus, Tokyo, Japan) or a microscope (Axioskop 2 plus; Zeiss, Jena, Germany) equipped with an Olympus C-4040ZOOM photographic device. For sectioning, plant tissue was fixed and embedded in LR White (Plano, Wetzlar, Germany) according to Schoof et al. (2000), and 2.5-μm thin sections were stained with 0.1% toluidine blue. Tissue for the observation of vasculature was cleared and visualized as described by Mattsson et al. (1999).
Analysis of the Cellular Localization of GFP-Fused Proteins
Transgenic plants expressing individual GFP fusion constructs were selected according to their phenotype and preanalyzed using an epifluorescence microscope before the confocal imaging. Primary leaves or roots were cut and mounted in water for microscopic observation. The subcellular localization of GFP fusion proteins was analyzed with a Zeiss cLSM 510 confocal laser scanning microscope using a C-Apochromat 63x/1.2 W Korr water immersion objective and a 488-nm argon laser in combination with a 505- to 530-nm bandpass filter set. Image acquisition and processing were performed using a Zeiss laser scanning microscope (LSM 510, version 3.0), SP3 software, and Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Thomas Schmülling, tschmue@zedat.fu-berlin.de.
Acknowledgments
This article is dedicated to the memory of Jeff Schell. We thank Hi-Gung Bae for help with confocal microscopy, Carola Scholz and Thomas Beiter for excellent technical assistance, and Catherine Scott-Taggart for proofreading. ARR5:GUS and CycB1:GUS transgenic seeds were kindly provided by Joe Kieber and Peter Doerner, respectively. We acknowledge financial support from the Deutsche Forschungsgemeinschaft (Schm 814/15-1 and Schm 814/17-1) and from the Grant Agency of the Academy of Science of the Czech Republic (A 6038002).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014928.
References
- Aloni, R. (1995). The induction of vascular tissues by auxin and cytokinin. In Plant Hormones: Physiology, Biochemistry and Molecular Biology, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 531–546.
- Armstrong, D.J. (1994). Cytokinin oxidase and the regulation of cytokinin degradation. In Cytokinins: Chemistry, Activity, and Function, D.W.S. Mok and M.C. Mok, eds (Boca Raton, FL: CRC Press), pp. 139–154.
- Bannai, H., Tamada, Y., Maruyama, O., Nakai, K., and Miyano, S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18, 298–305. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Cork, A., Williamson, R.E., and Gorst, J.R. (1995). STUNTED PLANT 1, a gene required for expansion in rapidly elongating but not in dividing cells and mediating root growth responses to applied cytokinin. Plant Physiol. 107, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H.-Q., Hawes, C., and Moore, I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Beemster, G.T.S., and Baskin, T.I. (2000). STUNTED PLANT 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 124, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková, E., Witters, E., Van Dongen, W., Kolář, J., Motyka, V., Brzobohatý, B., Van Onckelen, H., and Macháčková, I. (1999). Cytokinins in tobacco and wheat chloroplasts: Occurrence and changes due to light/dark treatment. Plant Physiol. 121, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao, R.P., Eklöf, J., Ljung, K., Marchant, A., Bennett, M., and Sandberg, G. (2002). Shoot derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29, 325–332. [DOI] [PubMed] [Google Scholar]
- Bieleski, R.L. (1964). The problem of halting enzyme action when extracting plant tissues. Anal. Biochem. 9, 431–442. [DOI] [PubMed] [Google Scholar]
- Bilyeu, K.D., Cole, J.L., Laskey, J.G., Riekhof, W.R., Esparza, T.J., Kramer, M.D., and Morris, R.O. (2001). Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 125, 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Santa Cruz, S., Hawes, C., Harris, N., and Oparka, K.J. (1996). Virus-mediated delivery of the green fluorescent protein to the endoplasmic reticulum of plant cells. Plant J. 10, 935–941. [Google Scholar]
- Casimiro, I., Marchant, A., Bhalerao, R.P., Beeckman, T., Dhooge, S., Swarup, R., Graham, N., Inzé, D., Sandberg, G., Casero, P.J., and Bennett, M.J. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575. [DOI] [PubMed] [Google Scholar]
- Coenen, C., and Lomax, T.L. (1997). Auxin-cytokinin interactions in higher plants: Old problems and new tools. Trends Plant Sci. 2, 351–356. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Custers, J.B.M., Snepvangers, S.C.H.J., Jansen, H.J., Zhang, L., and van Lookeren Campagne, M.M. (1999). The 35S-CaMV promoter is silent during early embryogenesis but activated during nonembryogenic sporophytic development in microspore culture. Protoplasma 208, 257–264. [Google Scholar]
- D'Agostino, I.B., Deruère, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Day, S.J., and Lawrence, P.A. (2000). Measuring dimensions: The regulation of size and shape. Development 127, 2977–2987. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., de Almeida-Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inzé, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., Van Der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M.S., Jacqmard, A., Kilby, N.J., and Murray, J.A.H. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner, P., Jørgensen, J.-E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380, 520–523. [DOI] [PubMed] [Google Scholar]
- Ehness, R., and Roitsch, T. (1997). Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Eklöf, S., Åstot, C., Moritz, T., Blackwell, J., Olsson, O., and Sandberg, G. (1997). Auxin-cytokinin interactions in wild-type and transgenic tobacco. Plant Cell Physiol. 33, 225–235. [Google Scholar]
- Eklöf, S., Åstot, C., Sitbon, F., Moritz, T., Olsson, O., and Sandberg, G. (2000). Transgenic tobacco plants co-expressing Agrobacterium iaa and ipt genes have wild-type hormone levels but display both auxin- and cytokinin-overproducing phenotypes. Plant J. 23, 279–284. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., and von Heijne, G. (2001). Prediction of organellar targeting signals. Biochim. Biophys. Acta 1541, 114–119. [DOI] [PubMed] [Google Scholar]
- Faiss, M., Zalubilová, J., Strnad, M., and Schmülling, T. (1997). Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 12, 401–415. [DOI] [PubMed] [Google Scholar]
- Faure, J.-D., and Howell, S.H. (1999). Cytokinin perception and signal transduction. In Biochemistry and Molecular Biology of Plant Hormones, P.J.J. Hooykaas, M.A. Hall, and K.R. Libbenga, eds (Amsterdam: Elsevier Science), pp. 461–474.
- Frugis, G., Giannino, D., Mele, G., Nicolodi, C., Chiappetta, A., Bitonti, M.B., Innocenti, A.M., Dewitte, W., Van Onckelen, H., and Mariotti, D. (2001). Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 126, 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, S., and Amasino, R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. [DOI] [PubMed] [Google Scholar]
- Gatz, C., Frohberg, C., and Wendenburg, R. (1992). Stringent repression and homogenous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 2, 397–404. [DOI] [PubMed] [Google Scholar]
- Hawes, C., Saint-Jore, C., Martin, B., and Zheng, H.-Q. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP. Trends Plant Sci. 6, 245–246. [DOI] [PubMed] [Google Scholar]
- Hemerly, A.S., Ferreira, P., de Almeida Engler, J., Van Montagu, M., Engler, G., and Inzé, D. (1993). cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houba-Hérin, N., Pethe, C., d'Alayer, J., and Laloue, M. (1999). Cytokinin oxidase from Zea mays: Purification, cDNA cloning and expression in moss protoplasts. Plant J. 17, 615–626. [DOI] [PubMed] [Google Scholar]
- Hülskamp, M., Miséra, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Jacqmard, A., De Veylder, L., Segers, G., de Almeida Engler, J., Bernier, G., Van Montagu, M., and Inze, D. (1999). Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.J., and Schreiber, B.M.N. (1997). Role and function of cytokinin oxidase in plants. Plant Growth Regul. 23, 123–134. [Google Scholar]
- Laufs, P., Dockx, J., Kronenberger, J., and Traas, J. (1998). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Bhalerao, R.P., and Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474. [DOI] [PubMed] [Google Scholar]
- Logan, D.C., and Leaver, C.J. (2000). Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot. 51, 865–871. [PubMed] [Google Scholar]
- Mähönen, A.P., Bonke, M., Kaupinnen, L., Riikonen, M., Benfey, P.N., and Helariutta, Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Martin, R.C., Mok, M.C., and Mok, D.W. (1997). Protein processing and auxin response in transgenic tobacco harboring a putative cDNA of zeatin O-xylosyltransferase from Phaseolus vulgaris. Plant J. 12, 305–312. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., and Neuhaus, J.-M. (1999). Cis-elements of protein transport to the plant vacuoles. J. Exp. Bot. 50, 165–174. [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular system to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- McGaw, B.A., and Horgan, R. (1983). Cytokinin catabolism and cytokinin oxidase. Phytochemistry 22, 1103–1105. [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. [DOI] [PubMed] [Google Scholar]
- Mok, D.W.S., and Mok, M.C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118. [DOI] [PubMed] [Google Scholar]
- Mok, M.C. (1994). Cytokinins and plant development: An overview. In Cytokinins: Chemistry, Activity, and Function, D.W.S. Mok and M.C. Mok, eds (Boca Raton, FL: CRC Press), pp. 155–166.
- Morris, R.O., Bilyeu, K.D., Laskey, J.G., and Cheikh, N.N. (1999). Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem. Biophys. Res. Commun. 255, 328–333. [DOI] [PubMed] [Google Scholar]
- Motyka, V., Faiss, M., Strnad, M., Kamínek, M., and Schmülling, T. (1996). Changes in cytokinin content and cytokinin oxidase activity in response to derepression of ipt gene transcription in transgenic tobacco calli and plants. Plant Physiol. 112, 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyka, V., Vaňková, R., Čapková, V., Petrášek, J., Kamínek, M., and Schmülling, T. (2003). Cytokinin-induced upregulation of cytokinin oxidase activity in tobacco includes changes in enzyme glycosylation and secretion. Physiol. Plant. 117, 11–21. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nogué, F., Grandjean, O., Craig, S., Dennis, E., and Chaudhury, A. (2000). Higher levels of cell proliferation rate and cyclin CycD3 expression in the Arabidopsis amp1 mutant. Plant Growth Regul. 32, 275–283. [Google Scholar]
- Ori, N., Juarez, M.T., Jackson, D., Yamaguchi, J., Banowetz, G.M., and Hake, S. (1999). Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 11, 1073–1080. [PMC free article] [PubMed] [Google Scholar]
- Prinsen, E., Van Dongen, W., Esmans, E., and Van Onckelen, H. (1998). Micro and capillary liquid chromatography-tandem mass spectrometry: A new dimension in phytohormone research. J. Chromatogr. 826, 25–37. [Google Scholar]
- Prinsen, E., Van Laer, S., Öden, S., and Van Onckelen, H. (2000). Auxin analysis. In Methods in Molecular Biology, Vol. 141: Plant Hormone Protocols, G.A. Tucker and J.A. Roberts, eds (Totowa, NJ: Humana Press), pp. 49–65. [DOI] [PubMed]
- Redig, P., Schmülling, T., and Van Onckelen, H. (1996. a). Analysis of cytokinin metabolism in ipt transgenic tobacco by liquid chromatography-tandem mass spectrometry. Plant Physiol. 112, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redig, P., Shaul, O., Inze, D., Van Montagu, M., and Van Onckelen, H. (1996. b). Levels of endogenous cytokinins, indole-3-acetic acid and abscisic acid during the cell cycle of synchronized tobacco BY-2 cells. FEBS Lett. 391, 175–180. [DOI] [PubMed] [Google Scholar]
- Reed, R.C., Brady, S.R., and Muday, G.K. (1998). Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 118, 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., Muday, G., and Estelle, M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, H., Frank, M., Werner, T., Strnad, M., and Schmülling, T. (1999). Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 18, 357–363. [DOI] [PubMed] [Google Scholar]
- Schmülling, T., Werner, T., Riefler, M., Krupkova, E., Bartrina, Y., and Manns, I. (2003). Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 116, 241–252. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Schwacke, R., Schneider, A., Van Der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flügge, U.I., and Kunze, R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fate. Genes Dev. 7, 787–795. [DOI] [PubMed] [Google Scholar]
- Skoog, F., and Miller, C.O. (1957). Chemical regulation of growth and organ formation in plant tissue cultures in vitro. Symp. Soc. Exp. Biol. 11, 118–131. [PubMed] [Google Scholar]
- Smart, C.M., Scofield, S.R., Bevan, M.W., and Dyer, T.A. (1991). Delayed leaf senescence in tobacco plants transformed with tmr, a gene for cytokinin production in Agrobacterium. Plant Cell 3, 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Hülskamp, M., Gendreau, E., and Höfte, H. (1998). Endoreduplication and development: Rule without dividing? Curr. Opin. Plant Biol. 1, 498–503. [DOI] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, M.M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNA. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Groll, U., and Altmann, T. (2001). Stomatal cell biology. Curr. Opin. Plant Biol. 4, 555–560. [DOI] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98, 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.H., Yu, H., and Goh, C.J. (2003). Functional characterisation of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid. Plant Mol. Biol. 51, 237–248. [DOI] [PubMed] [Google Scholar]