Abstract
Lungs are exposed to the outside environment and therefore to toxic and infectious agents or allergens. This may lead to permanent activation of innate immune response elements. A Disintegrin And Metalloproteinases (ADAMs) and ADAMs with Thrombospondin motifs (ADAMTS) are proteinases closely related to Matrix Metalloproteinases (MMPs). These multifaceted molecules bear metalloproteinase and disintegrin domains endowing them with features of both proteinases and adhesion molecules. Proteinases of the ADAM family are associated to various physiological and pathological processes and display a wide spectrum of biological effects encompassing cell fusion, cell adhesion, "shedding process", cleavage of various substrates from the extracellular matrix, growth factors or cytokines... This review will focus on the putative roles of ADAM/ADAMTS proteinases in airway diseases such as asthma and COPD.
Introduction
The lung is continuously exposed to the outside environment and various potential aggressions such as noxious and infectious agents or allergens. The innate immune responses are permanently activated in this particular organ. Moreover, secretory materials such as surfactant and mucous also contribute to host defense against inflammation. Among airway diseases, asthma and COPD (Chronic Obstructive Pulmonary Disease) appear to be growing public health concerns worldwide and the number of listed asthmatic and COPD patients still increases over time.
Asthma is a complex clinically-defined syndrome mainly characterized by symptoms (wheezing, cough, breathlessness) and airway obstruction. Hallmarks of asthma are mainly airway hyperresponsiveness caused by a wide variety of stimuli and airway inflammation involving eosinophils and mast cells. Moreover, an asthma-associated remodeling of the airways including extensive changes in the extracellular matrix has been characterized. The main changes reported are a subepithelial fibrosis, a smooth muscle hypertrophy, a glandular metaplasia in the bronchial epithelium, and the deposition of extracellular matrix components throughout the airway wall. These features are very often associated with altered behaviour of airway structural cells including epithelial cells or fibroblasts [1,2].
COPD is characterized by a progressive airway obstruction mainly linked to tobacco consumption and/or toxic fumes and other environmental factors. COPD patients also display profound modifications of the extracellular matrix leading to an airway remodeling including collagen fibers deposition in the bronchial and bronchiolar walls, mucous hyperplasia, and smooth muscle cell hypertrophy [3-5].
As the key role of extracellular matrix and soluble mediators has been unveiled, there is accumulating evidences demonstrating the crucial role played by matrix metalloproteinases (MMPs) in lung diseases [6,7]. These aspects have been largely discussed in previous reviews [8,9]. The present review focuses on another subfamily of proteinases also belonging to the metzincins (zinc-bearing proteinases) and structurally related to MMPs: the ADAMs (A Disintegrin And Metalloproteinase) [10-14]. ADAM proteinases have been described as "signalling scissors" since they are associated to shedding processes of key factors implicated in physiological as well as in pathological activities [15]. This shedding process is quite interesting as it appears as an emerging concept that could be implicated in airway diseases. Indeed, ADAM-17 has been defined as the prototypical TNF-α convertase enzyme [16]. Besides this very well known example, many other sheddase activities have been reported and can address many physiological processes such as the regulation of cell proliferation by cleavage of membrane-bound heparin-binding epidermal growth factor (HB-EGF) [17]. Some cell receptors including the low-affinity immunoglobulin E receptor (CD23) can also be targeted by sheddases. Indeed, ADAM-10 appears to be the main sheddase for CD23 leading to increased levels of its soluble form [18,19]. The literature emerging in the last years suggests that ADAMs scissors-function plays a crucial role in airway diseases.
In the present review, after a brief general description of ADAM proteins, we discuss the implications of these proteinases in various physiological and pathological processes. The potential contribution of ADAM/ADAMTS proteins to asthma pathology will be described as well as ADAMs/ADAMTS' involvement in COPD.
Structural features of ADAMs
To date, about 40 members of the ADAM family have been described in different species (for a constantly updated database, see http://people.virginia.edu/~jw7g/Table_of_the_ADAMs.html and http://degradome.uniovi.es/). Twenty-five ADAMs are expressed in Homo sapiens while thirty-five members are expressed in Mus musculus. Together with ADAMTS (ADAMs with Thrombospondin motifs type I) and SVMPs (Snake Venom Metalloproteinases), ADAM proteinases constitute the subfamily of adamalysins [12] which belongs to the superfamily of metzincins. This superfamily also includes astacins, matrixins (also referred to as matrix metalloproteinases), serralysins and pappalysins [20,21]. Those metzincins are characterized by (1) a catalytic site containing a consensus sequence (HEXXHXXGXXH) in which three histidine residues coordinate a zinc ion and (2) by a conserved methionine residue forming a "Met-turn" beneath the active zinc site. This "Met-turn" provides a hydrophobic environment for the zinc ion and the three ligating histidine residues at the catalytic centre of the enzyme [22,23].
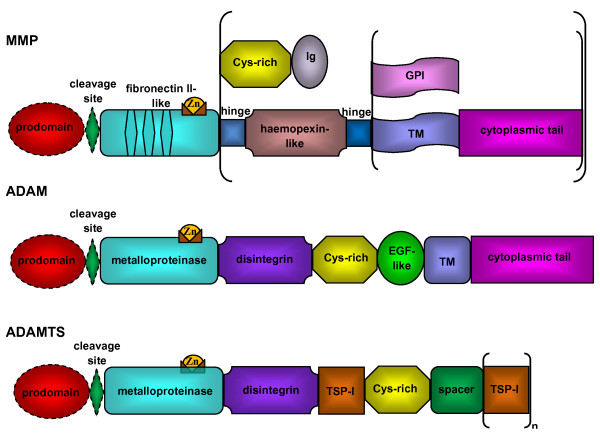
Structure of ADAMs and ADAMTS is highly conserved and involves metalloproteinase and disintegrin domains endowing them with features of both proteinases and adhesion molecules [11,13]. As illustrated in figure 1, the detailed structure of ADAMs is far more complex than that of MMPs. Domains shared with MMPs are the prodomain maintaining the catalytic site inactive and the metalloproteinase domain containing the Zinc binding site. ADAM activation mechanisms are mostly similar to MMP's activation and generally imply the prodomain removal from the precursor protein via a proprotein convertase of furin type [24]. However, maturation of some ADAMs, such as ADAM-8 and ADAM-28 occurs as an autocatalytic process [25,26]. The metalloproteinase domain with its catalytic consensus site is active in only about half of ADAM proteinases. The following domains are characteristic of ADAMs and include a disintegrin domain mediating cell-cell, cell-matrix interactions via the interaction with integrins; a cystein-rich domain implicated in cell adhesion; an epidermal growth factor (EGF)-like domain and a cytoplasmic tail involved in various intracellular signalization pathways [11].
Figure 1.
Structural organization of MMPs, ADAMs and ADAMTS. The typical structure of MMP is made of a prodomain, a furin cleavage site (all MT-MMPs, MMP-21,-23, and -28), a catalytic metalloproteinase domain with fibronectin type II repeats (MMP-2, MMP-9), a linker peptide and a haemopexin domain (except for MMP-7, -26, and -23), a linker peptide, a transmembrane domain and cytoplasmic tail (MMP-14, -15, -16, -24) or glycosylphosphatidylinositol (GPI) anchor (MMP-17, -25). MMP-23 bears C-terminal cysteine-rich (Cys-rich) and Ig-like (Ig) domains and its propeptide lacks a cystein switch motif. Common structure of ADAMs is a prodomain, a cleavage site (by a furin or furin-like proprotein convertase except for ADAM-8 and ADAM-28 which use an autocatalytic process), a metalloproteinase domain, a disintegrin domain, a cysteine-rich region (Cys-rich), an epidermal-growth factor repeat (EGF-like), a transmembrane domain (TM) and a cytoplasmic tail. ADAMTS do not possess a transmembrane domain (TM) but bear a various number of thrombospondin type I motifs (TSP-1) at their C-terminal extremity.
Although the structure of ADAM and ADAMTS proteinases is closely related, ADAMTS molecules are characterized by a various number of thrombospondin type one motifs (TSP-1) at their C-terminal end and the absence of transmembrane and cytoplasmic domains [10,27] (figure 1). In the C-terminal extremity, different types of modules have been described for some of the ADAMTS. All these data are regularly updated on http://www.lerner.ccf.org/bme/apte/adamts.
The metalloproteinase system is controlled by endogenous physiological inhibitors ("Tissue Inhibitors of Metalloproteinases" or TIMPs) which are small proteins with molecular weights ranging from 21 to 28 kDa. These inhibitors display six disulfide bonds in their structure forming a rigid conformation which is mandatory for their biological activity. TIMPs are able to inhibit proteinase activity of several members of the ADAM family [28-31]. N-terminal domain of TIMPs and more specifically the "functional binding edge" is interacting with the catalytic domain of the ADAM proteinase [32]. The interaction of the catalytic site-bound Zinc atom with a cystein present in the N-terminal extremity of TIMP leads to an inactivation of ADAMs. This process has been described for ADAM-17 or ADAMTS-4 interacting with TIMP-3 [30,32]. More recently, a novel method of purification using sodium chlorate has confirmed that C-terminal domain of ADAMTS-4 and -5 and more particularly their TS-domains favors the interaction with the N-terminal domain of TIMP-3 [33].
When taking into consideration the complex multi-domain structure of ADAMs and ADAMTS, one can anticipate their implication in many physiological and pathological processes. From these complex structural features and bearing in mind that only half of proteins of this family display a catalytic activity, one can expect that functions of ADAMs and ADAMTS will not be restricted to the cleavage of extracellular matrix or mediators but will embrace various functions including the regulation of cell-cell and cell-matrix interactions. Although these ADAM/ADAMTS functions are not yet as much discerned as those of MMPs, a real interest from the scientific community has emerged these last years, specifying not only the exact structure of these proteins, but also identifying new features involving ADAMs in health and diseases. In this review, we are discussing known and potential implications of ADAMs in lung homeostasis as well as in its deregulation.
Implication of ADAMs and ADAMTS in physiological and pathological processes
Since ADAM proteinases are defined as multi-domain proteins, studies have focused their attention on the multiple functions that can be ascribed to these proteinases. ADAMs have been described in various physiological processes such as egg fertilization, myogenesis, cell fate determination but also in diverse pathological processes.
Physiological processes
Properties attributed to ADAMs are evidently crucial when one considers their structural organization. We will present hereafter selected examples illustrating the diversity of biological processes that can be affected by ADAM proteins. Most of ADAMs are membrane-bound proteins and can assist cell fusion, cell adhesion, peptidic mediators processing, linked or not to plasma membrane. They also play a key role in some intracellular signaling pathways. The final picture is rendered even more complex since alternative splicing can induce variations in the C-terminal region of membrane-bound ADAMs and thereby give rise to different cytosolic tails or secreted proteins [34].
Some ADAMs appear essential in cell fusion processes. It is worth underlining that the two first identified ADAMs (ADAM-1 and -2) were recognized as fertilin-alpha and -beta in 1987 [35] since they could induce the fusion of the sperm with the egg. This process is mediated through the interaction of the disintegrin domain of ADAM-2 present on the sperm with integrin α6β1 beared on the egg surface [36]. Moreover, ADAM proteins are key enzymes in embryonic development since ADAM-10 is able to cleave NOTCH protein and consequently regulate central nervous system development [37]. ADAMs also contribute to intracellular signalling processes and have the ability to interact with tyrosine kinases and some components of the cytoskeleton through their cytoplasmic domain [11]. The disintegrin domain of some ADAMs is able to regulate cell adhesion through interaction with various integrins. For instance ADAM-15 is described as a novel component of adherens junctions [38]. Importantly, as stated earlier, ADAMs/ADAMTS are able to cleave membrane-bound growth factors, cytokines and proteoglycans, leading to the detachment of mature soluble forms. This process is largely referred to as sheddase activity. So far, the most studied sheddases are ADAM-17 and ADAM-10 responsible for the cleavage of pro-TNF and CD23, respectively [16,18,39]. ADAMTS proteinases also display a catalytic activity. Indeed, ADAMTS-4 and ADAMTS-5 are able to cleave aggrecan [40,41], ADAMTS-2 processes type I, II and III procollagen chains [42].
Pathological processes
Dysregulation of ADAMs expression has been reported in different types of pathologies such as cancer, osteoarthritis, neurodegenerative inflammation or asthma. In most studies, an overexpression of these proteinases has been described and is linked to a dysregulation of tissue homeostasis sometimes leading to a specific pathological phenotype. ADAMs might therefore be considered as potential candidates to target in a therapeutic setting. For instance, ADAM-17 expression is increased in breast cancer tissues and its expression is higher in advanced-grade than low-grade tumors. Patients displaying a huge expression of this proteinase have a shorter overall survival than those with a low expression of ADAM-17 [43] suggesting that ADAM-17 might be a good target to predict the outcome of cancer development. ADAMTS-4 and ADAMTS-5 are implicated in osteoarthritis development since ADAMTS-4/-5 double-knockout animals are less affected than wild-type mice [44,45]. Alzheimer's disease is characterized by beta amyloid deposition in the brain. ADAM-10 acts as an alpha-secretase and thereby cleaves the amyloid precursor to release a soluble component. Many authors have hypothesized that overexpression of ADAM-10 might have beneficial effects on the pathological deposition of amyloid protein [46,47] since ADAM-10 overexpressing mice display reduced susceptibility to amyloid deposition [47].
The complex structure of ADAMs also suggests that these enzymes may be functionally relevant to different steps linked to asthma pathogenesis. Indeed, the active metalloproteinase domain of some ADAMs might be important to shed growth factors and cytokines, contributing in this way to the control of inflammation which is a hallmark of asthma pathology. Disintegrin domain might also act in concert with cystein-rich region to interfere with pro-inflammatory cytokines [48].
These data illustrate how much ADAMs are multifunctional proteins and suggest that these proteinases may serve as mediators during the progression of asthmatic pathology but also COPD (table 1).
Table 1.
ADAMs/ADAMTS modulation in airway diseases.
| ADAMs | Modulation | Type of airway disease | Type of study | Reference |
|---|---|---|---|---|
| ADAM-8 | ↗ | asthma | human | [51,57,74] |
| ↗ | asthma | mouse | [58-60,73][Paulissen et al, submitted] | |
| ADAM-9 | ↗ | asthma | human | [51] |
| ADAM-10 | ↗ | asthma | mouse | [73] |
| ADAM-12 | ↗ | asthma | human | [51] |
| ADAM-17 | ↗ | asthma | mouse | [73] |
| ↗ | COPD | rat | [80] | |
| ADAM-28 | ↗ | asthma | mouse | [73] |
| ADAM33 | ↗ | asthma | human | [57,71,82] |
| SNP | COPD | human | [79,83] | |
| ADAMTS-1 | ↘ | asthma | human | [51] |
| ADAMTS-12 | SNP | asthma | human | [84] |
| ADAMTS-15 | ↘ | asthma | human | [51] |
| ↗ | asthma | mouse | [73] | |
SNP: Single Nucleotide Polymorphism
Expression of ADAMs and ADAMTS in the lung
In the lung, different cell types can express different classes of proteinases. Some structural cells from bronchial tree are able to produce enzymes belonging to the metzincin superfamily that are important in regulation processes and in the cascade leading to inflammation. However, some data - especially those concerning the expression of ADAMs in lung tissues - are more recent and rather incomplete [49] (table 2). In lung tissue, an expression of ADAM-8, -9, -10, -12, -15, -17 and ADAMTS-1, -2, and TS-12 has been observed [50] with a modulation of ADAM-12 and ADAMTS-1 in tumors [50]. In sputum cells, an expression of ADAM-8, -9, -10, -12, -15, -17 and ADAMTS-1, TS-15 has been reported [51]. Moreover, epithelial cells have been shown to express ADAM-9, -10, -12, -15, -17 and ADAMTS-1 with an exception for immortalized bronchial epithelial cells (BEAS-2B) which do not express ADAM-12 [50]. Another epithelial cell line (A549, an alveolar epithelial cell line) was shown to express ADAM-19 and ADAMTS-9 [52]. Whereas mesenchymal cells such as fibroblasts and smooth muscle cells abundantly express ADAM-33 [53-55], epithelial cells may also express this proteinase [49,56]. Although only some authors have reported that airway epithelial cells express ADAM-8 [57,58], all authors have agreed to confirm that inflammatory cells produce ADAM-8, a proteinase that has been suggested to be a key mediator in inflammatory processes [51,57-60].
Table 2.
ADAMs/ADAMTS expression in lung cell types.
| ADAMs | Lung cell types | Reference |
|---|---|---|
| ADAM-8 | epithelial cells | [49,57,58] |
| inflammatory cells | [51,57,59][Paulissen et al, submitted] | |
| smooth muscle cells | [57](-) [58](+) | |
| ADAM-9 | epithelial cells | [49,85] |
| inflammatory cells | [49] | |
| smooth muscle cells | [49] | |
| ADAM-10 | epithelial cells | [49] |
| smooth muscle cells | [49] | |
| ADAM-12 | inflammatory cells | [50] |
| smooth muscle cells | [50] | |
| ADAM-17 | epithelial cells | [49,86] |
| inflammatory cells | [49] | |
| smooth muscle cells | [49] | |
| ADAM-19 | epithelial cells | [49,52] |
| endothelial cells | [49] | |
| inflammatory cells | [49] | |
| smooth muscle cells | [49] | |
| ADAM-28 | epithelial cells | [87] |
| ADAM-33 | epithelial cells | [57](+) [88](-) |
| endothelial cells | [49] | |
| fibroblasts | [54,88] | |
| inflammatory cells | [49,57] | |
| smooth muscle cells | [55,57,82,88] | |
Contribution of ADAM and ADAMTS proteinases to the asthmatic phenotype
In some individuals, an inflammatory reaction occurs in the lungs after exposure to specific allergens. Following a single allergen exposure, an early-phase reaction is produced in pulmonary tissues followed by a late-phase reaction. The early-phase reaction is characterized by the activation of mast cells and macrophages and the release of various mediators including histamine and eicosanoids while the late-phase reaction consists of recruitment of eosinophils, CD4+ T cells, basophils and neutrophils. Moreover, T helper cells amplify the inflammatory response via the release of Th2 cytokines. Following repetitive exposure to allergens, a chronic inflammation develops with associated tissue alterations such as mucus hypersecretion, vascular leakage, smooth muscle contraction, and bronchial hyperresponsiveness [61]. Asthma is associated to an airway remodeling that includes 1) a subepithelial fibrosis which appears as a pathognomonic feature of asthmatic bronchi, 2) changes in extracellular matrix composition with an absence of classical components of basement membrane (mainly collagen IV and laminin) and a fragmentation of elastic fibers, 3) a goblet cells hyperplasia and 4) increased angiogenesis [62].
Over the last years, the attention has risen about the roles of ADAM proteinases in processes leading to the asthmatic phenotype described above (see figure 2). ADAM-33 was one of the first ADAM proteinases to be identified as an asthma susceptibility gene after an ambitious study based on a vast genome screening [53]. An association of ADAM-33 gene polymorphism with the hyperresponsiveness linked to the asthmatic pathology has now been confirmed by many studies [63-66]. However, these data need to be clarified since not all authors report such a link between asthma and ADAM-33 [67,68]. These studies linking asthma and variants in ADAM-33 gene are summarized in table 3. Discrepancies between published studies can be explained by the diversity of studied populations and the complexity of this gene subject to alternative splicing processes. Moreover, important differences of statistical power of all these studies might also account for some of the reported differences between cohorts. Molecular mechanisms and exact roles of ADAM-33 in the pathological process leading to asthma are therefore not yet fully elucidated. While it was reported that ADAM-33 expression is mainly detectable in smooth muscle cells and in fibroblasts, authors have recently shown that ADAM-33 is also expressed by other cell types including endothelial cells [49,69]. ADAM-33 therefore might play a key role in asthma-associated airway remodeling since the purified catalytic domain of this proteinase provokes an increased development of the vascular network in asthmatic patients [70]. An argument to speculate for a possible key role of ADAM-33 in asthma physiopathology is the increased ADAM-33 expression reported after stimulation by some Th2 cytokines (IL-4 and IL-13) [71]. In humans, the expression of ADAM-33 was reported to be correlated to disease severity. Indeed, severe asthmatics display higher levels of ADAM-33 expression in their bronchial biopsies when compared to mild asthmatics or controls. Moreover, these asthmatics exhibit ADAM-33 staining in epithelial, submucosal and smooth muscle cells as demonstrated by immunohistochemistry [57]. This overexpression of ADAM-33 in the airways of asthmatics was also confirmed in animal models. Indeed, ADAM-33 levels were reported to increase in lungs of mice after allergen exposure [71]. Nevertheless, the demonstration of ADAM-33 implication in pathological processes leading to an asthma phenotype is still not fully accomplished. Indeed, phenotypes obtained in ADAM-33 KO mice did not suggest that the absence of ADAM-33 actually modulates baseline or allergen-induced airway responsiveness [72].
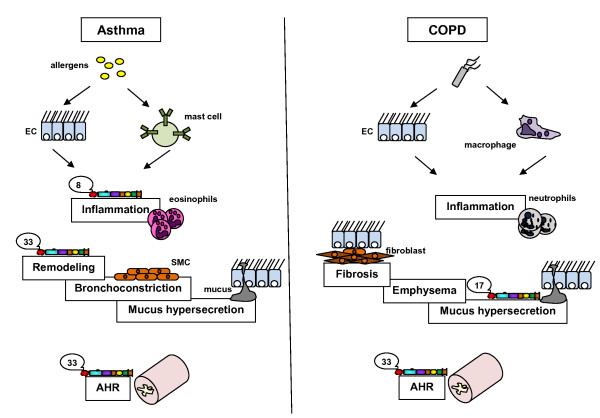
Figure 2.
Intervention of ADAM/ADAMTS proteinases in asthma and COPD. Succinctly, in asthma, inhaled allergens provoke the degranulation of sensitized mast cells and the activation of epithelial cells (EC) while in COPD, inhaled cigarette smoke activates epithelial cells and macrophages. After a first challenge in both diseases, an inflammatory reaction occurs resulting in the recruitment of eosinophils and CD4+ T cells for asthma, neutrophils and CD8+ T cells for COPD. Following a chronic inflammation, tissue alterations such as mucus hypersecretion, bronchoconstriction appear in asthma while small airway fibrosis, alveolar destruction (emphysema) and mucus hypersecretion occur in COPD. An airway hyperresponsiveness is linked to both diseases. However, it is reversible in asthma but not in COPD. ADAM-8 plays a role in asthma-related inflammation while ADAM-33 is associated to remodeling processes and hyperresponsiveness associated to asthma. In COPD, ADAM-17 acts on mucus hypersecretion process while ADAM-33 is associated with COPD-related hyperresponsiveness.
Table 3.
ADAM-33 polymorphism studies in human populations.
| Type of study | Population | Linkage asthma | Studied polymorphisms | Reference |
|---|---|---|---|---|
| FBAS; CC; LDT | UK Caucasian | yes | 135 | [53] |
| US Caucasian | ||||
| FBAS; CC | Hispanic | no | 6 | [68] |
| CC | African from US | yes | 8 | [65] |
| White from US | ||||
| Hispanic from US | ||||
| Dutch white | ||||
| FBAS; CC | German | yes | 15 | [64] |
| CC | Korean | yes | 5 | [89] |
| FBAS | North American * | no | 17 | [90] |
| LDT | Dutch Caucasian | yes | 8 | [91] |
| LDT | UK | yes | 17 | [92] |
| CC | German white * | no | 10 | [93] |
| CC | Japanese | yes | 14 | [66] |
| FBAS | Japenese * | yes | 23 | [63] |
| CC | Australian Caucasian | yes | 10 | [94] |
| CC | Northeast Chinese | no | 3 | [67] |
| FBAS | European-American * | no | ND | [95] |
| hispanic * | no | ND | [95] | |
| allele frequency | Japenese | yes | 10 | [96] |
| CC | Northeast Chinese | yes | 6 | [97] |
| allele frequency | Thai | yes | 8 | [98] |
| CC | Northeast Chinese | yes | 6 | [99] |
| FBAS; CC | UK | yes | 4 | [100] |
Family- based association study: FBAS; Case-control study: CC; linkage disequilibrium test: LDT; ND: no determined; *: children
ADAM-8 is another member of the ADAM family potentially associated to asthma. The first report to suggest an ADAM-8 implication in asthma was published in 2004 [59]. This microarray study has shown that ADAM-8 expression is increased in mice exposed to allergens [59]. In 2008, another microarray study has confirmed the involvement of ADAM-8 in an acute model of asthma, mimicking the inflammation found in human airways, while no difference was found in the chronic model of asthma mimicking human airway remodeling [73]. Moreover, ADAM-8 mRNA levels are increased in sputum cells from asthmatic patients when compared to healthy subjects [51]. An immunohistochemistry targeting ADAM-8 has shown an elevated production of this proteinase in bronchial biopsies from asthmatics related to disease severity as reported for ADAM-33 [57]. A genomic study has recently reported a link between ADAM-8 single nucleotide polymorphisms and asthma in humans [74]. As membrane-bound CD23 is processed by ADAM-8 leading to ectodomain cleavage and resulting in the release of a soluble form of CD23 (sCD23), the low-affinity IgE receptor, ADAM-8 could take part to the cascade of events leading to asthma phenotype [51]. ADAM-8 has already been described to be a sheddase for CD23 [18,75]. The proteolytic release of CD23 from cells is likely to be a key event in allergic asthma. ADAM-8 also cleaves important effectors in asthma pathology such as pro-TNF-α and L-selectin [75,76]. Moreover, ADAM-8 is involved in macrophages activation [75]. The pharmacological delivery of IL-4 or IL-13 as well as use of mice transgene overexpressing these interleukins enhance ADAM-8 levels when mice are exposed to allergens suggesting that ADAM-8 depends not only from allergens but also from Th2 cytokines [59]. Other authors have studied the effects of an overexpression of a soluble form of ADAM-8 by liver tissue and did not find any difference regarding asthma phenotype [60]. Recently, we demonstrated that ADAM-8 is overexpressed in lungs from mice experimentally exposed to allergens and that the depletion of ADAM-8 by the use of KO animals or by immunodepletion dramatically decrease airway inflammation after allergen exposure (Paulissen et al, submitted). It is also worth noting that these ADAM-8 depleted animals do not display developmental abnormalities as described by Kelly et al [77]. Taken together, these data strongly suggest that ADAM-8 is a key mediator in asthma. Further studies should be performed in order to unveil the exact mechanisms implicating ADAM-8 in this disease.
Besides ADAM-8 overexpression, a modulation of RNA levels of ADAM-9, ADAM-12, ADAMTS-1 and ADAMTS-15 has been demonstrated in induced sputum from asthmatic patients [51]. Recently, a genomic study has demonstrated that many ADAM and ADAMTS proteinases such as ADAM-10,-17,-28 and ADAMTS-4, -9,-15 are also overexpressed in chronic asthma [73]. However, further studies might be led to explore their potential role in asthma-related pathology.
All these data highlight the implication of ADAM proteinases in asthma pathogenesis and suggest that new therapeutic strategies based on the inhibition of certain members of this proteinases family could be investigated.
Contribution of ADAM and ADAMTS proteinases in COPD
Chronic obstructive pulmonary disease (COPD) is characterized by a destruction of the lung parenchyma leading to alveolar wall destruction (emphysema) and important structural alterations in bronchial walls such as epithelial metaplasia or airway wall fibrosis [4]. The major risk factor for COPD is the inhalation of cigarette smoke. Despite the improvement of therapeutic strategies and a better understanding of this disease, the morbidity and mortality related to COPD are still significant. Matrix metalloproteinases such as MMP-9 and MMP-12 which have been reported to be modulated in airway secretions from COPD patients might contribute to disease progression and exacerbations by their catalytic activity. However, despite their potential importance in this disease, only few data are available concerning ADAM proteinases involvement in COPD (see figure 2).
ADAM-33 has also been identified as a susceptibility gene for COPD since single nucleotide polymorphisms (SNPs) observed in this gene are associated with a higher risk for developing COPD [78]. ADAM-33 has recently been reported to be linked to airway hyperresponsiveness and airway inflammation in the general population suffering from COPD [79].
Data describing higher ADAM-17 (TACE for TNF-alpha converting enzyme) production in lung tissues from rats exposed to tobacco in a COPD model as compared to control animals support the implication of ADAM proteinases in this obstructive lung pathology [80]. Moreover, siRNA (small interfering RNA) raised against ADAM-17 mRNA as well as metalloproteinase inhibitors (GM-6001 and TNF-alpha inhibitor 1), prevent smoking- induced mucin overproduction in human airway epithelial cells (NCI-H292 cells) [81].
Conclusions
Many peptidic mediators secreted in the lung by both structural as well as inflammatory cells are implicated in physiological processes and their overexpression or inhibition is in many cases part of intrinsic pathological mechanisms. ADAMs and ADAMTS proteins can cleave many of these factors and are therefore key mediators for the control of many biological processes in the lung. Among other activities, these proteinases are also active in the control of extracellular matrix homeostasis and cell migration. It seems therefore logical to set up some therapeutic strategies to target ADAM(TS) enzymes activity in obstructive airways diseases.
This review, aiming at summarizing some lung-related biological actions of ADAMs/ADAMTS, demonstrates to which extent these factors are important in both physiological and pathological processes in lung tissues. Many basic researches have still to be performed to clearly identify target proteinases that appear to play a direct role in pathogenesis as well as potential anti-target ADAMs whose inhibition could cause damages because they have a direct or indirect beneficial effect on lung physiology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GP drafted the manuscript. NR supervised the analysis of data and revised the manuscript. MMG, CC, FQC, SB, JH and MEH approved the final version of the manuscript. J-MF initiated the project. AN revised the manuscript critically. DDC initiated the project, was responsible to find grants, and approved the final version of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Genevieve Paulissen, Email: gpaulissen@ulg.ac.be.
Natacha Rocks, Email: nat.rocks@ulg.ac.be.
Maud M Gueders, Email: maud.gueders@ulg.ac.be.
Celine Crahay, Email: celine.crahay@ulg.ac.be.
Florence Quesada-Calvo, Email: fquesadacalvo@ulg.ac.be.
Sandrine Bekaert, Email: s.bekaert@ulg.ac.be.
Jonathan Hacha, Email: jonathan.hacha@ulg.ac.be.
Mehdi El Hour, Email: melhour@ulg.ac.be.
Jean-Michel Foidart, Email: jmfoidart@ulg.ac.be.
Agnes Noel, Email: agnes.noel@ulg.ac.be.
Didier D Cataldo, Email: didier.cataldo@ulg.ac.be.
Acknowledgements
The Communauté française de Belgique (Actions de Recherches Concertées), the Fonds de la Recherche Scientifique Médicale, the Fonds National de la Recherche Scientifique (F.N.R.S., Belgium), the Fonds spéciaux de la Recherche (University of Liège), the Fondation Léon Fredericq (University of Liège), the DGO6 from the «Région Wallonne» (Belgium), the European Union Framework Programs (FP-7)- Microenvimet n°201279, the Interuniversity Attraction Poles Program- Belgian Science Policy IUAP program #35 (Brussels, Belgium).
References
- Bousquet J, Chanez P, Lacoste JY, White R, Vic P, Godard P, Michel FB. Asthma: a disease remodeling the airways. Allergy. 1992;47(1):3–11. doi: 10.1111/j.1398-9995.1992.tb02242.x. [DOI] [PubMed] [Google Scholar]
- Cataldo D, Louis R, Godon A, Munaut C, Noel A, Foidart JM, Bartsch P. [Bronchial morphologic modification in asthma] Rev Med Liege. 2000;55(7):715–720. [PubMed] [Google Scholar]
- Celli BR, MacNee W. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23(6):932–946. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- Jeffery PK. Remodeling in asthma and chronic obstructive lung disease. Am J Respir Crit Care Med. 2001;164(10 Pt 2):S28–38. doi: 10.1164/ajrccm.164.supplement_2.2106061. [DOI] [PubMed] [Google Scholar]
- Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (COPD) Lancet. 2004;364(9434):613–620. doi: 10.1016/S0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- Lagente V, Boichot E. Role of matrix metalloproteinases in the inflammatory process of respiratory diseases. J Mol Cell Cardiol. 2009. in press . [DOI] [PubMed]
- Greenlee KJ, Werb Z, Kheradmand F. Matrix metalloproteinases in lung: multiple, multifarious, and multifaceted. Physiol Rev. 2007;87(1):69–98. doi: 10.1152/physrev.00022.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueders MM, Foidart JM, Noel A, Cataldo DD. Matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs in the respiratory tract: potential implications in asthma and other lung diseases. Eur J Pharmacol. 2006;533(1-3):133–144. doi: 10.1016/j.ejphar.2005.12.082. [DOI] [PubMed] [Google Scholar]
- Parks WC, Shapiro SD. Matrix metalloproteinases in lung biology. Respir Res. 2001;2(1):10–19. doi: 10.1186/rr33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386(Pt 1):15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Killar L, White J, Black R, Peschon J. Adamalysins. A family of metzincins including TNF-alpha converting enzyme (TACE) Ann N Y Acad Sci. 1999;878:442–452. doi: 10.1111/j.1749-6632.1999.tb07701.x. [DOI] [PubMed] [Google Scholar]
- Black RA, White JM. ADAMs: focus on the protease domain. Curr Opin Cell Biol. 1998;10(5):654–659. doi: 10.1016/S0955-0674(98)80042-2. [DOI] [PubMed] [Google Scholar]
- Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, Foidart JM, Noel A, Cataldo D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90(2):369–379. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8(12):929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385(6618):729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- Rocks N, Estrella C, Paulissen G, Quesada-Calvo F, Gilles C, Gueders MM, Crahay C, Foidart JM, Gosset P, Noel A, Cataldo DD. The metalloproteinase ADAM-12 regulates bronchial epithelial cell proliferation and apoptosis. Cell Prolif. 2008;41(6):988–1001. doi: 10.1111/j.1365-2184.2008.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, Broadway N, Hartmann D, Saftig P, Umland S, Sehara-Fujisawa A, Black RA, Ludwig A, Becherer JD, Conrad DH, Blobel CP. ADAM10 is a principal 'sheddase' of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006;7(12):1293–1298. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]
- Lemieux GA, Blumenkron F, Yeung N, Zhou P, Williams J, Grammer AC, Petrovich R, Lipsky PE, Moss ML, Werb Z. The low affinity IgE receptor (CD23) is cleaved by the metalloproteinase ADAM10. J Biol Chem. 2007;282(20):14836–14844. doi: 10.1074/jbc.M608414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt HB, Overgaard MT, Laursen LS, Weyer K, Sottrup-Jensen L, Oxvig C. Mutational analysis of the proteolytic domain of pregnancy-associated plasma protein-A (PAPP-A): classification as a metzincin. Biochem J. 2001;358(Pt 2):359–367. doi: 10.1042/0264-6021:3580359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawston TE, Wilson AJ. Understanding the role of tissue degrading enzymes and their inhibitors in development and disease. Best Pract Res Clin Rheumatol. 2006;20(5):983–1002. doi: 10.1016/j.berh.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX. Catalytic domain architecture of metzincin metalloproteases. J Biol Chem. 2009;284(23):15353–15357. doi: 10.1074/jbc.R800069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth FX, McKay DB, Bode W. The metzincins--topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 1995;4(5):823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres K, Anders A, Kojro E, Gilbert S, Fahrenholz F, Postina R. Tumor necrosis factor-alpha converting enzyme is processed by proprotein-convertases to its mature form which is degraded upon phorbol ester stimulation. Eur J Biochem. 2003;270(11):2386–2393. doi: 10.1046/j.1432-1033.2003.03606.x. [DOI] [PubMed] [Google Scholar]
- Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J. 2000;348(Pt 1):21–27. doi: 10.1042/0264-6021:3480021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomann U, Wildeboer D, Webster A, Antropova O, Zeuschner D, Knight CG, Docherty AJ, Lambert M, Skelton L, Jockusch H, Bartsch JW. The metalloprotease disintegrin ADAM8. Processing by autocatalysis is required for proteolytic activity and cell adhesion. J Biol Chem. 2002;277(50):48210–48219. doi: 10.1074/jbc.M203355200. [DOI] [PubMed] [Google Scholar]
- Jones GC, Riley GP. ADAMTS proteinases: a multi-domain, multi-functional family with roles in extracellular matrix turnover and arthritis. Arthritis Res Ther. 2005;7(4):160–169. doi: 10.1186/ar1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knauper V, Docherty AJ, Murphy G. TNF-alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435(1):39–44. doi: 10.1016/S0014-5793(98)01031-X. [DOI] [PubMed] [Google Scholar]
- Amour A, Knight CG, Webster A, Slocombe PM, Stephens PE, Knauper V, Docherty AJ, Murphy G. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473(3):275–279. doi: 10.1016/S0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- Wayne GJ, Deng SJ, Amour A, Borman S, Matico R, Carter HL, Murphy G. TIMP-3 inhibition of ADAMTS-4 (Aggrecanase-1) is modulated by interactions between aggrecan and the C-terminal domain of ADAMTS-4. J Biol Chem. 2007;282(29):20991–20998. doi: 10.1074/jbc.M610721200. [DOI] [PubMed] [Google Scholar]
- Wang WM, Ge G, Lim NH, Nagase H, Greenspan DS. TIMP-3 inhibits the procollagen N-proteinase ADAMTS-2. Biochem J. 2006;398(3):515–519. doi: 10.1042/BJ20060630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. Structural determinants of the ADAM inhibition by TIMP-3: crystal structure of the TACE-N-TIMP-3 complex. J Mol Biol. 2008;381(5):1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- Troeberg L, Fushimi K, Scilabra SD, Nakamura H, Dive V, Thogersen IB, Enghild JJ, Nagase H. The C-terminal domains of ADAMTS-4 and ADAMTS-5 promote association with N-TIMP-3. Matrix Biol. 2009;28(8):463–9. doi: 10.1016/j.matbio.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz RM, Karkkainen I, Huovila AP. Aberrant alternative exon use and increased copy number of human metalloprotease-disintegrin ADAM15 gene in breast cancer cells. Genes Chromosomes Cancer. 2004;41(4):366–378. doi: 10.1002/gcc.20102. [DOI] [PubMed] [Google Scholar]
- Primakoff P, Hyatt H, Tredick-Kline J. Identification and purification of a sperm surface protein with a potential role in sperm-egg membrane fusion. J Cell Biol. 1987;104(1):141–149. doi: 10.1083/jcb.104.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP. Fertilin beta and other ADAMs as integrin ligands: insights into cell adhesion and fertilization. Bioessays. 2001;23(7):628–639. doi: 10.1002/bies.1088. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16(2):209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham C, Levkau B, Raines EW, Herren B. ADAM15 is an adherens junction molecule whose surface expression can be driven by VE-cadherin. Exp Cell Res. 2002;279(2):239–247. doi: 10.1006/excr.2002.5606. [DOI] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385(6618):733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- Arner EC. Aggrecanase-mediated cartilage degradation. Curr Opin Pharmacol. 2002;2(3):322–329. doi: 10.1016/S1471-4892(02)00148-0. [DOI] [PubMed] [Google Scholar]
- Nagase H, Kashiwagi M. Aggrecanases and cartilage matrix degradation. Arthritis Res Ther. 2003;5(2):94–103. doi: 10.1186/ar630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colige A, Ruggiero F, Vandenberghe I, Dubail J, Kesteloot F, Van Beeumen J, Beschin A, Brys L, Lapiere CM, Nusgens B. Domains and maturation processes that regulate the activity of ADAMTS-2, a metalloproteinase cleaving the aminopropeptide of fibrillar procollagens types I-III and V. J Biol Chem. 2005;280(41):34397–34408. doi: 10.1074/jbc.M506458200. [DOI] [PubMed] [Google Scholar]
- McGowan PM, McKiernan E, Bolster F, Ryan BM, Hill AD, McDermott EW, Evoy D, O'Higgins N, Crown J, Duffy MJ. ADAM-17 predicts adverse outcome in patients with breast cancer. Ann Oncol. 2008;19(6):1075–1081. doi: 10.1093/annonc/mdm609. [DOI] [PubMed] [Google Scholar]
- Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem. 2002;277(25):22201–22208. doi: 10.1074/jbc.M200431200. [DOI] [PubMed] [Google Scholar]
- Majumdar MK, Askew R, Schelling S, Stedman N, Blanchet T, Hopkins B, Morris EA, Glasson SS. Double-knockout of ADAMTS-4 and ADAMTS-5 in mice results in physiologically normal animals and prevents the progression of osteoarthritis. Arthritis Rheum. 2007;56(11):3670–3674. doi: 10.1002/art.23027. [DOI] [PubMed] [Google Scholar]
- Prinzen C, Trumbach D, Wurst W, Endres K, Postina R, Fahrenholz F. Differential gene expression in ADAM10 and mutant ADAM10 transgenic mice. BMC Genomics. 2009;10:66. doi: 10.1186/1471-2164-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, Flamez P, Dequenne A, Godaux E, van Leuven F, Fahrenholz F. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004;113(10):1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkiewska A. Disintegrin-like/cysteine-rich region of ADAM 12 is an active cell adhesion domain. Exp Cell Res. 1999;252(2):423–431. doi: 10.1006/excr.1999.4632. [DOI] [PubMed] [Google Scholar]
- Dijkstra A, Postma DS, Noordhoek JA, Lodewijk ME, Kauffman HF, ten Hacken NH, Timens W. Expression of ADAMs ("a disintegrin and metalloprotease") in the human lung. Virchows Arch. 2009;454(4):441–449. doi: 10.1007/s00428-009-0748-4. [DOI] [PubMed] [Google Scholar]
- Rocks N, Paulissen G, Quesada Calvo F, Polette M, Gueders M, Munaut C, Foidart JM, Noel A, Birembaut P, Cataldo D. Expression of a disintegrin and metalloprotease (ADAM and ADAMTS) enzymes in human non-small-cell lung carcinomas (NSCLC) Br J Cancer. 2006;94(5):724–730. doi: 10.1038/sj.bjc.6602990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulissen G, Rocks N, Quesada-Calvo F, Gosset P, Foidart JM, Noel A, Louis R, Cataldo DD. Expression of ADAMs and their inhibitors in sputum from patients with asthma. Mol Med. 2006;12(7-8):171–179. doi: 10.2119/2006-00028.Paulissen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating DT, Sadlier DM, Patricelli A, Smith SM, Walls D, Egan JJ, Doran PP. Microarray identifies ADAM family members as key responders to TGF-beta1 in alveolar epithelial cells. Respir Res. 2006;7:114. doi: 10.1186/1465-9921-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR. et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418(6896):426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- Powell RM, Wicks J, Holloway JW, Holgate ST, Davies DE. The splicing and fate of ADAM33 transcripts in primary human airways fibroblasts. Am J Respir Cell Mol Biol. 2004;31(1):13–21. doi: 10.1165/rcmb.2003-0330OC. [DOI] [PubMed] [Google Scholar]
- Haitchi HM, Powell RM, Shaw TJ, Howarth PH, Wilson SJ, Wilson DI, Holgate ST, Davies DE. ADAM33 expression in asthmatic airways and human embryonic lungs. Am J Respir Crit Care Med. 2005;171(9):958–965. doi: 10.1164/rccm.200409-1251OC. [DOI] [PubMed] [Google Scholar]
- Yang Y, Haitchi HM, Cakebread J, Sammut D, Harvey A, Powell RM, Holloway JW, Howarth P, Holgate ST, Davies DE. Epigenetic mechanisms silence a disintegrin and metalloprotease 33 expression in bronchial epithelial cells. J Allergy Clin Immunol. 2008;121(6):1393–1399. doi: 10.1016/j.jaci.2008.02.031. 1399 e1391-1314. [DOI] [PubMed] [Google Scholar]
- Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, Ernst P, Lemiere C, Martin JG, Hamid Q. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119(4):863–871. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Onoda S, Hattori Y, Maitani Y, Sakai H, Misawa M. Upregulation of ADAM8 in the airways of mice with allergic bronchial asthma. Lung. 2009;187(3):179–185. doi: 10.1007/s00408-009-9145-7. [DOI] [PubMed] [Google Scholar]
- King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, Witte DP, Rothenberg ME. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol. 2004;31(3):257–265. doi: 10.1165/rcmb.2004-0026OC. [DOI] [PubMed] [Google Scholar]
- Matsuno O, Miyazaki E, Nureki S, Ueno T, Kumamoto T, Higuchi Y. Role of ADAM8 in experimental asthma. Immunol Lett. 2006;102(1):67–73. doi: 10.1016/j.imlet.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bousquet J, Jeffery PK, Busse WW, Johnson M, Vignola AM. Asthma. From bronchoconstriction to airways inflammation and remodeling. Am J Respir Crit Care Med. 2000;161(5):1720–1745. doi: 10.1164/ajrccm.161.5.9903102. [DOI] [PubMed] [Google Scholar]
- Bergeron C, Al-Ramli W, Hamid Q. Remodeling in asthma. Proc Am Thorac Soc. 2009;6(3):301–305. doi: 10.1513/pats.200808-089RM. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Ohtsuki Y, Tokunaga K, Yamaoka-Sageshima M, Ichikawa K, Aoki T, Shibasaki M, Arinami T. ADAM33 polymorphisms are associated with asthma susceptibility in a Japanese population. Clin Exp Allergy. 2006;36(5):602–608. doi: 10.1111/j.1365-2222.2006.02471.x. [DOI] [PubMed] [Google Scholar]
- Werner M, Herbon N, Gohlke H, Altmuller J, Knapp M, Heinrich J, Wjst M. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy. 2004;34(1):26–31. doi: 10.1111/j.1365-2222.2004.01846.x. [DOI] [PubMed] [Google Scholar]
- Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, Xu J, Bleecker ER, Meyers DA. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112(4):717–722. doi: 10.1016/S0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- Hirota T, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, Shirakawa T, Doi S, Fujita K, Suzuki Y, Nakamura Y, Tamari M. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin Exp Allergy. 2006;36(7):884–891. doi: 10.1111/j.1365-2222.2006.02522.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Liu QJ, Li JS, Li HC, Wei CH, Guo CH, Gong YQ. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet. 2006;33(4):303–306. doi: 10.1111/j.1744-313X.2006.00617.x. [DOI] [PubMed] [Google Scholar]
- Lind DL, Choudhry S, Ung N, Ziv E, Avila PC, Salari K, Ha C, Lovins EG, Coyle NE, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, Matallana H, Lilly CM, Salas J, Selman M, Boushey HA, Weiss ST, Chapela R, Ford JG, Rodriguez-Cintron W, Silverman EK, Sheppard D, Kwok PY, González Burchard E. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med. 2003;168(11):1312–1316. doi: 10.1164/rccm.200306-877OC. [DOI] [PubMed] [Google Scholar]
- Garlisi CG, Zou J, Devito KE, Tian F, Zhu FX, Liu J, Shah H, Wan Y, Motasim Billah M, Egan RW, Umland SP. Human ADAM33: protein maturation and localization. Biochem Biophys Res Commun. 2003;301(1):35–43. doi: 10.1016/S0006-291X(02)02976-5. [DOI] [PubMed] [Google Scholar]
- Puxeddu I, Pang YY, Harvey A, Haitchi HM, Nicholas B, Yoshisue H, Ribatti D, Clough G, Powell RM, Murphy G, Hanley NA, Wilson DI, Howarth PH, Holgate ST, Davies DE. The soluble form of a disintegrin and metalloprotease 33 promotes angiogenesis: implications for airway remodeling in asthma. J Allergy Clin Immunol. 2008;121(6):1400–1406. doi: 10.1016/j.jaci.2008.03.003. 1406 e1401-1404. [DOI] [PubMed] [Google Scholar]
- Jie Z, Jin M, Cai Y, Bai C, Shen Y, Yuan Z, Hu Y, Holgate S. The effects of Th2 cytokines on the expression of ADAM33 in allergen-induced chronic airway inflammation. Respir Physiol Neurobiol. 2009;168(3):289–294. doi: 10.1016/j.resp.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Chen C, Huang X, Sheppard D. ADAM33 is not essential for growth and development and does not modulate allergic asthma in mice. Mol Cell Biol. 2006;26(18):6950–6956. doi: 10.1128/MCB.00646-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Valentin E, Crahay C, Garbacki N, Hennuy B, Gueders M, Noel A, Foidart JM, Grooten J, Colige A, Piette J, Cataldo D. New asthma biomarkers: lessons from murine models of acute and chronic asthma. Am J Physiol Lung Cell Mol Physiol. 2009;296(2):L185–197. doi: 10.1152/ajplung.90367.2008. [DOI] [PubMed] [Google Scholar]
- Tremblay K, Lemire M, Potvin C, Tremblay A, Hunninghake GM, Raby BA, Hudson TJ, Perez-Iratxeta C, Andrade-Navarro MA, Laprise C. Genes to diseases (G2D) computational method to identify asthma candidate genes. PLoS ONE. 2008;3(8):e2907. doi: 10.1371/journal.pone.0002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourie AM, Coles F, Moreno V, Karlsson L. Catalytic activity of ADAM8, ADAM15, and MDC-L (ADAM28) on synthetic peptide substrates and in ectodomain cleavage of CD23. J Biol Chem. 2003;278(33):30469–30477. doi: 10.1074/jbc.M213157200. [DOI] [PubMed] [Google Scholar]
- Naus S, Reipschlager S, Wildeboer D, Lichtenthaler SF, Mitterreiter S, Guan Z, Moss ML, Bartsch JW. Identification of candidate substrates for ectodomain shedding by the metalloprotease-disintegrin ADAM8. Biol Chem. 2006;387(3):337–346. doi: 10.1515/BC.2006.045. [DOI] [PubMed] [Google Scholar]
- Kelly K, Hutchinson G, Nebenius-Oosthuizen D, Smith AJ, Bartsch JW, Horiuchi K, Rittger A, Manova K, Docherty AJ, Blobel CP. Metalloprotease-disintegrin ADAM8: expression analysis and targeted deletion in mice. Dev Dyn. 2005;232(1):221–231. doi: 10.1002/dvdy.20221. [DOI] [PubMed] [Google Scholar]
- van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Schouten JP, Boezen HM. A disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general population. Am J Respir Crit Care Med. 2005;172(3):329–333. doi: 10.1164/rccm.200411-1486OC. [DOI] [PubMed] [Google Scholar]
- Gosman MM, Boezen HM, van Diemen CC, Snoeck-Stroband JB, Lapperre TS, Hiemstra PS, Ten Hacken NH, Stolk J, Postma DS. A disintegrin and metalloprotease 33 and chronic obstructive pulmonary disease pathophysiology. Thorax. 2007;62(3):242–247. doi: 10.1136/thx.2006.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju CR, Xia XZ, Chen RC. Expressions of tumor necrosis factor-converting enzyme and ErbB3 in rats with chronic obstructive pulmonary disease. Chin Med J (Engl) 2007;120(17):1505–1510. [PubMed] [Google Scholar]
- Shao MX, Nakanaga T, Nadel JA. Cigarette smoke induces MUC5AC mucin overproduction via tumor necrosis factor-alpha-converting enzyme in human airway epithelial (NCI-H292) cells. Am J Physiol Lung Cell Mol Physiol. 2004;287(2):L420–427. doi: 10.1152/ajplung.00019.2004. [DOI] [PubMed] [Google Scholar]
- Ito I, Laporte JD, Fiset PO, Asai K, Yamauchi Y, Martin JG, Hamid Q. Downregulation of a disintegrin and metalloproteinase 33 by IFN-gamma in human airway smooth muscle cells. J Allergy Clin Immunol. 2007;119(1):89–97. doi: 10.1016/j.jaci.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Sadeghnejad A, Ohar JA, Zheng SL, Sterling DA, Hawkins GA, Meyers DA, Bleecker ER. Adam33 polymorphisms are associated with COPD and lung function in long-term tobacco smokers. Respir Res. 2009;10:21. doi: 10.1186/1465-9921-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Hoffjan S, Hayes MG, Schneider D, Nicolae R, Heinzmann A, Jerkic SP, Parry R, Cox NJ, Deichmann KA, Ober C. Fine mapping and positional candidate studies on chromosome 5p13 identify multiple asthma susceptibility loci. J Allergy Clin Immunol. 2006;118(2):396–402. doi: 10.1016/j.jaci.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Shintani Y, Higashiyama S, Ohta M, Hirabayashi H, Yamamoto S, Yoshimasu T, Matsuda H, Matsuura N. Overexpression of ADAM9 in non-small cell lung cancer correlates with brain metastasis. Cancer Res. 2004;64(12):4190–4196. doi: 10.1158/0008-5472.CAN-03-3235. [DOI] [PubMed] [Google Scholar]
- Booth BW, Sandifer T, Martin EL, Martin LD. IL-13-induced proliferation of airway epithelial cells: mediation by intracellular growth factor mobilization and ADAM17. Respir Res. 2007;8:51. doi: 10.1186/1465-9921-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidl ID, Huber G, Eichmann K. An ADAM family member with expression in thymic epithelial cells and related tissues. Gene. 2002;283(1-2):163–170. doi: 10.1016/S0378-1119(01)00871-X. [DOI] [PubMed] [Google Scholar]
- Umland SP, Garlisi CG, Shah H, Wan Y, Zou J, Devito KE, Huang WM, Gustafson EL, Ralston R. Human ADAM33 messenger RNA expression profile and post-transcriptional regulation. Am J Respir Cell Mol Biol. 2003;29(5):571–582. doi: 10.1165/rcmb.2003-0028OC. [DOI] [PubMed] [Google Scholar]
- Lee JH, Park HS, Park SW, Jang AS, Uh ST, Rhim T, Park CS, Hong SJ, Holgate ST, Holloway JW, Shin HD. ADAM33 polymorphism: association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy. 2004;34(6):860–865. doi: 10.1111/j.1365-2222.2004.01977.x. [DOI] [PubMed] [Google Scholar]
- Raby BA, Silverman EK, Kwiatkowski DJ, Lange C, Lazarus R, Weiss ST. ADAM33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol. 2004;113(6):1071–1078. doi: 10.1016/j.jaci.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, Zheng SL, Meyers DA, Bleecker ER, Postma DS. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34(5):757–760. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- Simpson A, Maniatis N, Jury F, Cakebread JA, Lowe LA, Holgate ST, Woodcock A, Ollier WE, Collins A, Custovic A, Holloway JW, John SL. Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33) predict impaired early-life lung function. Am J Respir Crit Care Med. 2005;172(1):55–60. doi: 10.1164/rccm.200412-1708OC. [DOI] [PubMed] [Google Scholar]
- Schedel M, Depner M, Schoen C, Weiland SK, Vogelberg C, Niggemann B, Lau S, Illig T, Klopp N, Wahn U, von Mutius E, Nickel R, Kabesch M. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res. 2006;7:91. doi: 10.1186/1465-9921-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedda MA, Duffy DL, Bradley B, O'Hehir RE, Thompson PJ. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet. 2006;14(9):1027–1036. doi: 10.1038/sj.ejhg.5201662. [DOI] [PubMed] [Google Scholar]
- Hersh CP, Raby BA, Soto-Quiros ME, Murphy AJ, Avila L, Lasky-Su J, Sylvia JS, Klanderman BJ, Lange C, Weiss ST, Celedón JC. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Respir Crit Care Med. 2007;176(9):849–857. doi: 10.1164/rccm.200704-592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami T, Jinnai N, Nakajima T, Sekigawa T, Hasegawa T, Suzuki E, Inoue I, Gejyo F. ADAM33 polymorphisms are associated with aspirin-intolerant asthma in the Japanese population. J Hum Genet. 2007;52(1):66–72. doi: 10.1007/s10038-006-0081-6. [DOI] [PubMed] [Google Scholar]
- Su D, Zhang X, Sui H, Lu F, Jin L, Zhang J. Association of ADAM33 gene polymorphisms with adult allergic asthma and rhinitis in a Chinese Han population. BMC Med Genet. 2008;9 doi: 10.1186/1471-2350-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongngarm T, Jameekornrak A, Limwongse C, Sangasapaviliya A, Jirapongsananuruk O, Assawamakin A, Chaiyaratana N, Luangwedchakarn V, Thongnoppakhun W. Association between ADAM33 polymorphisms and asthma in a Thai population. Asian Pac J Allergy Immunol. 2008;26(4):205–211. [PubMed] [Google Scholar]
- Zhang X, Su D, Sui H, Jin L, Lu F, Zhang J. Association of ADAM33 gene polymorphisms with adult concomitant allergic rhinitis and asthma in Chinese Han population. Mol Biol Rep. 2009;36(6):1505–1509. doi: 10.1007/s11033-008-9343-z. [DOI] [PubMed] [Google Scholar]
- Blakey JD, Sayers I, Ring SM, Strachan DP, Hall IP. Positionally cloned asthma susceptibility gene polymorphisms and disease risk in the British 1958 Birth Cohort. Thorax. 2009;64(5):381–387. doi: 10.1136/thx.2008.102053. [DOI] [PubMed] [Google Scholar]