Abstract
Animals and plants evolved systems to permit non-cell-autonomous trafficking of RNA, whereas DNA plays a cell-autonomous role. In plants, plasmodesmata serve as the conduit for this phenomenon, and viruses have evolved to use this pathway for the spread of infectious nucleic acids. In this study, a plant DNA virus was used to explore the constraints imposed on the movement of DNA through this endogenous RNA trafficking pathway. The combined properties of the geminivirus-encoded movement protein and plasmodesmata were shown to impose a strict limitation on the size of the viral genome at the level of cell-to-cell movement. Size-increased viral genome components underwent homologous and nonhomologous recombination to overcome this strict limitation. Our results provide insights into the genetic mechanisms that underlie viral evolution and provide a likely explanation for why relatively few types of plant DNA viruses have evolved: they would have had to overcome the constraints imposed by an endogenous system operating to ensure that DNA acts in a cell-autonomous manner.
INTRODUCTION
A paradigm is emerging in which RNA acts beyond the cellular sites of transcription (Fire et al., 1998; Jorgensen et al., 1998; Fire, 1999; Lucas et al., 2001; Hannon, 2002; Wu et al., 2002). In plants, a role for non-cell-autonomous RNA has been demonstrated with respect to long-distance signaling that is associated with post-transcriptional gene silencing (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998; Fagard and Vaucheret, 2000; Vance and Vaucheret, 2001), virus-induced gene silencing (Lindbo et al., 1993; Voinnet and Baulcombe, 1997; Ruiz et al., 1998; Baulcombe, 1999), and development (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Kim et al., 2001; Lucas et al., 2001; Jorgensen, 2002). In animals, RNA interference has similarly been shown to act in a non-cell-autonomous manner (Fire et al., 1998; Timmons et al., 2001), and the genes involved in the transmission of gene silencing are being elucidated at present (Winston et al., 2002). In plants, plasmodesmata serve as the conduit for the cell-to-cell trafficking of RNA-protein complexes (Fujiwara et al., 1993; Noueiry et al., 1994; Rojas et al., 1997; Lough et al., 1998).
It is now generally accepted that plasmodesmata evolved the capacity for the movement of endogenous proteins and ribonucleoprotein complexes to exert non-cell-autonomous control over developmental processes (Lucas et al., 1993, 1995; Zambryski and Crawford, 2000; Nakajima et al., 2001; Haywood et al., 2002; Kim et al., 2002; Lee et al., 2003). At the organismal level, the special features of the plant vascular system, and in particular the specialized cell types of the phloem, allow for the long-distance movement of information macromolecules. Here, plasmodesmata also serve as both the pathway and the site(s) of regulation for the entry and exit of such molecules, and recent studies have demonstrated the long-distance translocation of a complex set of endogenous RNAs (Ruiz-Medrano et al., 1999; Kim et al., 2001).
Plant viruses also have evolved, or acquired, the capacity to use this plasmodesmata pathway for the cell-to-cell spread of their infectious nucleic acids (Carrington et al., 1996; Gilbertson and Lucas, 1996; Ghoshroy et al., 1997; Lazarowitz and Beachy, 1999). Generally, these viral genomes are composed of RNA, although in a few cases the infectious agent has been shown to be DNA (Hull, 2002). Plant viruses also have gained the capacity to use the vascular pathway to establish a systemic infection. An extensive literature documents the capacity of hundreds of RNA viruses to establish systemic infection in plants; by comparison, the number of plant DNA viruses is rather limited (Hull, 2002). This situation is in marked contrast to that of viruses that infect animals, in which the viral genomes are distributed more equally between RNA and DNA (van Regenmortel et al., 2000). This difference between plant and animal viruses may reflect the evolution of a plant-specific plasmodesmata pathway that uses RNA as the non-cell-autonomous signaling macromolecule (Lucas et al., 2001). Thus, within the context of somatic plant tissues, DNA became confined to a cell-autonomous role, whereas RNA evolved to function at both the cell-autonomous and non-cell-autonomous levels. The mechanism(s) by which such segregation of function was established may reflect either a simple difference in size (chromosomes versus ribonucleoproteins) and/or cellular location (nucleus versus cytoplasm) or a more specific process(es) founded on molecular recognition.
Insight into the nature of the mechanisms used by plants to allow the selective trafficking of nucleic acids has come from studies using plant viruses as experimental systems (Deom et al., 1992; Lucas and Gilbertson, 1994; Lazarowitz and Beachy, 1999). Plant DNA viruses offer a unique tool for testing the hypothesis that a specific RNA recognition mechanism prevents the trafficking of cell-autonomous DNA. Both DNA and RNA plant viruses use two distinct mechanisms for cell-to-cell movement. The de novo formation of tubules that contain virus-like particles (Linstead et al., 1988; van Lent et al., 1991; Ritzenthaler et al., 1995; Storms et al., 1995) likely reflects a mechanism by which some DNA (e.g., members of the Caulimoviridae family [Hohn, 1999]) and RNA (e.g., members of the Comoviridae family [Lomonossoff and Shanks, 1999]) viruses circumvent the need to use the endogenous plasmodesmata pathway for nucleic acid trafficking and detection by the RNA recognition system. Other viruses form a nucleoprotein complex that is compatible with the endogenous plasmodesmata pathway. In this situation, virus-encoded movement proteins facilitate this process through the formation of a complex that traffics through plasmodesmata (Deom et al., 1992; Lucas and Gilbertson, 1994; Lazarowitz and Beachy, 1999) and avoids detection by the endogenous RNA/DNA surveillance/recognition system (Vance and Vaucheret, 2001). Although it might have been relatively straightforward for the RNA viruses to exploit this endogenous pathway, DNA viruses would have had to overcome the additional challenge based on their different molecular structures.
Plant DNA viruses that use tubule formation to spread have monopartite, double-stranded genomes in the size range of 7.5 to 8.0 kb (Rothnie et al., 1994; Hull, 2002). By contrast, DNA viruses that traffic cell to cell via the plasmodesmata pathway have single-stranded genomes (often multipartite) (e.g., members of the Geminiviridae family) in which the individual components are ∼2.5 to 3 kb (Stanley, 1991; Timmermans et al., 1994; Hanley-Bowdoin et al., 1999). Although this difference in genome size has been attributed to limitations imposed by encapsidation (Stanley, 1991), studies with geminivirus capsid protein (CP) mutants failed to support this notion (Etessami et al., 1989; Klinkenberg et al., 1989; Elmer and Rogers, 1990; Sudarshana et al., 1998). This raised the possibility that the fundamental principle dictating the retention of the geminiviral genome size reflects the requirements for exploitation of the endogenous RNA trafficking system. Evidence to support this concept was provided by studies demonstrating that geminivirus movement proteins (BC1 and BV1) have the capacity to recognize DNA on the basis of size and form (Rojas et al., 1998). Additional evidence for the operation of a selection mechanism based on genome size has come from experiments demonstrating that size-increased geminiviral DNA components revert to genome size during systemic infection (Etessami et al., 1989; Elmer and Rogers, 1990).
The aim of this study was to test the hypothesis that the fundamental principle responsible for the strict maintenance of geminiviral genome (DNA) size reflects the functional limitations imposed by the plant's endogenous RNA trafficking pathway. Our findings provide strong support for this concept, because we demonstrated that the selection process for viral genome size occurred during the initial cell-to-cell movement of the infectious DNA. The identification of this site was accomplished through the examination of each step in the infection process using a combination of genetic, molecular, and cellular approaches.
RESULTS
Production of Bean dwarf mosaic virus DNA-A Size-Increased Clones
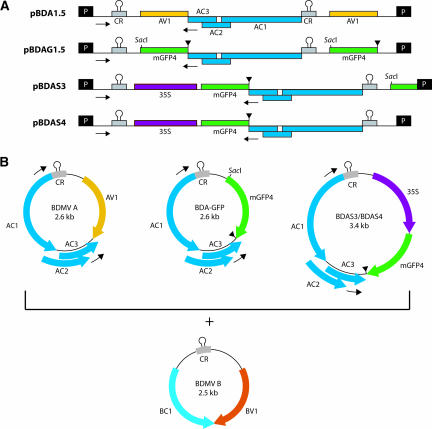
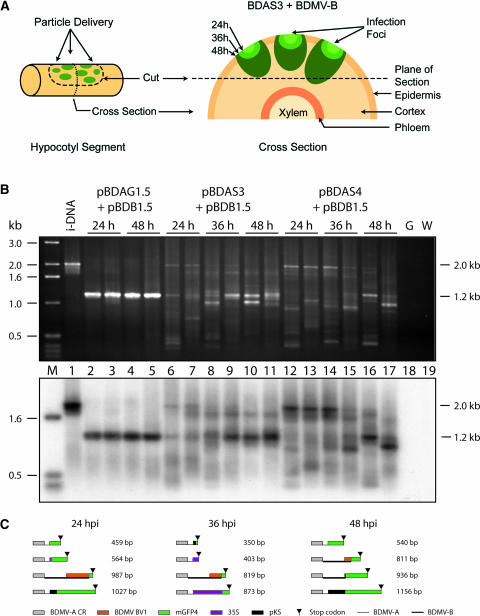
To examine the cellular mechanism(s) underlying the strict maintenance of geminiviral genome size, we used the bipartite begomovirus Bean dwarf mosaic virus (BDMV). A series of size-increased BDMV DNA-A (BDMV-A) clones was engineered to identify the cellular boundary at which genome size reversion occurs. For our studies, we used the reporter gene green fluorescent protein (mGFP4) (Haseloff et al., 1997). We had shown previously that mGFP4 could be exchanged for the AV1 (CP) gene of BDMV-A without altering the genome size (Sudarshana et al., 1998). This construct, here referred to as BDA-GFP, served as an effective reporter for viral cell-to-cell and systemic spread (Sudarshana et al., 1998; Wang et al., 1999; Garrido-Ramirez et al., 2000). Figure 1A shows the plasmid maps of pBDA1.5 and pBDAG1.5, which are multimeric clones of wild-type BDMV-A and BDA-GFP, respectively. During the infection process, genome-sized (∼2.6 kb) BDMV-A or BDA-GFP is generated from these clones (Figure 1B). This occurs by a process referred to as replicational release. In this process, the viral replication-associated protein (AC1) binds to the common region, which contains the origin of replication, and mediates the synthesis of nascent viral sense single-stranded DNA molecules that are released at the next common region (Figure 1A).
Figure 1.
Schematic illustration of the Bean dwarf mosaic virus (BDMV) DNA-A and DNA-B Components Used in Genome Size-Reversion Studies.
(A) Linear maps are shown for the following constructs: pBDA1.5, a multimeric clone of wild-type BDMV DNA-A (BDMV-A); pBDAG1.5, a multimeric clone of BDMV-A in which the AV1 (CP) gene was replaced with mGFP4; pBDAS3, a size-increased BDMV-A containing a 35S-mGFP4 cassette and having a truncated copy of mGFP4 outside of the common region; and pBDAS4, a size-increased BDMV-A identical to pBDAS3 except that it lacks the truncated mGFP4.
(B) Circular maps of the BDMV-A components released from the multimeric clones shown in (A). Each component was coinoculated (by particle bombardment) into plants along with pBDB1.5, which releases wild-type BDMV-B.
AC1 to AC3, AV1, BV1, and BC1 are viral genes; CR, common region, showing a highly conserved stem loop; mGFP4, a modified form of GFP; P, plasmid cloning vector (pBluescript KS+ [pKS]); 35S, promoter from Cauliflower mosaic virus. Inverted triangles indicate the mGFP4 stop codon/bidirectional transcriptional terminator, and arrows indicate the positions of the size-reversion primer pair. Note that SacI is adjacent to the mGFP4 start codon. The BDMV movement proteins BV1 and BC1 mediate the nuclear-cytoplasmic export and cell-to-cell trafficking of viral DNA, respectively.
Two size-increased clones, pBDAS3 and pBDAS4, were generated by placing mGFP4 under the control of the 35S promoter (Figure 1A). These clones would be predicted to yield an ∼3.4-kb recombinant BDMV-A component by replicational release (Figure 1B). Here, pBDAS3 was designed to optimize the capacity for genome size reversion by the inclusion of a partial copy of mGFP4 in the recombinant plasmid, thereby potentiating homologous recombination. By contrast, the absence of this partial mGFP4 sequence from the pBDAS4 recombinant plasmid would allow size reversion only by means of nonhomologous recombination. We reasoned that pBDAS3 would be efficient in reversion and, thus, would produce infectious forms of BDMV-A to allow the identification of the preferred modes of genome size reversion (i.e., homologous versus nonhomologous recombination). Finally, both pBDAS3 and pBDAS4 were engineered such that, when size reversion occurred, only the 35S-mGFP4 sequences, which are nonessential for virus replication and movement, could be deleted to generate a wild-type-sized infectious DNA-A component.
Reversion of Size-Increased BDMV-A Components Detected in Systemically Infected Tissues
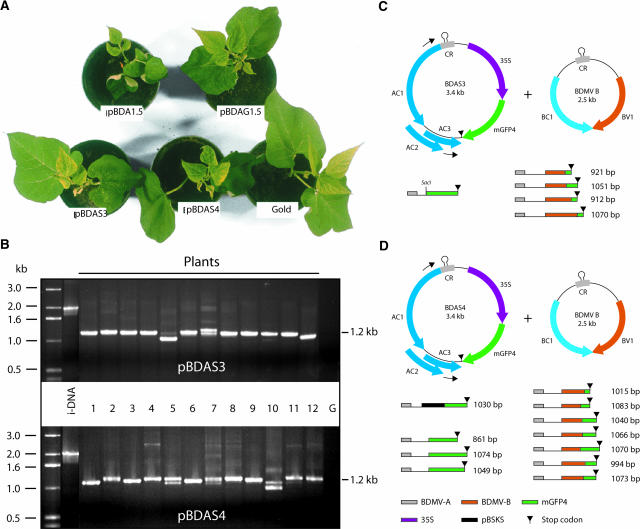
Genome size reversion could reflect selection imposed during (1) replication, (2) nuclear-cytoplasmic transport (mediated by BV1), (3) cell-to-cell movement through plasmodesmata (mediated by BC1), and/or (4) entry/exit into the phloem translocation stream. Figure 2 shows the results of experiments performed to determine whether BDMV-A genome size reversion was a requirement for systemic movement. Plants coinoculated with each of the BDMV-A recombinant plasmids shown in Figure 1A and pBDB1.5 developed symptoms of BDMV infection, demonstrating the infectious nature of these size-increased constructs (Figure 2A). The rate of infectivity was equivalent in all cases (data not shown), whereas symptom severity for pBDAG1.5, pBDAS3, and pBDAS4 was attenuated slightly or comparable to that induced by wild-type BDMV (pBDA1.5).
Figure 2.
Size Reversion of the BDMV Genome Detected in Systemically Infected Tissue.
(A) Disease symptoms displayed by young bean plants coinoculated with each of the indicated DNA-A components and pBDB1.5 or gold particles (control). Plants were photographed 15 days after bombardment.
(B) PCR analysis of DNA extracted from systemically infected tissue demonstrating BDMV genome size reversion. Tissues were collected from 12 seedlings (1 to 12) each inoculated with pBDAS3 or pBDAS4, and the size-reversion primer pair was used to direct the amplification of reverted (∼1.2 kb) and nonreverted (∼2 kb) DNA-A fragments. i-DNA served as a control for the nonreverted genome size. G, gold.
(C) and (D) Sequence analyses of PCR-amplified products illustrating the underlying nature of the genome size reversion for DNA-A components derived from pBDAS3 and pBDAS4 in the systemically infected plants examined in (B). CR, common region.
To assess for genome size reversion, total DNA was extracted from systemically infected leaves and used in PCR analyses. A primer pair was designed to direct amplification across the region of the viral genome containing the heterologous sequences, where recombination was most likely to occur. This size-reversion primer pair was used to generate DNA-A fragments for characterization by agarose gel electrophoresis and sequence analysis. The size-reversion primer pair directed the amplification of the predicted 1.2-kb DNA fragment from wild-type BDMV-infected plants (data not shown). In the absence of size reversion, this primer pair would direct the amplification of a 2.0-kb fragment (e.g., from input DNA [i-DNA]). As illustrated in Figure 2B, genome size reversion was detected in all plants infected with the size-increased constructs, based on the presence of PCR-amplified fragments in the range of 1.0 to 1.2 kb. With respect to pBDAS3, equivalent-sized fragments were detected in 9 of the 12 plants examined (Figure 2B). Furthermore, sequence analysis established that eight of these nine plants represented revertants generated by homologous recombination of the two GFP sequences contained within the input plasmid (Figure 2C). (Note the presence of two classes of homologous recombinant DNA-A components, both having functional GFP genes.) Analysis of the remaining four fragments revealed that reversion had occurred through nonhomologous recombination between the DNA-A and DNA-B components. Collectively, these results demonstrate the action of a strong selection mechanism for the generation of genome-sized components at some stage during the systemic infection process.
Results obtained with pBDAS4, which lacks the capacity for homologous recombination, provided further support for the notion that the virus encounters an explicit barrier that can be overcome only by the generation of a genome within a very narrow size range. As with pBDAS3, all pBDAS4 fragments fell within the 1.0- to 1.2-kb size class; by contrast, however, each of the PCR-generated fragments exhibited a slightly different size. Sequence analysis further supported this observation (Figure 2D) and revealed a difference in the nature of the reverted molecules. In this situation, of the 12 revertants examined, nonhomologous recombination occurred within the i-DNA (2), within pBDAS4 (2), or between the DNA-A and DNA-B components (8). Finally, during analysis of the PCR products, we observed faint bands that were generally larger than the fragment corresponding to the i-DNA. Because an increase in genome size seems unlikely, in the context of the present experiments, these bands were considered to represent spurious PCR products.
Genome Size Reversion Not Detected during Replication in Single Cells
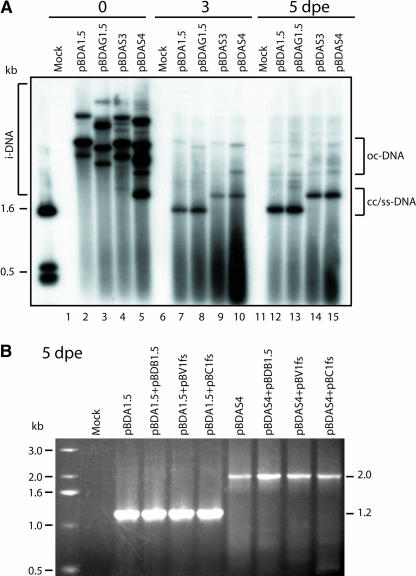
To identify the site(s) at which genome size reversion occurred, we next conducted protoplast replication experiments; this system allows for the assessment of the role of replication and/or nuclear-cytoplasmic transport in this process. Replication in protoplasts of the BDMV-A components, released from pBDA1.5, pBDAG1.5, pBDAS3, and pBDAS4, were examined by DNA gel blot hybridization analysis with a mixture of these recombinant plasmids as probe. As shown in Figure 3A, newly replicated viral DNA forms were detected 3 days after electroporation for all four constructs, and by 5 days after electroporation, the i-DNA was no longer detectable. The difference in size between the newly replicated components derived from pBDAS3 and pBDAS4 versus pBDA1.5 and pBDAG1.5 suggested that detectable levels of genome size reversion did not occur with the size-increased DNA-A component from either pBDAS3 or pBDAS4. Our finding that pBDAS3 failed to display evidence of genome size reversion in single cells demonstrates that the capacity for homologous recombination does not favor the production of size-reverted recombinant DNA-A components in this experimental system. These results demonstrate that the geminivirus replication complex can generate larger than genome-sized DNA; however, because the amount of the size-increased DNA components was reduced, compared with that of wild-type-sized BDMV-A and BDA-GFP, there appears to be an effect of genome size on the efficacy of replication.
Figure 3.
Viral Genome Size Reversion Was Not Detected in Protoplast Transfection Assays.
(A) DNA gel blot hybridization analysis of total nucleic acids extracted from protoplasts transfected with the indicated viral constructs at 0, 3, and 5 days after electroporation (dpe). The bracket at left delimits multiple forms of the uncut, double-stranded, circular-input plasmid DNA (i-DNA); the brackets at right indicate newly replicated open-circular (oc) and covalently closed (cc) double-stranded DNA/single-stranded (ss) DNA. Mock represents total nucleic acids extracted from protoplasts electroporated with buffer.
(B) PCR analysis with the size-reversion primer pair of total nucleic acids extracted from protoplasts transfected with either pBDA1.5 (wild-type BDMV-A) or the size-increased pBDAS4 alone or with the indicated DNA-B constructs (pBV1fs is a frameshift mutant of BDMV BV1, and pBC1fs is a frameshift mutant of BDMV BC1). For control experiments, note the presence of the expected 1.2-kb fragment amplified from wild-type DNA-A; for BDAS4 treatments, only the 2-kb fragment was amplified.
A role for nuclear-cytoplasmic transport of nascent viral DNA in the genome size-reversion process was explored through experiments in which pBDA1.5 or pBDAS4 was electroporated alone or coelectroporated with pBDB1.5 (to provide BV1 and BC1 movement proteins), pBV1fs (a BV1 frameshift mutant), or pBC1fs (a BC1 frameshift mutant). Experiments performed with wild-type BDMV-A (from pBDA1.5) served as the control (Figure 3B), because they demonstrated the presence of DNA fragments of the expected size (∼1.2 kb). Parallel studies conducted with the size-increased DNA-A component (from pBDAS4) revealed the presence of an ∼2.0-kb fragment for all treatments, irrespective of the presence of functional BV1 and/or BC1. Thus, we found no evidence for genome size reversion under these experimental conditions, a finding consistent with our DNA gel blot data (Figure 3A). These results suggested that the primary selection pressure for genome size reversion was not at the level of (1) viral replication, (2) BV1-mediated nuclear-cytoplasmic transport of nascent viral DNA, or (3) intracellular interactions between BV1, BC1, and the viral DNA.
Genome Size Reversion Detected at the Level of Cell-to-Cell Movement
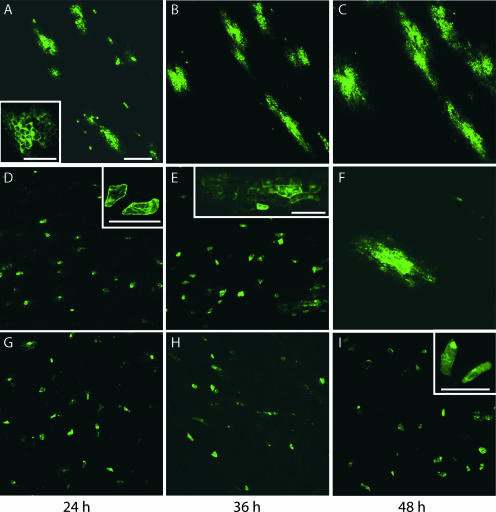
A role for the non-cell-autonomous RNA trafficking pathway as a determinant for genome size reversion was examined next. To this end, we used the loss of GFP fluorescence, which occurs as a consequence of genome size reversion, as a marker for this event. Here, it is important to stress that our previous experiments have demonstrated that in the bean hypocotyl system used for these studies, GFP serves as a cell-autonomous reporter (i.e., free GFP does not move cell to cell) (Sudarshana et al., 1998). Figure 4 illustrates the pattern of GFP fluorescence detected in epidermal cells of bean hypocotyls that were cobombarded with pBDAG1.5, pBDAS3, or pBDAS4 and pBDB1.5; tissues were examined 24, 36, and 48 h after bombardment. The pattern of BDMV-GFP (BDA-GFP plus BDMV-B) cell-to-cell movement, as revealed by GFP fluorescence, served as a control for these experiments. Significant viral replication and movement were detected by 24 h after bombardment (Figure 4A), as indicated by the presence of multicellular foci of epidermal cells in which GFP fluorescence was detected. These infection foci continued to expand until 48 h after bombardment (Figures 4B and 4C). Table 1 presents the quantitative data associated with these experiments and demonstrates that, by 48 h after bombardment, >50% of the infection foci showed extensive cell-to-cell movement. Infection sites that failed to expand with time presumably reflect the situation in which the bombarded cells received only the DNA-A component.
Figure 4.
Genome Size Reversion Occurs during the Process of Cell-to-Cell Movement.
(A) to (C) Epifluorescence images of bean hypocotyl epidermal cells collected after infection with BDA-GFP plus BDMV-B. Within 24 h (A), the GFP-tagged virus (wild-type genome size) displayed extensive cell-to-cell movement, as indicated by foci of cells containing GFP, and these sites of infection expanded with time (36 h [B] and 48 h [C]).
(D) to (F) Equivalent images collected after infection with size-increased BDAS3 plus BDMV-B. Note that size reversion, as revealed by the expanding foci of cells containing GFP, was detected by 36 h after infection (E).
(G) to (I) Equivalent images collected after infection with size-increased BDAS4 plus BDMV-B. Note that GFP fluorescence was confined predominantly to single cells.
Bar in (A) = 500 μm (common to all); bars in insets = 50 μm.
Table 1.
Viral Cell-to-Cell Movement Detected in Epidermal Cells of Bean Hypocotyls Coinoculated with the Indicated BDMV-A Constructs and BDMV-B
| Time Course
|
|||
|---|---|---|---|
| Input DNA a | 24 h after Bombardment | 36 h after Bombardment | 48 h after Bombardment |
| pBDAG1.5 + pBDB1.5 | 148/431 (34.3) b | 169/324 (52.1) | 63/108 (58.3) |
| pBDAS3 + pBDB1.5 | 19/386 (4.9) | 35/376 (9.3) | 36/327 (11.0) |
| pBDAS4 + pBDB1.5 | 46/616 (7.5) | 31/311 (10.0) | 25/332 (7.5) |
Recombinant plasmids were introduced into epidermal cells of bean hypocotyls by particle bombardment.
Values represent the number of foci at which GFP expression was not confined to single cells (numerator) and the total number of infection foci examined (denominator). The percentage of infection foci showing GFP expression not limited to single cells is shown in parentheses. For BDMV-GFP, foci expanded with time and by 48 h after bombardment were composed of >50 cells; for BDAS3, foci expanded as per BDMV-GFP; for BDAS4, foci did not expand with time and were limited to two to five epidermal cells.
The experimental system used to further elucidate the dynamics of the size-reversion process is illustrated in Figure 5A. Infected bean hypocotyl tissues were dissected surgically to separate the outer epidermal and cortical layers (infected tissues; identified based on GFP fluorescence) from the inner cortical and vascular tissues (noninfected cells). As illustrated in Figure 5B, DNA extracts prepared from these tissues were used in the PCR with the size-reversion primer pair, and the products were analyzed by gel electrophoresis and DNA gel blot hybridization with a mixture of pBDAG1.5, pBDAS3, and pBDAS4 probes. Consistent with the results presented in Figures 4A to 4C, in these control experiments only the ∼1.2-kb wild-type genome-sized fragment was amplified from tissues infected with BDMV-GFP. The identity of these fragments as being derived from these viral constructs was confirmed by DNA gel blot hybridization analysis.
Figure 5.
Dynamics of Genome Size Reversion Revealed by Analysis of Infected Bean Hypocotyl Tissue.
(A) Schematic illustration of the method by which epidermal/cortical tissues were collected for PCR analysis.
(B) The top gel shows PCR analysis with the size-reversion primer pair of total nucleic acids extracted from epidermal and cortical cells of hypocotyls inoculated with either pBDAG1.5 (BDA-GFP) or the size-increased pBDAS3/4, together with pBDB1.5. G, hypocotyls inoculated with gold particles only; W, control PCR conducted with water. The bottom gel shows DNA gel blot hybridization analysis of this gel probed with a mixture of pBDAG1.5, pBDAS3, and pBDAS4 under high-stringency conditions.
(C) Sequence analysis of PCR-amplified products illustrating the underlying nature of the size-reverted DNA-A components derived from pBDAS4 in epidermal/cortical cells of infected hypocotyls. CR, common region; hpi, hours post inoculation.
In experiments performed with pBDAS3, the construct optimized for size reversion (mediated by homologous recombination), the time course for cell-to-cell movement was delayed initially compared with that for BDMV-GFP (Figures 4A and 4D, Table 1). Detection of GFP was confined almost exclusively to single cells at 24 h after bombardment, likely reflecting both virus-associated gene expression and the inability of the viral genome to move through plasmodesmata. In the examples in which GFP was detected in foci, these were limited to two to three cells in nature. Examination of hypocotyls at 36 h after bombardment indicated the presence of clusters of GFP-expressing epidermal cells (Figure 4E, Table 1); some of these foci then continued to expand in a pattern similar to that observed for BDMV-GFP (Figures 4C and 4F). Such expanding foci likely reflected the generation of genome size-reverted DNA-A recombinant molecules that contained a functional GFP gene; molecular evidence for such revertants was provided previously by our analysis of BDMV-A components in systemically infected tissues (Figure 2C). The PCR data presented in Figure 5B provide further supporting evidence for the notion that size reversion occurs during cell-to-cell movement. Here, it is important to note the time-dependent progression from the size-increased to the size-reverted forms; at 24 h after bombardment, approximately equal amounts of size-increased and size-reverted forms were detected, whereas by 48 h after bombardment, the size-reverted form predominated. Furthermore, the products that emerged from this selection process, although varying slightly in size, converged toward that of the wild-type genome-sized fragment (∼1.2 kb). Together, these findings are fully consistent with the involvement of homologous recombination.
In the case of pBDAS4, which lacks the capacity for homologous recombination, size reversion to a functional DNA-A component would necessarily generate a preponderance of molecules with dysfunctional GFP open reading frames. Results in support of this hypothesis are presented in Figures 4G to 4I and Table 1 (where GFP detection was confined predominantly to single cells); additional supporting evidence is contained in the data presented in Figure 2D. In the rare cases in which GFP was observed in more than one cell, these foci did not increase with time and were restricted to two to five cells. PCR analysis of the DNA extracted from these infected tissues provided further insight into the mechanism underlying the selection process for this construct. In contrast to the results obtained for pBDAS3, the pBDAS4 size-increased fragment was detected for almost the entire period of the experiment (Figure 5B). In addition, although a similar pattern of PCR fragments was obtained from both pBDAS3 and pBDAS4 infected tissues, the convergence toward wild-type genome size appeared to be delayed in the pBDAS4 experiment.
To explore the derivation of movement-competent DNA-A components from pBDAS4, representative PCR-amplified fragments from hypocotyl tissues were sequenced and compared with those obtained from systemically infected tissues. In contrast to the uniformly sized fragments detected in systemically infected tissue (Figure 2D), fragments generated from infected hypocotyl tissues exhibited a wide range of sizes (Figure 5B). The complete absence of this range of fragments from systemically infected tissues further underscores the site and nature of the selection process. Whereas extensive nonhomologous recombination occurred in the infected epidermal cells, generating a wide spectrum of recombinant DNA-A components (Figure 5C), only the approximately genome-sized molecules were detected in the systemic tissues (Figure 2B). These results likely reflect selection acting on the wide spectrum of recombinant DNA-A components during cell-to-cell trafficking into the surrounding tissue, an event that occurred well before viral entry into the vascular system. Together, these results support the hypothesis that the process of cell-to-cell movement functions as the mechanism underlying genome size reversion.
Movement Protein Mutants Confirm the Requirement of Cell-to-Cell Trafficking for Genome Size Reversion
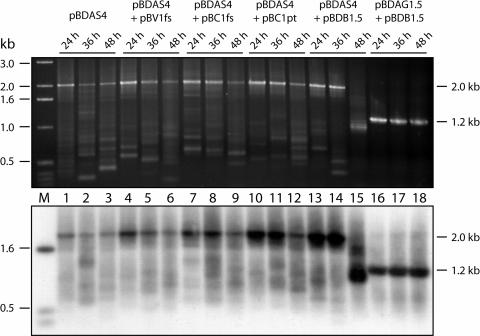
In previous studies, genetic, molecular, and cellular analyses revealed the essential roles for BV1 and BC1 in the cell-to-cell movement of the infectious form of BDMV (Noueiry et al., 1994; Sudarshana et al., 1998). Biochemical studies further demonstrated that both BV1 and BC1 recognize DNA on the basis of size, with high affinity for genome-sized molecules (Rojas et al., 1998). These findings were used to design experiments to test the hypothesis that functional BV1 and BC1 play essential roles in the mechanism of selectivity for the cell-to-cell trafficking of genome-sized recombinant DNA-A components. A control experiment was first performed in which bean hypocotyls were bombarded with pBDAS4 alone, and PCR and DNA gel blot analyses were conducted to establish the pattern of fragments associated with the recombinant DNA-A components generated during replication per se. The results displayed in Figure 6 demonstrate the presence, over the course of the experiment, of an ∼2.0-kb fragment that was derived from either the i-DNA or the release and replication of the size-increased component. Also apparent in these pBDAS4 data is the presence of numerous, smaller-sized fragments generated from recombinant DNA-A components produced during replication. Because these recombinant components were produced in the absence of BDMV movement proteins (i.e., BV1, BC1, and the BDMV CP, which has partial redundancy of BV1 function), they likely were generated in and retained within the nucleus of individual infected cells.
Figure 6.
Functional Cell-to-Cell Movement Protein (BC1) and Nuclear Export Factor (BV1) Are Essential for the Selection and Spread of Size-Reverted DNA-A.
PCR analysis with the size-reversion primer pair of total nucleic acids extracted from epidermal and cortical cells of hypocotyls inoculated with the indicated plasmids. pBV1fs is a frameshift mutant of BDMV BV1, pBC1fs is a frameshift mutant of BDMV BC1, and pBC1pt is a point mutation in BC1 (Asp-78) that renders BC1 dysfunctional (Noueiry et al., 1994). M, molecular mass marker.
The requirement for both BV1 and BC1 was next investigated using a series of BDMV-B mutants. Analysis of hypocotyls cobombarded with pBDAS4 and pBV1fs (which lacks BV1 but produces a functional BC1) showed a pattern of fragments equivalent to that observed for pBDAS4 alone. Parallel experiments were performed with pBDAS4 and either pBC1fs or pBC1pt; these mutants produce functional BV1 and either lack BC1 or produce a dysfunctional BC1, respectively. In both cases, the PCR fragment patterns observed were equivalent to those obtained with pBDAS4 alone and pBDAS4 plus pBV1fs (Figure 6). By contrast, when pBDAS4 was cobombarded with pBDB1.5 (which generates wild-type BDMV-B that provides wild-type BV1 and BC1), a different fragment pattern emerged during the course of the experiment. In this case, by 48 h after bombardment, the size-increased fragment had disappeared, and the sizes of the resulting fragments had converged on the expected genome-sized fragment of ∼1.2 kb. Lastly, in the control experiment performed using pBDAG1.5 and pBDB1.5, only the genome-sized fragment was detected, consistent with the release and replication of the wild-type-sized DNA-A component. Together, these experiments provided support for the hypothesis that both BV1 and BC1 are required for the selection and cell-to-cell movement of genome-sized recombinant DNA-A components.
BC1 Preferentially Mediates the Cell-to-Cell Movement of Genome-Sized DNA
Microinjection experiments were next performed to determine whether BC1 exhibits a preference in trafficking for genome-sized DNA. Circular double-stranded DNA molecules of various sizes were introduced into mesophyll cells of mature Nicotiana benthamiana leaves (Rojas et al., 1997). This form of DNA was chosen because a number of lines of evidence suggest that it represents an infectious form with the capacity to move cell to cell through plasmodesmata (Noueiry et al., 1994; Rojas et al., 1998). The results from these experiments are presented in Table 2. Control microinjection experiments were first performed to establish the functional nature of purified recombinant BC1. Consistent with our previous studies (Noueiry et al., 1994), BC1 mediated both its own cell-to-cell movement and the diffusion of 10-kD F-dextran through the dilated plasmodesmata microchannels. As expected, in these tissues, when injected alone, 10-kD F-dextran remained confined to the target cell (Table 2).
Table 2.
BDMV BC1 Mediates Cell-to-Cell Movement of DNA in a Size-Dependent Manner
| Cell-to-Cell Movement c
|
|||||
|---|---|---|---|---|---|
| Injected Agent a | Coinjected Agent | Size b (kb) | Total | Movement | Extent |
| 10-kD F-dextran | – d | – | 12 | 0 | 0 |
| BC1 | 10-kD F-dextran | – | 13 | 13 | 3 to 4 |
| BC1-OG | – | – | 10 | 10 | 3 to 4 |
| pSP72 | – | 2.5 | 12 | 0 | 0 |
| pSP72 | BC1 | 2.5 | 16 | 16 | 3 to 4 |
| pKS | – | 3.0 | 12 | 0 | 0 |
| pKS | BC1 | 3.0 | 12 | 12 | 2 to 4 |
| pSP64-V1 | – | 3.4 | 12 | 0 | 0 |
| pSP64-V1 | BC1 | 3.4 | 22 | 20 | 1 |
| pBDA | – | 5.5 | 11 | 0 | 0 |
| pBDA | BC1 | 5.5 | 17 | 4 | 1 |
| pCGN1547 | – | 14.4 | 10 | 0 | 0 |
| pCGN1547 | BC1 | 14.4 | 14 | 0 | 0 |
Details for injected and coinjected agents are as follows: 10-kD F-dextran, fluorescein isothiocyanate–labeled 10-kD dextran (1.0 mM); BC1 and BC1-OG, recombinant protein and Oregon Green–labeled BC1 (both at 2 μg/μL). All DNA probes were labeled with the fluorescent dye TOTO-1 and used at a concentration of 50 ng/μL.
Approximate size of DNA molecules used in microinjection experiments.
Data are presented as total number of injections, number of injections in which the probe moved into the surrounding mesophyll cells, and the extent of movement, as indicated by the number of cells through which the probe moved out from the injected cell.
–, Not applicable.
A series of plasmid DNA molecules of different sizes (circular and double stranded) were labeled with the fluorescent dye TOTO-1 or Cy3 (Noueiry et al., 1994; Lough et al., 1998) and then used in microinjection experiments. When injected alone, these labeled DNA molecules remained in the target cell (Table 2). Coinjection of unlabeled BC1 with pSP72 (2.5 kb) resulted in extensive cell-to-cell movement of the DNA in a manner equivalent to that observed with BC1-OG or unlabeled BC1 plus 10-kD F-dextran. A similar result was obtained when pBluescript KS+ (3.0 kb) was introduced into mesophyll cells together with BC1. Interestingly, the capacity of BC1 to mediate the movement of pSP64-V1 (3.4 kb) was reduced in terms of both efficacy and extent, suggesting that BC1 has a preference for DNA molecules that fall within the 2.5- to 3.0-kb size class. This notion was supported by the finding that BC1 was highly inefficient at mediating the movement of pBDA (5.5 kb) and incapable of trafficking pCGN1547 (14.4 kb; Table 2). These results provide strong support for the hypothesis that BC1 preferentially recognizes, binds, and traffics genome-sized DNA.
DISCUSSION
The aim of the present study was to test the hypothesis that the fundamental principle responsible for the strict maintenance of geminiviral genome (DNA) size reflects the functional limitations imposed by the plant's endogenous RNA trafficking pathway. Our findings provide strong support for this concept, because we demonstrated that the selection process for viral genome size occurred during the initial cell-to-cell movement of the infectious DNA. The identification of this site was accomplished through the examination of each step in the infection process using a combination of genetic, molecular, and cellular approaches.
Consistent with previous studies with other bipartite geminiviruses (Etessami et al., 1989; Hayes et al., 1989; Klinkenberg et al., 1989; Elmer and Rogers, 1990), during the establishment of systemic infection, the size-increased BDMV-A components underwent genome size reversion. Examination of the molecular nature and size of the revertants revealed the presence of viral DNA components that fell within a very narrow size range (Figure 2). This result clearly underscores the action of a precise mechanism involved in genome size reversion. Sequence analysis provided insight into the genetic mechanism by which these revertants were generated; this involved both homologous and nonhomologous recombination as well as template switching between the DNA-A and DNA-B components (Figures 2C and 2D). These results are consistent with previous reports on the mechanism of geminivirus recombination (Etessami et al., 1989; Elmer and Rogers, 1990; Stanley, 1991; Azzam et al., 1994). Furthermore, the detection of plasmid sequences in some revertants indicates that this process likely occurred in the inoculated cells before or during the replicational release of the viral DNA component from the input plasmids (Figures 1 and 2). These results implicated either replication or nuclear-cytoplasmic export as a potential mechanism for the selection of size-reverted viral genomes.
Indubitably, replication must play a central role in the process of genome size reversion through the generation of revertants. However, the critical question regards the extent to which replication contributes to the selection of genome-sized revertants. Experiments conducted using both protoplast (cell) and epidermal (cellular) assays provided evidence that genome-sized revertants were not replicated preferentially at the single cell level. Inspection of the data from our protoplast assays (Figure 3) demonstrated that the size-increased DNA-A components (BDAS3 and BDAS4) remained unaltered in size during the course of these experiments (5 days after electroporation). Compelling evidence that replication per se does not preferentially generate size revertants in protoplasts was provided by PCR analysis in which we were unable to detect evidence of size reversion (Figure 3B). The fact that reversion was not detected even in the case of pBDAS3, a construct optimized for homologous recombination, strengthens the notion that revertants must be present at levels undetectable by PCR analysis. Collectively, these results establish the fact that the viral replication machinery can replicate these size-increased components and, furthermore, that this process does not select for genome-sized revertants. These findings are consistent with previous studies indicating that size-increased geminivirus constructs replicate in protoplasts or cell cultures without undergoing size reversion (Stanley, 1991; Palmer et al., 1999).
BDMV BV1 plays a pivotal role in the infection process by mediating the export of nascent viral DNA to the cytoplasm: BV1 mutants are defective in both nuclear export (Noueiry et al., 1994) and cell-to-cell movement of the infectious DNA (Sudarshana et al., 1998). In protoplast assays, size reversion was not detected in the presence of BV1 alone or BV1 plus BC1 or in the absence of BV1 (Figure 3B). Thus, nuclear-cytoplasmic transport is not the step at which genome-sized revertants are selected.
Two steps of the infection process remain at which the selection of genome-sized revertants could occur: cell-to-cell movement or long-distance transport in the phloem. A test for the role of cell-to-cell movement in this process was provided by our studies conducted with epidermal/cortical cells. Here, the wild-type genome-sized BDA-GFP served as the control to establish both the time frame for viral cell-to-cell movement (Figures 4A to 4C) and the nature of the PCR-amplified fragments associated with the infection process (Figure 5B). In this context, limited/delayed cell-to-cell movement occurred for both pBDAS3 and pBDAS4, as reflected by the limited detection of GFP signal (Figure 4, Table 1); this finding implicated cell-to-cell movement as the initial site for size reversion. We further examined the size reversion process by extraction and analysis of DNA obtained from these same tissues (Figure 5B), which revealed the time frame in which revertants were generated. Unlike with the control, a wide range of recombinant DNA-A components were observed from the earliest time point (Figure 5), suggesting that recombination occurs by a size-independent process. Interestingly, this range of revertants was not detected in the protoplast assay, even at 5 days after electroporation, demonstrating an important influence of the status of the inoculated cell on recombination.
The eventual establishment of infection foci equivalent to the wild-type by pBDAS3-derived components was highly correlated with the appearance and accumulation of genome-sized fragments. These findings are fully consistent with the involvement of homologous recombination in the generation of a BDA-GFP–like component (Figure 2). The relatively low number of these infection foci, compared with those established by pBDAG1.5, likely reflects the generation of revertants lacking a functional GFP open reading frame (i.e., products of nonhomologous recombination) (Figures 2C and 4F, Table 1). Additionally, the wide range of non-genome-sized revertants also may serve to sequester the BDMV movement proteins and/or to form dysfunctional movement complexes that interfere with the cell-to-cell movement of genome-sized complexes.
Viral genetics provided a powerful tool to further test the hypothesis that genome size reversion occurs at the level of cell-to-cell movement and requires functional BC1. Selection for genome-sized revertants within epidermal/cortical cells did not occur with BDAS4 alone or in the presence of either BC1 or BV1 (Figure 6). However, replication of BDAS4 in the presence of BDMV-B (providing both BV1 and BC1) resulted in the selection of genome-sized revertants. The need for a movement-competent BC1 in this selection process (Figure 6) further confirmed that cell-to-cell movement represents the pivotal step in genome size reversion. In this regard, it is important to note that size reversion also requires functional BV1, because this viral movement protein is essential for the export of nascent DNA into the cytoplasm. However, BV1 is not the primary factor responsible for the selection of genome-sized revertants, because it binds and facilitates the nuclear export of greater than genome-sized molecules (Noueiry et al., 1994; Rojas et al., 1998). Furthermore, because size reversion did not occur in the presence of the movement-defective BC1 point mutant, it is unlikely that these findings could be explained by a defect in BV1–BC1 interaction.
Microinjection-based studies provided another line of evidence for the role of BC1 in the size-selection process. These data provided direct evidence that BC1 mediates the efficient cell-to-cell movement of DNA that falls within a narrow size range mapping to wild-type BDMV genome size (Table 2). Even though BC1 can bind to 3.4- and 5.5-kb DNA (Rojas et al., 1998), its capacity to mediate the movement of these molecules is limited because they were detected only in cell(s) adjacent to the target cell. Such limited movement provides an explanation for the observed three- to five-cell foci detected in BDAS4 experiments (Table 1). Alternatively, this finding may reflect the limited movement of non-replication-competent genome-sized recombinant DNA-A components containing 35S-GFP.
These studies provide us with a foundation to develop a model for the manner in which geminiviruses evolved to use (1) the endogenous scavenging system involved in the identification and trafficking of DNA from the cytoplasm to nucleus and (2) the host cell-to-cell trafficking pathway on which plant non-cell-autonomous RNA molecules traffic. In our model, DNA typically is excluded from entry into this non-cell-autonomous pathway. The selection process likely reflects differences in nucleic acid structure and/or the size of the macromolecule. In an evolutionary context, this screening system may have evolved to prevent the movement of small chromosomal fragments that are present in the cytoplasm generated during cell division. After reconstitution of the nuclear envelope, any such endogenous DNA fragments would be recognized by the scavenging system and returned to the nucleus. Plant DNA viruses use this cytoplasmic DNA recognition system to facilitate the nuclear accumulation and subsequent replication of their genomes and either avoided (tubule-forming DNA viruses) or evolved a mechanism that permits viral DNA to access the non-cell-autonomous RNA trafficking pathway. This process of cell-to-cell movement required the evolution of specialized movement proteins to facilitate DNA entry into this plasmodesmata pathway.
During the course of the evolution of this capacity to infect plants, it is clear that limitations were imposed on geminiviral genome size. The question to be addressed now concerns the underlying process(es) that exerted this size selection. Our results, based on BDMV, allow us to discount a direct role for replication and nuclear-cytoplasmic transport in this process. Several independent lines of evidence established that the process of cell-to-cell movement imposed an intense selection on the size of DNA molecules that could move through plasmodesmata. Here, the essential role of BC1 in this process presumably reflects a requirement for compatibility between the nature and size of the BC1-DNA complex and the limitation(s) imposed by the plasmodesmata.
The scenario described below is advanced to explain the events that underlie geminiviral genome size reversion. Within the inoculated cell, a wide range of recombinant molecules is generated in the nucleus, and these molecules are exported to the cytoplasm through the action of BV1. The DNA binding capacity of BC1 allows it to recognize and traffic molecules in the 2.5- to 5.5-kb range. Smaller molecules (<2 kb) do not form stable BC1-DNA complexes (Rojas et al., 1998) and thus are subjected to the endogenous DNA recognition system. Large DNA molecules (>5.5 kb) form a complex whose dimensions preclude passage through the tortuous pathway presented by the microchannels of the plasmodesmata (Lucas and Gilbertson, 1994). The corollary to this condition is that, as with any size-fractionation process, although a range of molecules can passage (2.5 to 5.5 kb), there must exist an optimal size class (∼2.5 to 3.0 kb) that is trafficked efficiently by BC1. Thus, the combined properties of BC1 and plasmodesmata ensure the progressive selection of recombinant DNA-A components that converge on the genome size. This selection process likely is accomplished during one to two cycles of replication and movement into adjacent cells, resulting in the enrichment of genome-sized revertants; this is followed by the rapid systemic spread of these revertant(s).
Our studies provide a plausible explanation for why viral evolution has led to a relatively limited diversity of plant DNA viral types. The underlying principle for this phenomenon appears to be based on the existence of an endogenous system that operates to ensure that DNA acts in a cell-autonomous manner. Analysis of the manner in which size-increased components underwent reversion yielded important insight into the complexity of the interaction between the virus and the host cells. The absence of revertants in transfected protoplasts demonstrated that this system lacked an essential endogenous component(s) required for BDMV to engage in recombination. This missing component(s) clearly is present in the context of the bean epidermal/cortical tissue. Identification of this component(s) would provide a valuable tool to further our studies of recombination and geminivirus evolution. Finally, a basic understanding of how biological systems evolved the capacity to regulate the non-cell-autonomous movement of nucleic acids will provide new insight into the viral evolutionary process and may lead to the development of novel approaches for the control of invasive nucleic acid species.
METHODS
Viral Clones and DNAs
The recombinant plasmids pBDA1, pBDA1.5, pBDB1.5, pBDAG1.5 (formerly named pBDMVA-mGFP1.5 [Sudarshana et al., 1998]), pBV1fs, and pBC1fs, which contained various full-length clones of BDMV DNA-A (BDMV-A; 2.6 kb) and DNA-B (2.5 kb) components, were as described previously (Gilbertson et al., 1991; Noueiry et al., 1994; Hou et al., 1998). The plasmid vectors pBluescript KS+ (pKS; Stratagene, La Jolla, CA) and pCGN1547 (McBride and Summerfelt, 1990) were used in microinjection studies.
Two size-increased BDMV-A constructs, pBDAS3 and pBDAS4, were generated through the deletion of the CP and its promoter; both constructs would yield recombinant DNA-A components of 3.4 kb (Figure 1). To engineer these constructs, a HindIII-EcoRI fragment, containing the 35S:mGFP4 gene and the nopaline synthase 3′ terminator, was excised from pBIN-mGFP4 (Haseloff et al., 1997) and ligated into HindIII-EcoRI–digested pKS. The resulting clone was first digested with ApaI-EcoRI, and the 1.8-kb fragment was ligated with a pBDA1.5-derived 635-bp BglII-ApaI fragment (containing the common region and flanking sequences) into pKS digested with BamHI-EcoRI. Finally, a 0.6-kb ClaI fragment from this clone was replaced by a ClaI BDMV-A monomer excised from pBDAG1.5 to generate pBDAS3, a multimeric infectious clone having a SacI site as a marker within the partial mGFP4 gene (Figure 1A). Plasmid pBDAS3 was digested partially with ApaI to remove an ∼0.45-kb fragment containing the partial mGFP4 gene and religated to generate pBDAS4 (Figure 1A).
Particle Bombardment of Bean Seedlings with Plasmid DNAs
All plasmid DNAs were prepared with the Plasmid Mega Kit (Qiagen, Chatsworth, CA) or the Quantum Prep Plasmid Midiprep Kit (Bio-Rad, Hercules, CA). Common bean (Phaseolus vulgaris cv Topcrop) seedlings were bombarded with gold particles coated with various DNA preparations with a PDS S-1000 particle acceleration device (DuPont, Wilmington, DE) as described (Sudarshana et al., 1998). Inoculated seedlings were either maintained (48 h) on moist filter paper in Petri dishes in darkness or planted in soil and grown in a controlled-environment chamber (Conviron PGR15; 250 μmol·m−2·s−1, 16-h photoperiod, 30°C day/22°C night, RH 60%).
Analysis of Plants Infected with BDMV Constructs
Plants were observed for symptom development from 10 to 15 days after bombardment. For PCR analysis, DNA was extracted from the youngest trifoliolate leaves according to Rojas et al. (1993). A BDMV-A fragment was amplified with the primers OBDAV2550 (5′-AGTAAG- AGAGCACTGTGGA-3′) and OBDAC1160 (5′-GATTCGCAACAGACAGAT-3′), hereafter referred to as the size-reversion primer pair. PCR parameters were as follows: 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 3 min, followed by 72°C for 10 min with eLONGase (Life Technologies, Rockville, MD). PCR products were purified using the Qiaquick PCR Purification Kit (Qiagen, Valencia, CA) and sequenced using the size-reversion primers. Sequence analyses were performed using the Basic Local Alignment Search Tool (BLAST) program. Detection of GFP at the whole-plant level was performed with a hand-held, long-wavelength UV lamp (UVP, Upland, CA). At the tissue/cellular level, GFP was detected with a Nikon Optiphot-2 microscope (Tokyo, Japan) (Sudarshana et al., 1998).
Protoplast Assay for the Size Reversion of BDMV DNA-A Constructs
Suspension cultured cells from tobacco (Nicotiana tabacum cv Xanthi nc) were used for the preparation of protoplasts (Hou et al., 1998). For each construct tested, 10 μg of DNA was electroporated into 3 × 106 protoplasts at 290 V and 490 μF for 8 ms (Hou et al., 1998). Transfected protoplasts were incubated at 25°C in darkness, and aliquots were removed at 0, 3, and 5 days after electroporation. Total genomic DNA was extracted from protoplasts/cells and analyzed by DNA gel blot hybridization with a 32P-labeled probe (a mixture of pBDA1.5, pBDAG1.5, pBDAS3, and pBDAS4) under high-stringency conditions (Hou et al., 1998) or by PCR with the size-reversion primer pair.
Hypocotyl Assay for the Size Reversion of BDMV DNA-A Constructs
Bean hypocotyls were bombarded with the appropriate plasmid DNA using the method described above, and tissue was examined at 24, 36, and 48 h after bombardment. Fluorescence analysis for GFP expression was performed with a Bio-Rad upright confocal laser scanning microscope (model MRC1024), and regions containing GFP were excised with a razor blade such that only epidermal and cortical cells were obtained. Total genomic DNA was extracted, PCR was conducted with the size-reversion primer pair, and then DNA gel blot hybridization analysis was performed with a 32P-labeled probe (Figure 5: pBDAG1.5, pBDAS3, and pBDAS4; Figure 6: pBDAG1.5 and pBDAS4). PCR-amplified DNA fragments were cloned using the TOPO-TA Cloning Kit (Invitrogen, Carlsbad, CA) and sequenced.
Protein Preparation, Microinjection, and Confocal Laser Scanning Microscopy
BDMV BV1 and BC1 were expressed in and purified from Escherichia coli (Noueiry et al., 1994), renatured, labeled with Oregon Green (Molecular Probes, Eugene, OR), and then used in microinjection studies (Rojas et al., 1997) that were performed on detached, mature leaves of Nicotiana benthamiana. Movement of the fluorescent probes out of target mesophyll cells was observed and recorded using a Leica confocal laser scanning microscope (model TCS-4D; Heidelberg, Germany) equipped with a Narishige four-dimensional hydraulic micromanipulator system (Noueiry et al., 1994).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Maria R. Rojas, mrrojas@ucdavis.edu.
Acknowledgments
This research was supported in part by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (9500838 and 2002-01418 to R.L.G. and W.J.L.) and the National Science Foundation (IBN 99-00539 and IBN 03-15174).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015057.
References
- Azzam, O., Frazer, J., de la Rosa, D., Beaver, J.S., Ahlquist, P., and Maxwell, D.P. (1994). Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology 204, 289–296. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Kasschau, K.D., Mahajan, S.K., and Schaad, M.C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom, C.M., Lapidot, M., and Beachy, R.N. (1992). Plant virus movement proteins. Cell 69, 221–224. [DOI] [PubMed] [Google Scholar]
- Elmer, S., and Rogers, S.G. (1990). Selection for wild type size derivatives of tomato golden mosaic virus during systemic infection. Nucleic Acids Res. 18, 2001–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etessami, P., Watts, J., and Stanley, J. (1989). Size reversion of African cassava mosaic virus coat protein deletion mutants during infection of Nicotiana benthamiana. J. Gen. Virol. 70, 277–289. [DOI] [PubMed] [Google Scholar]
- Fagard, M., and Vaucheret, H. (2000). (Trans)gene silencing in plants: How many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167–194. [DOI] [PubMed] [Google Scholar]
- Fire, A. (1999). RNA-triggered gene silencing. Trends Genet. 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S.Q., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T., Giesman-Cookmeyer, D., Ding, B., Lommel, S.A., and Lucas, W.J. (1993). Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the red clover necrotic mosaic virus movement protein. Plant Cell 5, 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Ramirez, E.R., Sudarshana, M.R., Lucas, W.J., and Gilbertson, R.L. (2000). Bean dwarf mosaic virus BV1 protein is a determinant of the hypersensitive response and avirulence in Phaseolus vulgaris. Mol. Plant-Microbe Interact. 13, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Ghoshroy, S., Lartey, R., Sheng, J., and Citovsky, V. (1997). Transport of protein and nucleic acids through plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 27–50. [DOI] [PubMed] [Google Scholar]
- Gilbertson, R.L., Faria, J.C., Hanson, S.F., Morales, F.J., Ahlquist, P., Maxwell, D.P., and Russell, D.R. (1991). Cloning of the complete DNA genomes of four bean-infecting geminiviruses and determining their infectivity by electric-discharge particle-acceleration. Phytopathology 81, 980–985. [Google Scholar]
- Gilbertson, R.L., and Lucas, W.J. (1996). How do plant viruses traffic on the ‘vascular highway’? Trends Plant Sci. 1, 260–268. [Google Scholar]
- Hanley-Bowdoin, L., Settlage, S.B., Orozco, B.M., Nagar, S., and Robertson, D. (1999). Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18, 71–106. [PubMed] [Google Scholar]
- Hannon, G.J. (2002). RNA interference. Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, R.J., Coutts, R.H.A., and Buck, K.W. (1989). Stability and expression of bacterial genes in replicating geminivirus vectors in plants. Nucleic Acids Res. 17, 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.), S303.–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn, T. (1999). Plant pararetroviruses–caulimoviruses: molecular biology. In Encyclopedia of Virology, 2nd ed., A. Granoff and R.G. Webster, eds (San Diego, CA: Academic Press), pp. 1281–1285.
- Hou, Y.M., Paplomatas, E.J., and Gilbertson, R.L. (1998). Host adaptation and replication properties of two bipartite geminiviruses and their pseudorecombinants. Mol. Plant-Microbe Interact. 11, 208–217. [Google Scholar]
- Hull, R. (2002). Matthews' Plant Virology, 4th ed. (San Diego, CA: Academic Press).
- Jorgensen, R.A. (2002). RNA traffics information systemically in plants. Proc. Natl. Acad. Sci. USA 99, 11561–11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L.S., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279, 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Yuan, Z.A., Cilia, M., Khalfan-Jagani, Z., and Jackson, D. (2002). Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4103–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. [DOI] [PubMed] [Google Scholar]
- Klinkenberg, F.A., Ellwood, S., and Stanley, J. (1989). Fate of African cassava mosaic virus coat protein deletion mutants after agroinoculation. J. Gen. Virol. 70, 1837–1844. [DOI] [PubMed] [Google Scholar]
- Lazarowitz, S.G., and Beachy, R.N. (1999). Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., Rojas, M.R., Gomez-Ospina, N., Staehelin, L.A., and Lucas, W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299, 392–396. [DOI] [PubMed] [Google Scholar]
- Lindbo, J.A., Silvarosales, L., Proebsting, W.M., and Dougherty, W.G. (1993). Induction of a highly specific antiviral state in transgenic plants: Implications for regulation of gene-expression and virus-resistance. Plant Cell 5, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstead, P.J., Hills, G.J., Plaskitt, K.A., Wilson, I.G., Harker, C.L., and Maule, A.J. (1988). The subcellular location of the gene 1 product of cauliflower mosaic virus is consistent with a function associated with virus spread. J. Gen. Virol. 69, 1809–1818. [Google Scholar]
- Lomonossoff, G.P., and Shanks, M. (1999). Comoviruses (Comoviridae). In Encyclopedia of Virology, 2nd ed., A. Granoff and R.G. Webster, eds (San Diego, CA: Academic Press), pp. 285–291.
- Lough, T.J., Shash, K., Xoconostle-Cázares, B., Hofstra, K.R., Beck, D.L., Balmori, E., Forster, R.L.S., and Lucas, W.J. (1998). Molecular dissection of the mechanism by which potexvirus triple gene block proteins mediate cell-to-cell transport of infectious RNA. Mol. Plant-Microbe Interact. 11, 801–814. [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 and its mRNA through plant plasmodesmata. Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Ding, B., and van der Schoot, C. (1993). Plasmodesmata and the supracellular nature of plants. New Phytol. 125, 435–476. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., and Gilbertson, R.L. (1994). Plasmodesmata in relation to viral movement within leaf tissues. Annu. Rev. Phytopathol. 32, 387–411. [Google Scholar]
- Lucas, W.J., Yoo, B.-C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2, 849–857. [DOI] [PubMed] [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14, 269–276. [DOI] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311. [DOI] [PubMed] [Google Scholar]
- Noueiry, A.O., Lucas, W.J., and Gilbertson, R.L. (1994). Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76, 925–932. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, K.E., Thomson, J.A., and Rybicki, E.P. (1999). Generation of maize cell lines containing autonomously replicating maize streak virus-based gene vectors. Arch. Virol. 144, 1345–1360. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C., Schmit, A.C., Michler, P., Stussi-Garaud, C., and Pinck, L. (1995). Grapevine fanleaf nepovirus P38 putative movement protein is located on tubules in-vivo. Mol. Plant-Microbe Interact. 8, 379–387. [Google Scholar]
- Rojas, M.R., Gilbertson, R.L., Russell, D.R., and Maxwell, D.P. (1993). Use of degenerate primers in the polymerase chain-reaction to detect whitefly-transmitted geminiviruses. Plant Dis. 77, 340–347. [Google Scholar]
- Rojas, M.R., Noueiry, A.O., Lucas, W.J., and Gilbertson, R.L. (1998). Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form- and size-specific manner. Cell 95, 105–113. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R., Zerbini, F.M., Allison, R.F., Gilbertson, R.L., and Lucas, W.J. (1997). Capsid protein and helper component proteinase function as potyvirus cell-to-cell movement proteins. Virology 237, 283–295. [DOI] [PubMed] [Google Scholar]
- Rothnie, H.M., Chapdelaine, Y., and Hohn, T. (1994). Pararetroviruses and retroviruses: A comparative review of viral structure and gene expression strategies. Adv. Virus Res. 44, 1–67. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T., Voinnet, O., and Baulcombe, D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cázares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Stanley, J. (1991). The molecular determinants of geminivirus pathogenesis. Semin. Virol. 2, 139–149. [Google Scholar]
- Storms, M.M.H., Kormelink, R., Peters, D., van Lent, J.W.M., and Goldbach, R.W. (1995). The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology 214, 485–493. [DOI] [PubMed] [Google Scholar]
- Sudarshana, M.R., Wang, H.L., Lucas, W.J., and Gilbertson, R.L. (1998). Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol. Plant-Microbe Interact. 11, 277–291. [Google Scholar]
- Timmermans, M.C.P., Das, O.P., and Messing, J. (1994). Geminiviruses and their uses as extrachromosomal replicons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 79–112. [Google Scholar]
- Timmons, L., Court, D.L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. [DOI] [PubMed] [Google Scholar]
- Vance, V., and Vaucheret, H. (2001). RNA silencing in plants: Defense and counterdefense. Science 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- van Lent, J., Storms, M., van der Meer, F., Wellink, J., and Goldbach, R. (1991). Tubular structures involved in movement of cowpea mosaic virus are also formed in infected cowpea protoplasts. J. Gen. Virol. 72, 2615–2623. [DOI] [PubMed] [Google Scholar]
- van Regenmortel, M.H.V., Fauquet, C.M., Bishop, D.H.L., Carstens, E.B., Estes, M.K., Lemon, S.M., Maniloff, J., Mayo, M.A., McGeoch, D.J., Pringle, C.R., and Wickner, R.B. (2000). Virus Taxonomy: Classification and Nomenclature of Viruses. Seventh Report of the International Committee on Taxonomy of Viruses (San Diego, CA: Academic Press).
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wang, H.L., Sudarshana, M.R., Gilbertson, R.L., and Lucas, W.J. (1999). Analysis of cell-to-cell and long-distance movement of a bean dwarf mosaic geminivirus-green fluorescent protein reporter in host and nonhost species: Identification of sites of resistance. Mol. Plant-Microbe Interact. 12, 345–355. [Google Scholar]
- Winston, W.M., Molodowitch, C., and Hunter, C.P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459. [DOI] [PubMed] [Google Scholar]
- Wu, X., Weigel, D., and Wigge, P.A. (2002). Signaling in plants by intercellular RNA and protein movement. Genes Dev. 16, 151–158. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.-L., Monzer, J., Yoo, B.-C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283, 94–98. [DOI] [PubMed] [Google Scholar]
- Zambryski, P., and Crawford, K. (2000). Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16, 393–421. [DOI] [PubMed] [Google Scholar]