Abstract
Our previous studies showed that a member of the YABBY gene family, FILAMENTOUS FLOWER (FIL), plays a role in specifying the abaxial side tissues in the development of lateral organs such as cotyledons, leaves, young flower buds, and flower organs. We examined the expression pattern of FIL and found a temporal change of expression domains in the developmental process of the floral meristem. We also examined the cis control regions by constructing a series of transgenic plants that carry green fluorescent protein under the control of the FIL promoter with several types of deletions, base changes, and tandem repeats and showed that the unique expression pattern is dependent on at least two cis-acting elements in the 5′ regulatory region. One element proximal to the FIL gene would be responsible for the expression of both the abaxial and adaxial sides, and the other element of the 12-bp sequence would work to repress expression on the adaxial side.
INTRODUCTION
Morphogenesis, in which axes are produced in multicellular organisms, is a basic strategy for their construction. In studying the development of organisms, axes generally are observed from early embryonic stages, and these axes basically determine the body pattern (Martinez-Arias and Lawrence, 1985). In early Drosophila embryos, the stem cells destined to be the head, breast, and abdomen are located at their correct positions according to the anterior-posterior and dorsal-ventral axes (St. Johnston and Nüsslein-Volhard, 1992). In the early embryogenesis of Arabidopsis, populations of stem cells are situated at the poles of the apical-basal axis. The apical stem cells located at the shoot meristem produce the aerial body, whereas basal stem cells at the root meristem generate the roots. If development along these axes becomes disordered during embryogenesis, the morphology of the multicellular body is completely destroyed and chaotic. Therefore, studying the axes of multicellular organisms is important to developmental biologists.
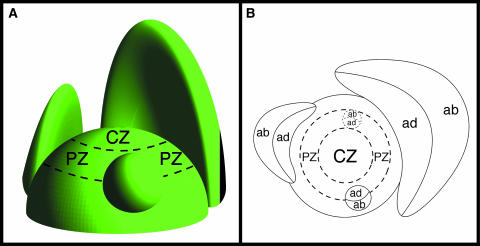
The adaxial-abaxial axis that is established determines the developmental pattern for generating leaves, flowers, and floral organs in the lateral regions of the plant shoot apex. The adaxial side of the organs is that near the stem apex (from a lateral view of the plant), and the side away from the apex is the abaxial side (Figure 1). For example, the adaxial surface is the top side of the leaf and the abaxial surface is the under side. Morphogenic analysis revealed that dome-shaped leaf primordia emerge at the peripheral zone of the shoot apex and that they enlarge gradually along an axis that is perpendicular to their adaxial-abaxial axis (Figure 1B). As their development continues, they become flattened, and their juxtaposition around the apex maintains the adaxial-abaxial polarity of the shoot. The leaves then grow larger in breadth and length, their expanse facilitating the interception of sunlight.
Figure 1.
Scheme of a Shoot Apical Meristem.
A leaf primordium is formed in the peripheral zone. The primordium emerges as a small dome and develops into flat leaves. The abaxial and adaxial sides are distinguished in the primordia. ab, abaxial side; ad, adaxial side; CZ, central zone; PZ, peripheral zone.
(A) Side view.
(B) Top view.
The Antirrhinum phantastica (phan) mutant produces filamentous leaves covered with abaxial leaf epidermis; however, ectopic patches of abaxial tissue are produced on the adaxial surface of leaves and petals (Waites and Hudson, 1995). Conversely, the dominant mutants Arabidopsis phabulosa-1d (phb-1d) and phavoluta (phv) generate filamentous leaves covered with an epidermis that has adaxial features (McConnell and Barton, 1998; McConnell et al., 2001). The Arabidopsis pinhead/zwille (pnh/zll) mutant, which appeared to have defects in the regulation of cells in the shoot apex, produced symmetric filamentous leaves. An expression analysis of PNH detected PNH mRNA in cells on the adaxial side of the leaf and in the vascular bundles (Moussian et al., 1998; Lynn et al., 1999). The Arabidopsis SERRATE (SE) gene carries a single C2H2-type zinc finger domain. The SE gene is expressed on the adaxial side of the leaf, and the mutant phenotype suggests that SE is necessary for normal shoot apical meristem (SAM) development through chromatin modification (Prigge and Wagner, 2001).
Cylindrical symmetric flowers are observed in the Arabidopsis filamentous flower (fil) mutant (Sawa et al., 1999a). The FIL gene is a member of the YABBY (YAB) gene family that carries a zinc finger domain and a high mobility group-related domain. FIL mRNA is expressed on the abaxial side of leaves, floral buds, and all floral organs (Sawa et al., 1999b; Siegfried et al., 1999). Other members of the YAB gene family also are expressed on the abaxial side of the lateral organs (Bowman and Smyth, 1999; Siegfried et al., 1999; Villanueva et al., 1999; Bowman, 2000). In the fil yab3 double mutant, the mutant phenotype is enhanced in that filamentous organs and an aberrant meristem are formed through derepression of the KNOX genes (Siegfried et al., 1999; Kumaran et al., 2002). In addition, ectopic adaxial-regulated expression of the FIL gene partially changes the adaxial epidermis of the leaves such that their cells have characteristics of the abaxial epidermis. This finding indicates that the FIL gene might determine the identity of cells on the abaxial side of lateral organs (Sawa et al., 1999b). The kanadi (kan) mutant is known as an enhancer mutant of CRABS CLAW, another member of the YAB gene family. The KAN gene also is expressed on the abaxial side of the leaf (Bowman and Smyth, 1999; Eshed et al., 2001; Kerstetter et al., 2001). The kan1 kan2 double mutant produces unexpanded narrow leaves (Eshed et al., 2001). These studies suggest that the establishment of adaxial-abaxial polarity in these mutants is in disorder and that the disturbance on the adaxial-abaxial axis might cause primordia to generate radially symmetrical lateral organs.
The establishment of axes is a very complex process. Surgical experiments have demonstrated that leaf primordia separated from the SAM at an early stage often produce filamentous leaves, whereas leaf primordia separated from the SAM at a late stage produce flat leaves that possess adaxial-abaxial polarity through normal development (Sussex, 1955). These results imply that adaxial-abaxial polarity plays an important role in producing lateral organs with a flat shape, which might be generated properly in the early leaf primordia by the reception of positional information from the SAM. A molecular study of PHB, PHV, and REVOLUTA/INTERFASCICULAR FIBERLESS genes showed that they encode homeodomain–Leu zipper and putative sterol/lipid binding (START) domains that are expressed specifically on the adaxial side of lateral organs. In phb and phv dominant mutants, aberrant expression of PHB and PHV mRNA is detected on the abaxial side. Mutations of phb and phv dominant mutants are found only in the START domain, which is suggested to be a receptor for the factor that controls the determination of the adaxial nature of lateral organs by the PHB and PHV genes (Zhong and Ye, 1999; McConnell et al., 2001). In Drosophila, morphogen gradients induce the expression of several genes in the important compartments along the body plan (Roth et al., 1989; Rushlow et al., 1989). However, there is no evidence that similar gradients exist in the SAM. The molecular mechanism for receiving positional information to determine the abaxial side of lateral organs is unknown.
Here, we report the timing of adaxial-abaxial polarity observed in the process of lateral organ development and analyze the role of cis-acting elements located in the regulatory region of the FIL gene.
RESULTS
Analysis of FIL Expression Using the Promoter:Green Fluorescent Protein Transgenic Plant
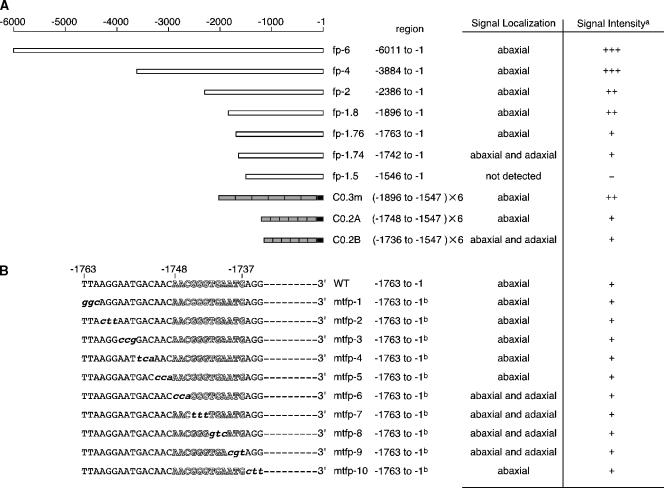
Spatially and temporally specific expression of FIL was examined using in situ RNA hybridization (Sawa et al., 1999b; Siegfried et al., 1999). To confirm the unique expression pattern, we constructed transgenic Arabidopsis plants that carried a series of FIL promoter:green fluorescent protein (GFP) constructs. We fused the longest 5′ regulatory region, up to −6011 from the putative initiation codon (Figure 2A), to FIL cDNA and transformed the fil-1 mutant. The transgene used complemented the fil mutation (data not shown). The GFP signal in the vegetative meristem, carrying fp-6 (Figure 2A), was first detected in a region expected to be a leaf primordium (P0 stage) (Figure 3A). It was unclear whether the GFP signal was observed on the abaxial side of the P0 primordium, because the exact size of the P0 primordium is unknown. However, the size of the GFP-expressing P0 primordium was much larger in the mtpf-6 line (Figures 3B1 and 3B2), which showed the GFP expression in leaf primordia tissues on both the abaxial and adaxial sides at later stages (Figure 2B).
Figure 2.
Functional Analysis of the 5′ Regulatory Region of FIL.
GFP signal localization and intensity were examined with 20 independent transgenic plants for each construct.
(A) A series of deletions in the 5′ regulatory region of the FIL gene. White boxes indicate the 5′ regulatory region connected to the GFP coding region and the 3′ terminus of nopaline synthase. Gray boxes in C0.3m, C0.2A, and C0.2B have six tandem repeats of regions at −1896 to −1547, −1748 to −1547, and −1736 to −1547, respectively, connected to the minimal promoter from −46 to −1 in the 35S promoter of Cauliflower mosaic virus (black boxes) (Benfey et al., 1989; Benfey and Chua, 1990).
(B) A series of base substitutions in the region from −1763 to −1734. The 5′ regulatory region from −1763 to −1 with or without the substitutions was connected to the GFP coding sequence and the 3′ terminus of nopaline synthase. A predicted putative Kr binding site exists in the region from −1763 to −1734 (white letters).
a Relative levels of GFP expression are denoted as follows: +++, strong; ++, medium; +, weak; −, not detected. b A point-mutated sequence exists in the region from −1763 to −1734, but the other sequence is identical to that of the wild type.
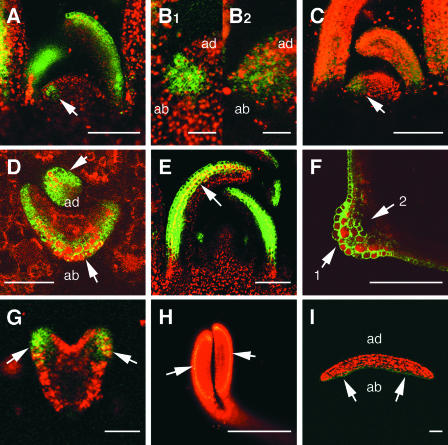
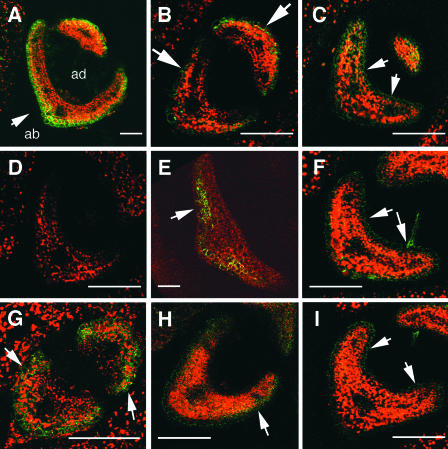
Figure 3.
FIL Expression in Leaves and Cotyledons.
GFP signals were examined in transgenic plants fp-6 ([A], [B1], and [D] to [I]) and mtfp-6 ([B2] and [C]) by confocal laser scanning microscopy. GFP signal and the autofluorescence of chloroplasts are shown in green and red, respectively; overlapped signal is shown in yellow and orange.
(A) Longitudinal section of a SAM. The GFP signal was found in cells on the abaxial side of leaves. The signal also was detected in a primordium (arrow).
(B1) Enlarged section of (A) indicated by the arrow.
(B2) Enlarged section of (C) indicated by the arrow.
The region of GFP expression in the mtfp-6 plant is larger than the region in the fp-6 plant shown in (B1) and (B2), indicating that the signal was located on the abaxial side in the P0 primordium of fp-6.
(C) Longitudinal section of a SAM of a mtfp-6 plant. The GFP signal was seen on both the abaxial and adaxial sides of leaves (see Figure 2B).
(D) Transverse section of a young leaf. A strong GFP signal was detected in three cell layers on the abaxial side (arrows).
(E) Longitudinal section showing the GFP signal in three cell layers on the abaxial side, from the bottom to the top of a leaf (arrow).
(F) Transverse section of a mature leaf. More than three cell layers expressed the GFP signal at the midrib (arrow 1). The GFP signal was not detected in the vascular bundle (arrow 2).
(G) The GFP signal was detected on the abaxial side of the cotyledon primordium in the heart-stage embryo (arrows).
(H) In an embryo at the bending-cotyledon stage, the GFP signal was detected in the abaxial epidermis of the cotyledons (arrows).
(I) Transverse section of a cotyledon of a 4-day-old seedling cotyledon showing the GFP signal on the abaxial epidermis (arrows).
ab, abaxial side; ad, adaxial side. Bars = 15 μm for (B1) and (B2), 50 μm for (D), (G), and (I), 100 μm for (A), (C), (E), and (H), and 200 μm for (F).
This observation strongly suggests that FIL expression was located on the abaxial side in the P0 primordium. In the P1-3 primordia, there are three FIL-expressing cell layers, including the epidermis. However, after growth of the leaf primordium, the number of cell layers expressing FIL increased gradually near the midrib, although the remainder of the leaf continued to have only three cell layers expressing FIL in the developing leaves (Figures 3D to 3E). It is worth noting that the GFP signal is absent in the cells of vascular bundles, although the vascular bundles are located at the boundary of the adaxial and abaxial sides (Figure 3F). When a leaf grows to 1 mm in length, the GFP signal starts to decrease, remains on the abaxial side of the petiole, and disappears completely after it grows to 1 cm in length. The GFP signal also was observed in primordia of other lateral organs, including cotyledons, cauline leaves, flowers, and floral organs. In embryos, FIL expression was detected on the abaxial side of developing cotyledons (Figures 3G and 3H). In embryos at the bending-cotyledon stage and in seedlings, FIL expression was detected in the epidermal tissues on the abaxial side of the cotyledons (Figures 3H and 3I).
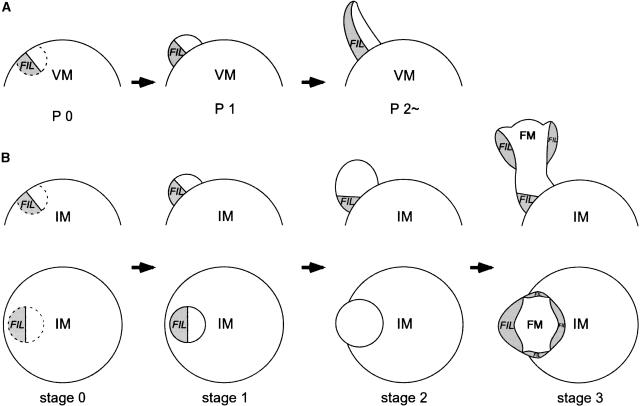
The pattern of GFP expression shows a unique change as the floral meristem develops. At the early stages, stages 0 and 1 (stage 0 means before a primordium has arisen; later stages are according to Smyth et al. [1990]), the expression was detected on the abaxial side of a flower primordium (Figures 4C, arrow 1, and 4D, arrow 1). This pattern confirms that a flower primordium forms as a lateral organ primordium. As the flower primordium grows to stage 2, however, the GFP signal disappeared when a top view of an inflorescence was examined. The signal reappeared in a floral meristem at stage 3 on the abaxial side of newly formed sepal primordia (Figures 4A and 4B) and continued to be present at later stages. When we examined longitudinal sections of a stage-2 meristem, the GFP signal was observed on the abaxial side at the bottom part of the meristem but not at the top of it (Figure 4C, arrows 2 and 3). In a floral meristem at stage 3, the signal at the bottom was weakened (Figure 4D, arrow 2), and strong GFP expression was observed on the abaxial side of sepal primordia initiated at the top of the meristem (Figure 4D, arrow 3). The GFP expression domains are summarized in Figure 5B. In later stages, GFP signals were observed on the abaxial side of successively formed primordia of other floral organs: petals, stamens, and carpels (Figures 4E and 4F).
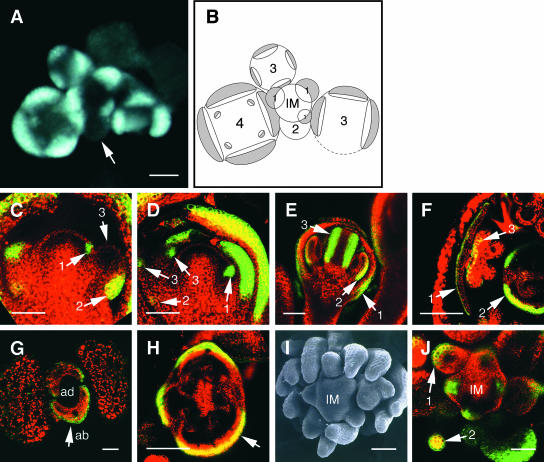
Figure 4.
Expression Analysis of FIL at the Reproductive Stage and in the fil Mutant.
(A) and (B) Transverse section of an fp-6 transgenic plant (see Figure 2A) examined by fluorescence microscopy (A) and its schematic view (B). Numbers in (B) indicate the stage of the floral buds. The GFP signal was observed in cells on the abaxial side of young flower primordia at stage 1. The signal was absent in the stage-2 flower primordium (arrow). In stage-3 and -4 flowers, the GFP signal was detected on the abaxial side of the newly formed primordia of sepals and petals. FIL-expressing regions are shown in gray.
(C) Arrow 1 indicates the GFP signal in the stage-0 primordium. In the stage-2 primordium, the signal was detected on the abaxial side of the basal region (arrow 2) but not in the upper region of the primordium (arrow 3).
(D) Longitudinal section of an inflorescence meristem. Arrow 1 shows the GFP signal on the abaxial side of a stage-1 flower primordium. Weak GFP expression remained on the abaxial side of the basal region of a stage-3 flower (arrow 2). GFP expression was induced on the abaxial side of sepal primordia (arrows 3).
(E) Longitudinal section of a stage-8 flower showing abaxial GFP expression detected in a sepal (arrow 1), a stamen (arrow 2), and a carpel (arrow 3).
(F) In a transverse section of a stage-12 flower, the GFP signal was observed in the petal epidermis on the abaxial side (arrow 1), on the abaxial side of the carpel (arrow 2), and on the abaxial side of the anther (arrow 3).
(G) Transverse section of a transgenic plant carrying fp-6 in the fil mutant. GFP was expressed on the abaxial side of the rosette leaves (arrow). The level and pattern of GFP expression in the fil mutant were the same as those in the wild type.
(H) Transverse section of a type-A flower of the fil mutant. Three sepals were formed in this flower, and the GFP signal was detected on the abaxial side of each (arrow).
(I) Top view of an inflorescence meristem of a fil plant showing a cluster of type-B filaments.
(J) Transverse section of a fil plant at the inflorescence meristem forming a cluster of type-B filaments. The GFP signal was observed on the abaxial side at the bottom of a filament (arrow 1) and on both the abaxial and adaxial sides in the upper region (arrow 2).
ab, abaxial side; ad, adaxial side; IM, inflorescence meristem. Bars = 20 μm for (C) to (F), (H), (I), and (J) and 50 μm for (A) and (G).
Figure 5.
Scheme of FIL Expression in the Vegetative Meristem and the Inflorescence Meristem.
(A) FIL expression in the vegetative meristem is shown in gray. In the vegetative meristem, FIL expression was detected first on the abaxial side of the precursor of a leaf primordium (P0) and continued to be present in cells on the abaxial side of the leaf primordium (P1) and in a young leaf (P2∼).
(B) FIL expression in the inflorescence meristem was observed on the abaxial side of a flower primordium at stages 0 and 1. The expression remained at the bottom of the primordium at stage 2, and FIL expression started on the abaxial side of sepal primordia formed at the top of a stage-3 flower.
FM, floral meristem; IM, inflorescence meristem; VM, vegetative meristem.
FIL Expression in the fil Mutant
To determine whether or not FIL expression is self-regulated, we transformed the fil-1 mutant with fp-6 constructs (Figure 2A). As with the wild type, the GFP signal was detected on the abaxial side of leaf primordia of the vegetative meristem (Figure 4G). We reported previously that the fil-1 mutant produces two kinds of aberrant flowers: type-A flowers with an aberrant number and arrangement of floral organs, and type-B filaments, which are peduncle-like filamentous structures. (Figure 4I) (Sawa et al., 1999a). In a type-A flower of the fil-1 mutant that carries the GFP construct, the GFP signal was detected on the abaxial side of the sepals, but the signal was less than the detectable level in other floral organs (Figure 4H). In a type-B filament, the GFP signal was observed on the abaxial side at the base of the filament (Figure 4J, arrow 1) but on both the abaxial and adaxial sides at the top (Figure 4J, arrow 2). These results suggest that FIL expression is not self-regulated and that the aberrant expression in the type-A flower and in the type-B filaments is the result of their deformed structures.
Deletion Analysis of the FIL Promoter
To clarify the mechanism responsible for the spatially and temporally specific expression of FIL, we examined the expression pattern in a series of transgenic plants with a deletion in the 5′ regulatory region of the FIL gene (Figure 2A).
In transverse sections of fp-6 and fp-4 transgenic plants, strong GFP signals were detected on the abaxial side of leaves (Figure 6A). In plants of fp-2, fp-1.8, and fp-1.76, the signals were observed on the abaxial side, but the signal strength was reduced (Figure 6B). In the fp-1.74 plants, which carry a 5′ regulatory region of −1742 to −1, however, the GFP signal was observed in tissues on both the abaxial and adaxial sides of the leaves and the signal intensity was weak (Figure 6C). When we trimmed the promoter region to −1546 (fp-1.5), the GFP signal was not detected (Figure 6D). These results strongly suggest that two cis-acting elements are responsible for the unique expression pattern of FIL. One element, which resides in a region between −1763 and −1743, might play a role in repressing the FIL expression on the adaxial side. Another element, located in a region between −1742 and −1547, might be responsible for FIL expression on both the abaxial and adaxial sides. When the two elements are present, FIL expression would be specific to tissues on the abaxial side. We then examined this model by constructing a transgenic line carrying six tandem repeats of the 5′ regulatory region at −1896 to −1547 connected to the 35S minimal promoter (Benfey et al., 1989; Benfey and Chua, 1990). As shown in Figure 6G, the GFP signal was located on the abaxial side, and the signal intensity was approximately the same as that of fp-2 and fp-1.8 (Figure 2A). The stepwise reduction of the GFP signal in the series of transgenic plants indicates that other cis-acting elements that determine the strength of gene expression reside between −3884 and −2387 and between −1896 and −1764.
Figure 6.
GFP Expression in Transgenic Plants Carrying the FIL 5′ promoter:GFP.
Transverse sections of young rosette leaves were examined by fluorescence microscopy. Green and red signals indicate GFP and autofluorescence of chloroplasts, respectively; overlapped regions are shown in yellow or orange.
(A) Transverse section of an fp-6 plant indicating that strong GFP expression was detected on the abaxial side (arrow).
(B) In the fp-1.76 plant, a weak GFP signal was expressed on the abaxial side (arrows).
(C) The fp-1.74 plant showed ectopic GFP expression on both the abaxial and adaxial sides. Note the green signal at the adaxial side (arrows).
(D) In the fp-1.5 plant, the GFP signal was not detected.
(E) The mtfp-1 plant showed a weak GFP signal on the abaxial side (arrow).
(F) Weak GFP expression was induced on both the abaxial and adaxial sides in the mtfp-6 plants. Arrows indicate the signal on the adaxial side.
(G) In the C0.3m plant, GFP was detected on the abaxial side (arrows).
(H) The C0.2A plant showed GFP on the abaxial side (arrow). The expression level was weaker than that of the C0.3m plant.
(I) A weak GFP signal was expressed on both the abaxial and adaxial sides in the C0.2B plant. Arrows show ectopic GFP expression on the adaxial side.
ab, abaxial side; ad, adaxial side. Bars = 50 μm.
cis-Acting Elements for the Abaxial Side–Specific Expression
To identify the cis-acting base sequence that controls abaxial side–specific expression, we made a series of constructs by replacing three continuous bases of the fp-1.76 promoter at −1763 to −1734 (Figure 2B) (Stanojevic et al., 1989; Tanaka et al., 2002) and examined the GFP signal in the transgenic plants. In mtfp-1, mtfp-2, mtfp-3, mtfp-4, mtfp-5, and mtfp-10, the GFP expression pattern and level were similar to those of fp-1.76 (Figure 6E). However, in mtfp-6, mtfp-7, mtfp-8, and mtfp-9, the GFP signal was detected on both the abaxial and adaxial sides (Figure 6F). These results strongly suggest that the 12-bp region at −1748 to −1737 controls the repression of FIL expression on the adaxial side. We then examined the role of the 12-bp sequence using two lines of transgenic plants, C0.2A and C0.2B. C0.2A and C0.2B had six tandem repeats. C0.2A had regions from −1748 to −1547 and C0.2B had regions from −1736 to −1547 connected to the 35S minimal promoter (Figure 2A). The GFP signal was on the abaxial side of the C0.2A plants (Figure 6H) but on both the abaxial and adaxial sides of the C0.2B plants (Figure 6I). The signal strength in the C0.2A and C0.2B lines was weak (Figure 2A). These results confirm that the 12-bp sequence from −1748 to −1547 is responsible for the abaxial side–specific expression of FIL, possibly by repressing the FIL expression on the adaxial side. It should be noted that an additional cis-regulatory element(s) that determines the expression level of FIL is located between −1896 and −1749, because the signal intensity in C0.3m plants was much stronger than that in C0.2A plants.
DISCUSSION
FIL Expression Is Localized in the Lateral Organ Primordia
The abaxial-regulated expression of FIL starts at the same time or immediately before the emergence of the primordia at the peripheral zone of the meristem and gradually decreases at late stages of organ development. As discussed above, the abaxial and adaxial sides of organ primordia are fixed by their juxtaposition to the shoot apex. It is postulated that the two sides would be established by receiving some positional information that would be transmitted from the apex to the organ primordia (Watanabe et al., 2003). This signaling system might work at very early stages of organ development, because our analysis showed that FIL is expressed in cells on the abaxial side of the precursor region of leaf primordia (Figures 3B1 and 3B2). It is not known, however, whether a similar signaling system operates in cotyledon development, because FIL expression in the developing cotyledons was detected in the heart-stage embryo, but the cotyledons are not formed from the SAM.
Our promoter:GFP analysis showed that the signal was detected on the abaxial side of young flower primordia at stages 0 to 1; however, expression was absent in the apical part of the primordia at stage 2, even though a residual signal was observed in the basal part. FIL expression reappeared on the abaxial side of newly formed sepal primordia at stage 3, when the top of the flower primordium develops into a flower meristem and starts to generate floral organs (Figure 5B). The unique expression pattern of FIL would coincide with the time at which the protuberance shifts from being an appendage of an inflorescence meristem to being a floral meristem.
In the fil mutant, a strong GFP signal was detected on the abaxial side of leaves (Figure 4G). This finding implies that the fil mutation does not influence abaxial-adaxial axis formation and that it might maintain FIL expression directly through positive feedback on the abaxial side of lateral organs. Another gene might induce FIL expression in the abaxial compartment. The expression pattern of the KAN gene is similar to that of FIL in the lateral organs (Eshed et al., 1999; Kerstetter et al., 2001). The kan1 kan2 double mutant produces abnormally narrow leaves and reduces the expression of FIL (Eshed et al., 2001). This finding suggests that KAN1 and KAN2 play a role in keeping the expression of FIL in the abaxial side tissues.
Two cis-Acting Elements Control the Abaxial Expression of FIL
Our promoter analysis of the 5′ regulatory region suggests that the abaxial side–specific expression of FIL is controlled by two different cis-acting elements: one element promotes expression on both the abaxial and adaxial sides of leaf primordia, and the other element (12 bp) suppresses expression on the adaxial side. In the phb and phv dominant mutants, the expression regions of PHB and PHV mRNA spread to the abaxial side and the expression level of the FIL mRNA is reduced greatly on the abaxial side (Siegfried et al., 1999; McConnell et al., 2001). These results imply that PHB and PHV play a role in keeping FIL expression on the abaxial side by repressing expression on the adaxial side. It is unknown what types of molecular mechanisms are at work, but a computer search of transcriptional factor binding sites predicted that the 12-bp sequence involves a putative binding site of a Drosophila protein, Krüppel (Kr). The Kr protein is known to repress gene expression and control the morphogenesis of the Drosophila embryo (Nüsslein-Volhard and Wieschaus, 1980; Stanojevic et al., 1989; Zhuo et al., 1990). In addition, the computer prediction indicates that there are three genes that encode Kr-like proteins in the Arabidopsis genome. It is possible that some Kr-like protein(s), which is expressed in cells on the adaxial side of lateral organ primordia, binds to the 12-bp region and represses FIL expression in a cell-autonomous manner.
Abaxial Expression Mechanism in YAB Genes
Using computer prediction, we found that the 5′ regulatory region of FIL, from −1896 to −1547 (C0.3m; Figure 2A), is highly similar to a region in the 5′ regulatory region of YAB3, from −1685 to −1373 (Figure 7) and that the putative Kr binding sequence also is observed in YAB3 (data not shown). Considering that the expression patterns of FIL and YAB3 overlap almost completely, these results strongly suggest that FIL and YAB3 were generated by gene duplication in recent times and that the spatially and temporally specific cis elements are preserved in the two genes. In the six members of the YAB gene family, five YAB genes, including FIL and YAB3, are known to be expressed in cells on the abaxial side, although the organs are different (Bowman and Smyth, 1999; Siegfried et al., 1999; Villanueva et al., 1999; Bowman, 2000). Recent studies showed that YAB genes are conserved in other plant species (Nagasawa et al., 2003). Although preliminary computer analysis did not show regions with high sequence identity with FIL, our study suggests that the spatially specific expression of the YAB gene family in plants involves a common system with two discrete regulatory elements, one that induces gene expression on both the abaxial and adaxial sides and another that represses expression on the adaxial side.
Figure 7.
Nucleotide Sequences of the 5′ Regulatory Region of FIL and YAB3.
The top sequence is from −1896 to −1547 of FIL, and the bottom sequence is from −1685 to −1373 of YAB3. The FIL sequence was used to make the C0.3m construct. Arrows indicate the sequence used to make C0.2A and C0.2B (see Figure 2A). Asterisks indicate nucleotides that are identical between FIL and YAB3. The box indicates the 12-bp sequence that is a putative Kr binding site.
METHODS
Construction of Transgenes
We screened the genome fragment that included the FIL coding region from the genomic library of the Arabidopsis thaliana Columbia wild type using colony hybridization and isolated the 8-kb region that included the FIL gene. The 4.7-kb region from SacI to BamHI of the 5′ regulatory region of the FIL promoter was ligated to the puc18 (Takara, Osaka, Japan) plasmid vector. The 1.3-kb region from the BamHI site to the putative initiation ATG of the FIL gene, in the 5′ regulatory region, with the SmaI site at the 3′ end was amplified by PCR using the KOD PCR enzyme (Toyobo, Osaka, Japan) and ligated into pBluescript II SK+ (Stratagene). The 4.7-kb region was placed into pBluescript II that contained the 1.3-kb region, and then we subcloned the 6-kb region from −6011 to −1 of the FIL 5′ regulatory region promoter (Figure 2A). The primers for fp-6 were 5′-TCTGGATCCACTTGTCAAGATTATGCAT-3′ and 5′-TTTCCCGGGCTTTTTTGTAAGAAGGGGAAAAATATTGG-3′. We generated deletion, point mutation, and gain-of-function series constructs by PCR using fp-6 as a template and deletion and point-mutated primers. To find the mutation site generated by PCR in the constructs used in this study, all regions then were amplified by PCR and checked by sequencing. Truncated and aberrant 5′ regulatory regions in the FIL promoter were ligated upstream of the GFP cDNA with the endoplasmic reticulum retention signal (Haseloff et al., 1997). Next, we ligated fused DNA into a binary vector, pARK5mcs (a gift from Meiji-Seika, Kaisha, Ltd., Tokyo, Japan).
Computer Programs Used in This Study
The FIL and YAB3 5′ regulatory regions were aligned using the wide-genome comparison program PipMaker (additional information about PipMaker is available at http://globin.cse.psu.edu/pipmaker) (Hardison et al., 1997). TRANSFAC databases, through TFSEARCH (Searching Transcription Factor Binding Sites, version 1.3), were used to search for transcriptional factor binding sites (TFSEARCH is located at http://150.82.196.184/research/db/TFSEARCH.html).
Sequencing
DNA fragments were subcloned into pBluescript II, and the puc18 (Takara) plasmid vector was sequenced using ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kits from Applied Biosystems (Foster City, CA) and an ABI Prism 3100 Genetic Analyzer from Applied Biosystems and Hitachi (Tokyo, Japan).
Plant Materials and Growth Conditions
We transformed Arabidopsis Columbia wild type using a vacuum transformation procedure (Bechtold et al., 1993) with Agrobacterium tumefaciens strain C58. Transgenic plants were selected on agar medium containing 50 μg/mL kanamycin. Seeds were dried and incubated at 4°C for 3 days. Plants were grown at 22°C in an air-conditioned room under continuous illumination of 50 to 100 μE·m−2·s−1.
Scanning Laser Microscopy
Young leaves of FIL promoter:GFP transgenic plants were fixed in 7% agarose (Gibco BRL) at 4°C for 15 min. Transverse and longitudinal sections (100 μm) then were made using a vibrating-blade microtome (Leica, Wetzlar, Germany) and placed into a drop of water on a covered glass slide. The sections were observed using a laser scanning microscope (Zeiss, Jena, Germany) at 488 nm for fluorescein isothiocyanate and 543 nm for rhodamine.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact K. Okada, kiyo@ok-lab.bot.kyoto-u.ac.jp.
Acknowledgments
We thank laboratory members at Kyoto University, Shinichiro Sawa (Tokyo University), and Tatsuya Sakai (RIKEN) for helpful discussions. This work was supported by a Research Fellowship from the Japan Society for the Promotion of Science, by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant-in-Aid for Science Research on Priority Area 14036220), and by funds from the Mitsubishi Foundation and from the Joint Studies Program for Advanced Studies of the Science and Technology Agency of Japan.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015214.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 326, 1194–1199. [Google Scholar]
- Benfey, P.N., and Chua, N.-H. (1990). The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250, 959–966. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.-H. (1989). The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8, 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L. (2000). The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 3, 17–22. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Oeltjen, J., and Miller, W. (1997). Long human-mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 7, 959–966. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Kumaran, M.K., Bowman, J.L., and Sundaresan, V. (2002). YABBY polarity genes mediate the repression KNOX homeobox genes in Arabidopsis. Plant Cell 14, 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias, A., and Lawrence, P.A. (1985). Parasegments and compartments in the Drosophila embryo. Nature 313, 639–642. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jurgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., and Nagato, Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard, C., and Wieschaus, E. (1980). Mutation affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, S., Stein, D., and Nüsslein-Volhard, C. (1989). A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59, 1189–1202. [DOI] [PubMed] [Google Scholar]
- Rushlow, C.A., Han, K., Manley, J.L., and Levine, M. (1989). The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell 59, 1165–1177. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Ito, T., Shimura, Y., and Okada, K. (1999. a). FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11, 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., and Okada, K. (1999. b). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic, D., Hoey, T., and Levine, M. (1989). Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature 341, 331–335. [DOI] [PubMed] [Google Scholar]
- St. Johnston, D., and Nüsslein-Volhard, C. (1992). The origin of pattern and polarity in the Drosophila embryo. Cell 68, 201–219. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1955). Morphogenesis in Solanum tuberosum L.: Experimental investigation of leaf dorsoventrality and orientation in the juvenile shoot. Phytomorphology 5, 286–300. [Google Scholar]
- Tanaka, K., Tsumaki, N., Kozak, C.A., Matsumoto, Y., Nakatani, F., Iwamoto, Y., and Yamada, Y. (2002). A Krüppel-associated box-zinc finger protein, NT2, represses cell-type-specific promoter activity of the α2(X1) collagen gene. Mol. Cell. Biol. 22, 4256–4267. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). Phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Watanabe, K., Matsumoto, N., Funaki, S., Tsugeki, R., and Okada, K. (2003). Axis-dependent regulation of lateral organs. In Morphogenesis and Pattern Formation in Biological Systems, T. Sekimura, S. Noji, N. Ueno, and P.K. Maini, eds (Tokyo, Japan: Springer-Verlag), pp. 165–176.
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, P., Stanojevie, D., Colgan, J., Han, K., Levine, M., and Manley, J.L. (1990). Activation and repression of transcription by the gap proteins hunchback and krüpell in cultured Drosophila cells. Genes Dev. 5, 254–264. [DOI] [PubMed] [Google Scholar]