Abstract
The AGAMOUS (AG) gene is necessary for stamen and carpel development and is part of a monophyletic clade of MADS-box genes that also includes SHATTERPROOF1 (SHP1), SHP2, and SEEDSTICK (STK). Here, we show that ectopic expression of either the STK or SHP gene is sufficient to induce the transformation of sepals into carpeloid organs bearing ovules. Moreover, the fact that these organ transformations occur when the STK gene is expressed ectopically in ag mutants shows that STK can promote carpel development in the absence of AG activity. We also show that STK, AG, SHP1, and SHP2 can form multimeric complexes and that these interactions require the SEPALLATA (SEP) MADS-box proteins. We provide genetic evidence for this role of the SEP proteins by showing that a reduction in SEP activity leads to the loss of normal ovule development, similar to what occurs in stk shp1 shp2 triple mutants. Together, these results indicate that the SEP proteins, which are known to form multimeric complexes in the control of flower organ identity, also form complexes to control normal ovule development.
INTRODUCTION
In angiosperms, the ovule is located within the pistil, which consists of one or several fused carpels. The unfertilized mature ovule is formed by a haploid embryo sac surrounded by one or two integuments. The ovule is connected to the maternal body through the funiculus. Several important events occur during ovule development: the ovule primordium has to be formed, ovule identity has to be specified, followed by pattern formation and morphogenesis. In recent years, the ovule, particularly of Arabidopsis, has emerged as a model system in which to study the genetic and molecular bases of organogenesis (Grossniklaus and Schneitz, 1998). Several genes have been identified that are involved in the initiation and development of structures as integuments or the gametophyte.
In Arabidopsis, ovules develop inside two fused carpels. AGAMOUS (AG) plays a primary role in specifying carpel formation, because ag mutants completely lack carpels (Yanofsky et al., 1990). However, when the ag mutant is combined with mutant alleles of the APETALA2 (AP2) gene, an ERF-type transcription factor, ectopic carpeloid structures, including ovules, are observed on the margins of the sepals (Bowman et al., 1991), indicating that carpeloid features can develop in the absence of AG activity. When the ag ap2 mutant was combined with mutant alleles of SHATTERPROOF1 (SHP1) and SHP2, forming the ag ap2 shp1 shp2 quadruple mutant, all carpeloid features, including ovules, were absent on the first-whorl organs (Pinyopich et al., 2003), showing that SHP1 and SHP2 are required for AG-independent carpel and ovule development.
shp1 and shp2 single mutants do not exhibit any phenotypic effect, and the double mutant by itself does not affect carpel identity, because the shp1 shp2 double mutant is disturbed only in dehiscence zone formation in the fruit, by which these mutant fruit are unable to shatter their seeds (Liljegren et al., 2000).
Interestingly, it was shown recently that SHP1 and SHP2 act redundantly with SEEDSTICK (STK; previously AGL11) in promoting ovule identity (Pinyopich et al., 2003). In the stk shp1 shp2 triple mutant, normal ovule and seed development was disrupted completely, with some of the ovules converted to leaf-like or carpel-like structures. The homeotic transformation of ovules into carpeloid structures was shown previously in petunia as a result of the cosuppression of two MADS-box genes, FLORAL BINDING PROTEIN7 (FBP7) and FBP11, which are homologous with STK (Angenent et al., 1995). Besides its role in ovule development, STK also is required for normal funiculus development and seed dispersal (Pinyopich et al., 2003).
Several studies have shown that MADS-box transcription factors act by the formation of multimeric complexes. The first example was reported for the Antirrhinum majus MADS-box transcription factors SQUAMOSA (SQUA), DEFICIENS (DEF), and GLOBOSA (GLO). Genetic evidence was obtained that SQUA together with DEF and GLO are needed for the establishment and maintenance of whorled phyllotaxy. The genetic interactions between SQUA, DEF, and GLO were confirmed at the molecular level by showing that the three MADS-box proteins form a ternary complex on the DNA (Egea-Cortines et al., 1999).
The formation of multimeric MADS-box protein complexes that promote flower organ development also was shown in Arabidopsis. Genetic studies showed that the closely related and functionally redundant MADS-box genes SEPALLATA1 (SEP1), SEP2, and SEP3 were necessary to determine the identity of petals, stamens, and carpels (Pelaz et al., 2000). Single sep mutants showed only subtle phenotypes, whereas the triple mutant produced indeterminate flowers composed of only sepals. This phenotype is strikingly similar to that of bc (ap3 ag or pi ag) double mutants, indicating that the SEP genes are in some way required for the activity of the class-B and -C organ identity genes. Experiments performed by Honma and Goto (2001) gave the molecular clue to the observed genetic interactions. They showed that in yeast and in vitro, the SEP3 protein establishes the interaction between AP1 and PISTILLATA (PI)/AP3 proteins and between AP3/PI and AG. Furthermore, overexpression studies in quadruple transgenic Arabidopsis plants overexpressing AP1-PI-AP3-SEP3 showed that vegetative leaves were converted to petal-like organs and that plants overexpressing PI-AP3-SEP3-AG had vegetative leaves converted to stamen-like organs. These data provided evidence that tetrameric MADS-box transcription factor complexes determine the identity of the floral organs, at least in petals and stamens.
Here, we report that STK, SHP1, and SHP2 overexpression in Arabidopsis results in homeotic conversions of sepals to carpeloid structures, including ovules. Interestingly, carpel formation also was observed when STK was overexpressed in the ag mutant, probably as a result of the ectopic expression of SHP1 and SHP2 induced by STK, indicating that AG activity is not needed to induce these homeotic transformations. This finding confirms that SHP1 and SHP2 are sufficient to promote carpel identity in the absence of AG. To investigate whether the redundancy between STK, SHP1, and SHP2 in promoting ovule identity and that between AG, SHP1, and SHP2 in promoting carpel identity is based on a biochemical interaction, we performed yeast two- and three-hybrid experiments. These experiments showed that STK cannot interact directly with AG, SHP1, and SHP2, although an interaction was obtained with SEP proteins. Genetic evidence for the role of SEP proteins in the formation of an ovule identity-promoting complex came from analysis of the SEP1/sep1 sep2 sep3 mutant, in which a large number of ovules lose their identity and are transformed into carpel- and leaf-like structures.
RESULTS
Ectopic Expression of STK in Transgenic Arabidopsis Plants Resulted in Ectopic Carpel and Ovule Formation
Previous experiments showed that the redundant petunia genes FBP7 and FBP11 were necessary for ovule identity determination and sufficient to induce ectopic ovule formation on sepals and petals (Colombo et al., 1995). STK is highly similar to these petunia genes and, together with SHP1 and SHP2, essential for ovule formation (Pinyopich et al., 2003). To determine whether ectopic STK expression also is sufficient to induce ectopic ovule formation, Arabidopsis plants were transformed with a chimeric gene construct in which the STK cDNA coding region was fused to the Cauliflower mosaic virus (CaMV) 35S double enhancer containing promoter (Benfey et al., 1990a, 1990b).
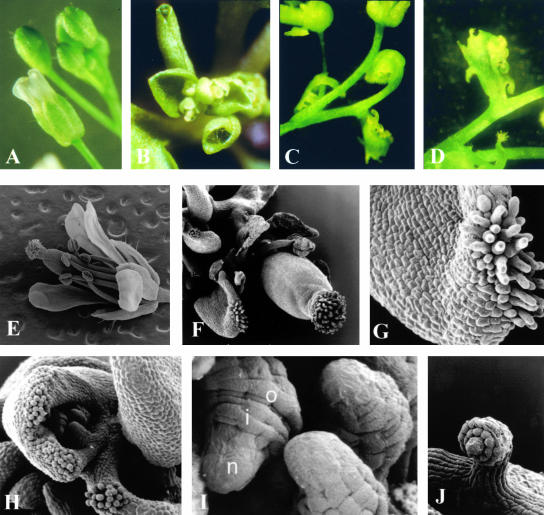
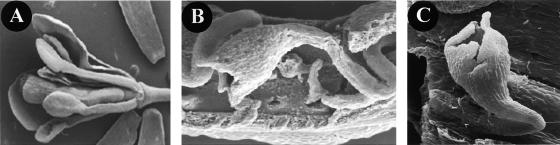
Sixty independent transgenic Arabidopsis lines were selected. Expression analysis of the transgene by reverse transcriptase–mediated (RT) PCR using RNA extracted from leaves revealed that of all the transgenic lines expressed STK ectopically (data not shown). Forty-five transgenic plants flowered extremely early (before the fourth leaf appeared), and the flowers of these plants had petals and stamens that were reduced in size; sometimes the petals were completely absent (Figure 1B). Furthermore, the first-whorl sepals were converted to carpeloid organs on which ovules developed, and in a few cases, the bract leaves were homeotically converted to carpeloid organs on which ovules developed (Figures 1C and 1D). To study these homeotic changes in more detail, scanning electron microscopy analysis was performed. As shown in Figures 1F and 1G, stigmatic papillae developed on the edges of the first-whorl organs. The carpeloid organs in the first whorl very often were folded, and on the inner site, ovule-like structures developed (Figure 1H). Structures that are typical for wild-type ovules, such as the developing inner and outer integuments, the funiculus, and the nucellus, could be recognized clearly in these ectopic ovules (Figures 1I and 1J).
Figure 1.
Flower Morphology of CaMV 35S::STK Transgenic Arabidopsis Plants.
Stereomicroscopic images are shown in (A) to (D), and scanning electron microscopy images are shown in (E) to (J).
(A) Wild-type flower.
(B) Inflorescence of a CaMV 35S::STK transgenic plant.
(C) and (D) Carpeloid bracts with ovules of a CaMV 35S::STK transgenic plant.
(E) Wild-type flower.
(F) Flower of a CaMV 35S::STK transgenic plant.
(G) Carpeloid sepal with stigmatic tissue.
(H) Curled carpeloid sepal with ectopic ovules.
(I) and (J) Ectopic ovules, with outer (o) and inner (i) integuments and nucellus (n).
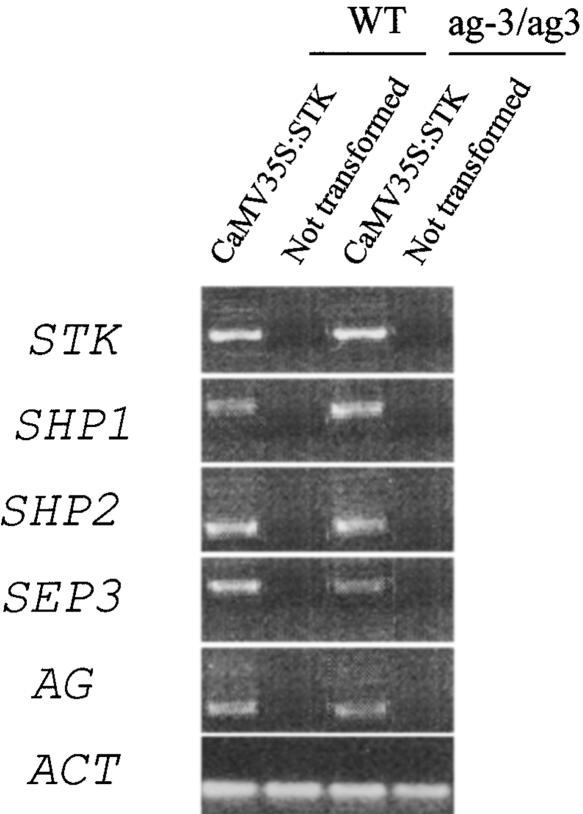
RT-PCR performed on leaves of the CaMV 35S::STK transgenic plants showed that ectopic STK expression resulted in the induction of ectopic SHP1, SHP2, AG, and SEP3 expression (Figure 2).
Figure 2.
Expression Analyses by RT-PCR of Genes Induced in Arabidopsis Wild-Type and ag-3 Mutant Plants Ectopically Expressing STK.
RT-PCR analysis using RNA extracted from bract leaves of wild-type (WT) and ag-3 mutant plants (ag/ag) in which STK was expressed ectopically (35S::STK). As a control, this analysis also was performed on plants that were not transformed. RT-PCR was performed using independent transformants, all of which gave similar results. Representative RT-PCR results are shown. In the CaMV 35S::STK plants, the SHP1, SHP2, SEP3, and AG genes all were induced. In the ag-3 mutant, the induced AG RNA encoded a nonfunctional transcript. ACT, control amplification on actin.
Ectopic expression of SHP1 and SHP2 (data not shown) resulted in the conversion of sepals to carpeloid structures, as described for STK, and the conversion of petals to stamenoid structures, confirming the data published by Liljegren et al. (2000). These experiments show clearly that ectopic STK, SHP1, and SHP2 activities are able to induce the carpel and ovule pathways in these transgenic plants.
Ectopic Expression of STK in the ag-3 Mutant
The carpeloid structures observed in the first-whorl floral organs of the STK, SHP1, and SHP2 overexpression plants resembled closely those observed in Arabidopsis plants in which AG was expressed ectopically (Mizukami and Ma, 1992). Furthermore, RT-PCR revealed that STK, when expressed ectopically, induced ectopic AG expression.
To understand whether the observed homeotic conversions were dependent on AG activity, we transformed ag-3 mutant plants (Bowman et al., 1989, 1991) with the construct for ectopic STK expression. In the ag-3 mutant flowers petals developed in place of stamens, and instead of carpels, four sepals arose that constituted the outer whorl of another inner ag flower (Figures 3A and 3B). The CaMV 35S::STK construct was used to transform ag-3/+ heterozygous plants, which were distinguished from wild-type plants using PCR primer-introduced restriction analysis (Jacobson and Moscovits, 1991). We analyzed 80 transgenic CaMV 35S::STK plants segregating for the ag mutant allele, all of which ectopically expressed STK.
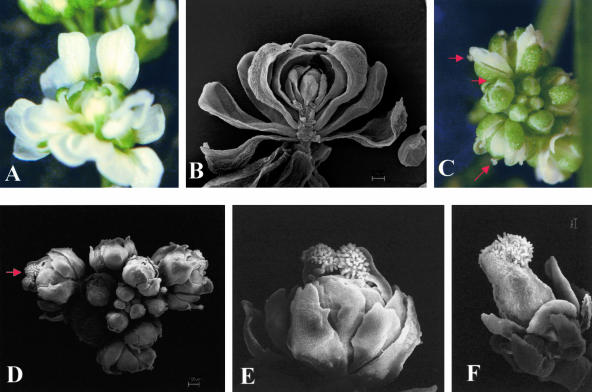
Figure 3.
Ectopic Expression of STK in the ag-3 Mutant.
Stereomicroscopic images are shown in (A) and (C), and scanning electron microscopy images are shown in (B) and (D) to (F).
(A) and (B) ag-3 mutant flower.
(C) and (D) Inflorescence of a transgenic ag-3 mutant plant ectopically expressing STK. Stigmatic tissues on top of the carpeloid sepals are indicated by the arrows.
(E) Close-up of a single flower of the inflorescence shown in (D). Stigmatic papillae are clearly visible.
(F) Flower of a transgenic ag-3 mutant plant ectopically expressing STK. A pistil-like structure develops in this flower.
Forty heterozygous plants (ag-3/+) ectopically expressing STK showed the same phenotype as those observed previously when wild-type plants were transformed: they were small as a result of extremely early flowering, and their sepals frequently were homeotically converted to carpeloid organs.
Twenty transgenic ag mutant (ag-3/ag-3) plants ectopically expressing STK were obtained and analyzed. These mutants flowered extremely early as well, maintaining a small size, and generally the flowers resembled typical ag-3 flowers. However, in two plants, homeotic conversion of the first-whorl sepals to carpeloid organs was observed (Figures 3C to 3E), suggesting that AG activity is not needed for the STK-induced homeotic transformations. Furthermore, in a flower of one of these plants, a pistil-like structure developed in the fourth whorl (Figure 3F). However, when we opened this pistil, no ovules were detected.
RT-PCR analyses performed on leaves of the transgenic plants that showed homeotic conversions (Figure 2) revealed that STK induced the ectopic expression of SHP1, SHP2, and SEP3, suggesting that the observed phenotypes could be promoted by SHP1 and SHP2 genes that are responsible for AG-independent carpel development (Pinyopich et al., 2003). Surprisingly, ovules were detected on none of the first-whorl organs that showed carpeloid features.
STK Interacts with AG in a Multimeric Complex
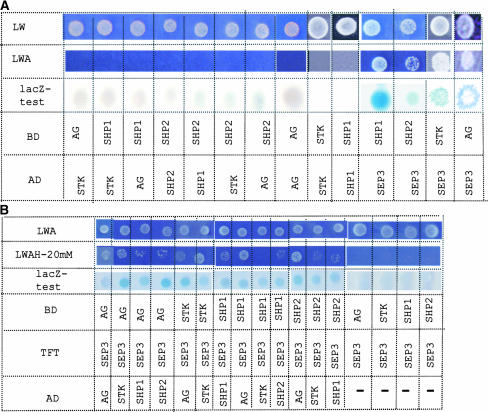
To investigate whether STK, SHP1, SHP2, and AG redundancy in promoting ovule and carpel formation was based on a biochemical interaction, we performed GAL4-based yeast two-hybrid experiments to assay the interactions between STK, AG, SHP1, and SHP2. The coding parts of the AG, SHP1, SHP2, and STK cDNAs were fused to the activation domain (AD) and binding domain (BD) and tested for interaction. In this assay, STK was not able to interact with AG, SHP1, and SHP2, and no interactions between SHP1, SHP2, and AG were observed (Figure 4A). Furthermore, none of the proteins was able to form homodimers.
Figure 4.
Interactions between MADS-Box Factors Detected by Yeast Two- and Three-Hybrid Assays.
(A) Yeast two-hybrid assays. MADS-box coding sequences were cloned in the pBD and pAD vectors. Yeast PJ69a was transformed with combinations of pBD and pAD constructs, and the presence of both vectors was confirmed by growth on dropout medium lacking Leu and Trp (LW). Interactions were assayed on selective dropout medium lacking Leu, Trp, and adenine (LWA) and by a β-galactosidase (lacZ) assay.
(B) Yeast three-hybrid assays. To determine whether SEP3 could bridge the interaction between STK, AG, SHP1, and SHP2, yeast three-hybrid assays were performed. SEP3 was cloned in the pTFT1 vector and transformed to yeast together with pAD and pBD vectors containing different combinations of MADS-box coding sequences. The presence of the three constructs was tested by plating on dropout medium lacking Leu, Trp, and adenine (LWA). Interactions were tested on dropout medium lacking Leu, Trp, adenine, and His (LWAH) with different concentrations of 3-aminotriazole. Only the results with 20 mM 3-aminotriazole are shown. Furthermore, interactions were tested by β-galactosidase (lacZ) assays.
Because previous studies showed that the SEP proteins interact with AP1, AP3, PI, and AG in the control of organ identity (Honma and Goto, 2001; Pelaz et al., 2001), we tested whether or not the SEP proteins could interact similarly with STK. These assays showed that SEP3 interacted with STK (Figure 4A). Because STK, SEP3, and AG all are expressed during ovule development, we wondered whether the STK and AG proteins, which do not appear to interact on their own, might form a multimeric complex that also includes SEP3. To test this notion, yeast three-hybrid experiments were performed by fusing the SEP3 protein with the nuclear localization signal of the TFT vector (Egea-Cortines et al., 1999). As presented in Figure 3B, yeast strain PJ69-4A was able to grow on selective medium only when all three proteins were expressed, showing that an interaction between STK and AG can be mediated by SEP3.
Furthermore, as shown in Figure 4A, SHP1 and SHP2 also were able to form heterodimers with SEP3. Therefore, we tested SEP3 and combinations of SHP1, SHP2, STK, and AG in the three-hybrid system. As shown in Figure 4B, SHP1 and SHP2 interacted, via SEP3, strongly with AG and weakly with STK. In fact, all combinations could be made using SEP3 as an intermediate, even interactions with themselves.
SEP Genes Are Required for Ovule Development
The data obtained in the yeast assays described above suggest that SEP proteins are necessary to form transcription factor complexes that control ovule development. However, previous studies have failed to reveal a role for the SEP genes during ovule development, because the sep triple mutant completely lacks carpels. To reveal genetic evidence that the SEP genes are involved in ovule development, we reduced SEP activity by examining SEP1/sep1 sep2 sep3 mutant plants. Interestingly, the ovules of these plants (Figure 5B) have a dramatic phenotype that closely resembles that observed in the stk shp1 shp2 mutant (Figure 5C). Normal ovule and seed development was disrupted, and some of the ovules were converted to leaf-like or carpel-like structures. These data demonstrate that the SEP genes are required for normal ovule development.
Figure 5.
Phenotypes of the SEP1/sep1 sep2 sep3 and stk shp1 shp2 Mutant Plants.
(A) A flower of a SEP1/sep1 sep2 sep3 plant. Flower development is normal in this mutant.
(B) Ovules of a SEP1/sep1 sep2 sep3 mutant plant. Ovule development is affected severely. Ovules are transformed into carpel- and leaf-like structures.
(C) Ovules of a stk shp1 shp2 plant. Ovule development is affected severely. The phenotype is similar to that shown in (B).
DISCUSSION
Ectopic Expression of STK, SHP1, and SHP2 Induce Carpel and Ovule Development in Arabidopsis
Ovule development is a complex process that can be divided into distinct phases, such as organ identity determination, pattern formation, and morphogenesis (Schneitz et al., 1995). A large number of Arabidopsis mutants have been identified that display altered ovule development. The analysis of these mutants and the cloning of their corresponding genes have started to provide some insight into the processes that control ovule morphogenesis. Until recently, the Arabidopsis genes that control ovule identity determination were not identified. The only information on this early phase in ovule development came from studies in petunia, in which two redundant MADS-box genes, FBP7 and FBP11, which specify ovule identity, were identified (Angenent et al., 1995; Colombo et al., 1995). Cosuppression of both genes resulted in the homeotic conversion of ovules to carpeloid structures, and ectopic expression of these genes induced ectopic ovule formation on sepals and petals.
Recently, it was shown that in Arabidopsis, ovule identity is promoted by three redundant genes: SHP1, SHP2, and STK (Pinyopich et al., 2003). In the shp1 shp2 stk triple mutant, all ovules are disturbed in their development, and some of them are homeotically transformed into carpeloid organs. Furthermore, it has been shown that an increased number of ectopic ovules were converted to carpeloid organs in the ag ap2 double mutant with respect to the ap2 single mutant (15%), indicating that AG also promotes ovule identity (Western and Haughn, 1999). To determine whether STK, SHP1, and SHP2, like FBP7 and FBP11, are able to induce ectopic ovule formation, we ectopically expressed these genes in Arabidopsis. These experiments showed that all of these genes induce homeotic conversions of sepals to carpeloid organs on which ovules developed.
This result is similar to that obtained in petunia. However, in petunia, homeotic conversions were restricted to the change of the sepal inner epidermis to placental cells on which ovules developed. No other pistil features were observed in the first-whorl organs of these transgenic petunia plants. Furthermore, occasionally, ovules also developed on the petals, without the development of any other detectable carpel features. This finding led to the conclusion that, at least in petunia, these two MADS-box genes were sufficient to induce ovule development without the presence of carpel structures. Although this might be true, molecular analysis showed that in the sepals and petals of these transgenic petunia plants, the class-C genes FBP6 and pMADS3 are induced (Colombo et al., 1995). That the induction of these genes did not result in severe homeotic conversions of sepals to carpeloid organs is not surprising, because the overexpression of FBP6 does not have any effect on sepals, and ectopic pMADS3 expression had only a very mild effect on sepals (Tsuchimoto et al., 1993; Kater et al., 1998). Our experiments with Arabidopsis show that ectopic ovule formation always is linked to the development of carpeloid structures.
STK, which is expressed specifically in the ovules, promotes ovule identity but is not a carpel identity gene, in contrast to SHP1 and SHP2, both of which are able to promote carpel and ovule identity (Pinyopich et al., 2003). The carpel structures that develop on the first-whorl organs of transgenic Arabidopsis plants that ectopically express STK are largely the result of the induction of AG. In the ag-3 mutant, ectopic expression of STK very rarely induced carpel formation in the first-whorl organs, indicating that AG is most efficient in inducing carpel formation. We never observed ovules on the carpeloid structures that developed in the ag-3 mutant overexpressing STK. This finding could be attributable to the fact that the most effective complex that promotes ovule identity contains AG (e.g., a complex consisting of AG-SEP3-STK). Our yeast three-hybrid assays are in agreement with this finding, because the stronger interactions were obtained when AG was part of the complex.
MADS-Box Proteins Form Complexes to Promote Ovule and Carpel Identity
To determine whether STK interacts with the carpel and ovule identity-promoting proteins AG, SHP1, and SHP2, yeast two-hybrid experiments were performed. These experiments revealed no interaction. Because it has been shown that SEP3 can mediate the interaction between AG and the class-B protein dimer AP3-PI (Honma and Goto, 2001), we tested STK, SHP1, and SHP2 for an interaction with SEP3. These experiments confirmed that SEP3 is able to interact with all of them. Subsequently, yeast three-hybrid experiments showed that ternary complexes can be formed using all possible combinations of SHP1, SHP2, STK, and AG as long as SEP3 is added as a mediator. Evaluation of the strength of the interactions in the yeast assays by observing growth on dropout medium with different concentrations of 3-aminotriazole and analyzing the lacZ color assay (data not shown) indicated that the interaction between AG, SEP3, and STK or AG, SEP3, and one of the SHP proteins was strong. On the other hand, the interaction (mediated by SEP3) between STK and SHP proteins and interactions such as STK-SEP3-STK, SHP-SEP3-SHP, and AG-SEP3-AG all were weaker.
The observed biochemical interactions could explain the observed redundancy between SHP1, SHP2, and STK in promoting ovule identity. AG and SEP probably form a stable complex with STK or one of the SHP proteins to promote ovule identity. Each of these complexes likely is enough to promote ovule identity. For instance, in the shp1 shp2 double mutant, the complex composed of STK-SEP-AG probably is sufficient to promote normal ovule development. When the STK, SHP1, and SHP2 genes all are inactive, the only possible complex that might be formed is an AG-SEP-AG complex, which presumably has reduced ovule-promoting activity.
The complexes composed of SHP and SEP proteins also likely are able to promote carpel identity in the absence of AG and AP2. In the ag single mutant, carpel formation is abolished completely. Because the SHP genes are induced by AG, these proteins are not present in this mutant to complement the absence of AG. However, in the ag ap2 mutant, the activity of SHP1, SHP2, and STK is observed in the first-whorl organs, indicating that ap2 directly or indirectly downregulates these genes. We could imagine that in the presence of these proteins in the first whorl of the ag ap2 mutant, complex formation between SEP and SHP proteins promotes carpel identity and complexes of SEP, SHP, and STK proteins promote ovule identity.
Ovule Development Is Affected in the SEP1/sep1 sep2 sep3 Mutant
SEP proteins have been shown to play key roles in the identity determination of petals and stamens by mediating the interaction between class-A and -B and class-B and -C, proteins, respectively (Pelaz et al., 2000; Honma and Goto, 2001). Furthermore, it has been proposed that they do not only function as mediators of protein interactions but also provide transactivation activity to the complex (Honma and Goto, 2001). Our protein interaction data show that SEP proteins are able to establish interaction between proteins that promote carpel and ovule identity. Genetic evidence for a role of SEP genes during ovule development was not obtained, because the sep triple mutant completely lacks carpels. Interestingly, genetic titration experiments in which SEP gene activity was reduced showed that when sep2 and sep3 activity was abolished completely and only one SEP1 allele was active, flowers developed rather normally but ovule development was affected severely, showing a clear role of SEP genes in ovule development.
The observed phenotype was very similar to that of the stk shp1 shp2 triple mutant, indicating that the genes with a major function in ovule development do not function without sufficient SEP activity. The protein complexes necessary for proper ovule development, including SHP1, SHP2, and STK, probably are not formed sufficiently because of the low abundance of SEP protein. The same result was obtained when plants were generated in which only one SEP2 allele was active, although one SEP3 allele was enough to establish normal ovule development (data not shown), suggesting that SEP3 is more efficient in promoting ovule identity. This difference in behavior between the three SEP genes indicates that they probably are not completely redundant and therefore that these three duplicated genes might be preserved during evolution. This idea is strengthened further by the fact that SEP3 expression is different from SEP1/2 expression (Flanagan and Ma, 1994; Savidge et al., 1995; Mandel and Yanofsky, 1998).
Evolutionary Conservation of STK–SEP Protein Interactions
In several species, STK homologs have been identified: the FBP7 and FBP11 genes in petunia (Angenent et al., 1995), OsMADS13 in rice (Lopez-Dee et al., 1999), and ZMM1 and ZAG2 in maize (Schmidt et al., 1993; Theissen et al., 1995). All of these STK homologs have more or less the same expression profile. Yeast two-hybrid protein interaction assays showed that the petunia proteins FBP7 and FBP11 interact with the SEP-like (AGL2-like) proteins FBP2, FBP5, and FBP9 (Immink et al., 2002). Interestingly, the rice MADS-box protein OsMADS13 also interacts with two SEP-like proteins, OsMADS24 and OsMADS45 (Favaro et al., 2002). The evolutionary conservation of the MADS-box protein partners was confirmed by yeast two-hybrid exchange assays, which showed that the protein partners of FBP7 interact with OsMADS13 and vice versa (Favaro et al., 2002). We also performed yeast two-hybrid exchange assays between STK and the partners of OsMADS13, and these partners were exchangeable (data not shown). These results suggest that SEP proteins already had, early in evolution, before the division between monocot and dicot plants occurred, a role in MADS-box protein complex formation. Interestingly, SEP proteins also seem to play a role in facilitating nuclear localization, because in petunia, the FBP7 and FBP11 proteins were transferred to the nucleus only in the presence of the SEP-like proteins FBP2, FBP5, and FBP9 (Immink et al., 2002). It will be interesting to investigate whether the nuclear localization of STK also is dependent on the interaction with SEP proteins.
METHODS
Plant Materials
Arabidopsis thaliana ecotype Columbia was used for the STK, SHP1, and SHP2 ectopic expression experiments. The plants were grown at 22°C under long-day conditions (16 h of light/8 h of dark) in a mixture composed of 2.5:0.5 soil:vermiculite. The ag-3 mutant ecotype Landsberg was provided by R. Sablowski (John Innes Centre, Norwich, UK). The genotype of the ag-3/+-derived plants was analyzed with a PCR-introduced restriction site polymorphism (Jacobson and Moscovits, 1991). DNA was amplified with the primers RSA-1 (5′-GTCGATTTCAGAAAATAAGAGCTC-3′) and RSA-5 (5′-GAAGTATTACCCGAATCCGCCCCAAGAAG-3′), and the product was digested with BslI. Fragments amplified from the wild-type allele were digested, whereas the ag-3 allele–derived PCR product was undigested. Fragments that differed by 20 bp were analyzed on a 3% agarose gel (2% low-melting-point agarose and 1% agarose).
Binary Constructs and Arabidopsis Transformation
For ectopic STK expression in wild-type and heterozygous Arabidopsis ag-3 mutant plants, the STK cDNA was cloned under the control of a double 35S promoter of Cauliflower mosaic virus (CaMV). The cDNA was amplified by PCR with the primers OL216 (5′-ATACCATGGGAAGAGGAAAGATAG-3′) and OL217 (5′-CGGGATCCAGATTATCCGAGATGAAGAA-3′) and cloned as a NcoI-BamHI fragment between the CaMV 35S promoter and a poly(A) terminator in a modified pUC19 vector. Introduced EcoRI and NcoI sites are underlined. The fragment containing the double CaMV 35S promoter, STK cDNA, and the poly(A) terminator was cloned as a AscI-PacI fragment in pCAMBIA1300. For ectopic SHP1 and SHP2 expression in Arabidopsis, the cDNAs were cloned under the control of a double CaMV 35S promoter. The SHP1 open reading frame was amplified with OL525 (5′-AATTCCAGCTGACCACCATGGAGGAAGGTGGGAGTAGTCAC-3′) and OL526 (5′-GATCCCCGGGAATTGCCATGTTACACAAGTTGAAGAGGAGGT-3′), and the open reading frame of SHP2 was amplified with OL531 (5′-AATTCCAGCTGACCACCATGGAGGGTGGTGCGAGTAATGAA-3′) and OL532 (5′-GATCCCCGGGAATTGCCATGTCAAACAAGTTGCAGAGGTGG-3′), and cloned in the Gateway overexpression vector pGD625 (derived from pGD120) (Immink et al., 2002) passing through pDONOR 201 (Life Technologies, Carlsbad, CA). Binary vectors were used to transform Agrobacterium tumefaciens C58C1/pMP90 (Koncz et al., 1984). Arabidopsis plants were transformed using the floral dip method described by Clough and Bent (1998).
Analysis of Arabidopsis Transformants
The seeds derived from the T0 35S::STK transformants were selected on medium containing 20 μg/mL hygromycin, and after 2 weeks, seedlings were transferred to soil. Hygromycin-resistant plants were analyzed for the presence of the CaMV 35S::STK transgene by PCR using primers based on the CaMV 35S sequence (OL212; 5′-CTCGGATTCCATTGCCCAGCTAT-3′) and on the STK sequence (OL218; 5′-TGGAGTTTTGAATCGTTTGGA-3′). The seeds obtained from the T0 35S::SHP1 and 35S::SHP2 transformants were selected on medium containing 50 μg/mL kanamycin, and after 2 weeks, resistant seedlings were transferred to soil. Kanamycin-resistant plants were analyzed for the presence of the transgene by PCR using the primers OL212 and OL526 for SHP1 and the primers OL212 and OL532 for SHP2.
STK, SHP1, SHP, AG, and SEP3 expression was assayed by RT-PCR. RNA was extracted from leaves (Kater et al., 1998) and retrotranscribed with RT-Superscript II (Life Technologies). STK, SHP1, SHP, AG, and SEP3 were amplified subsequently with specific primers and analyzed on agarose gels.
Scanning Electron Microscopy
Plant material was fixed overnight in 3% glutaraldehyde in 0.025 M phosphate buffer, pH 7, at 4°C, washed subsequently in 0.025 M phosphate buffer, pH 7, and incubated for 4 h in 1% osmic acid in 0.05 M phosphate buffer, pH 7. Samples were washed again in 0.05 M phosphate buffer, pH 7, dehydrated gradually in an ethanol series of 25, 50, 70, 85, 95, and 100%, and dried in liquid carbon dioxide. Samples then were covered with gold, placed in a Nanotech sputter coater, and observed with a LEO 1430 scanning electron microscope (LEO Electron Microscopy, Thornwood, NY).
Yeast Two- and Three-Hybrid Assays
The two- and three-hybrid assays were performed in Saccaromyces cerevisiae strain PJ69-4A (James et al., 1996) as described previously (Davies et al., 1996). pBD, pAD, and pTFT1 (Egea-Cortines et al., 1999) vector constructs were selected on Yeast Synthetic Dropout (YSD) medium lacking Leu, Trp, and adenine, respectively. Three-hybrid interactions were assayed on selective YSD medium lacking Leu, Trp, adenine, and His supplemented with different concentrations of 3-aminotriazole (1, 3, 5, 10, and 20 mM). β-Galactosidase tests were performed according to Davies et al. (1996).
Genes used for the yeast two- and three-hybrid assays were cloned in the Gateway vector GAL4 system (pDEST32 for binding domain fusions and pDEST22 for activation domain fusions) passing through pDONOR201 (Life Technologies). The cDNA of the genes was amplified by PCR with specific primers containing the attB1 and attB2 sequences for homologous recombination. Plasmids used as BD vectors were NOB249 for STK, NOB256 for AG, NOB250 for SHP1, and NOB284 for SHP2. Plasmids used as AD vectors were NOB246 for STK, NOB257 for AG, NOB247 for SHP1, NOB282 for SHP2, NOB283 for SEP1, and NOB245 for SEP3.
pTFT1 was digested with EcoRI-SalI. SEP3 cDNA was amplified with the primers OL340 (5′-CGGAATTCGGAAGAGGGAGAGTAGAATT-3′) and OL304 (5′-CGCTCGAGTCAAATAGAGTTGTTGTCATAAGGTAACC-3′), digested with EcoRI and XhoI, and subcloned in pTFT1. Introduced EcoRI and NcoI sites are underlined.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Lucia Colombo, lucia.colombo@unimi.it.
Acknowledgments
We thank Robert Sablowsky for providing the Arabidopsis ag-3 mutant. We thank Hans Sommer for the pTFT1 vector, Stefan de Folter for the pGD625 vector, Brendan Davies for the two-hybrid vector pBD-AGAMOUS, and Anna Gallard and Vittoria Brambilla for helping with some of the experiments. We are grateful to Giulio Melone, Simona Masiero, and Thomas Dresselhaus for their help with the scanning electron microscopy analysis. This work was supported by Ministero dell'Istruzione, dell'Università e della Ricerca (FIRB2002 and CNR Progetto strategico-legge 449/97), the European Community (REGIA Project QLRT-1999-876), and a grant from the National Science Foundation (to M.F.Y.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015123.
References
- Angenent, G.C., Franken, J., Busscher, M., van Dijken, A., van Went, J.L., Dons, H.J.M., and van Tunen, A.J. (1995). A novel class of MADS-box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.H. (1990. a). Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 9, 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, P.N., Ren, L., and Chua, N.H. (1990. b). Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J. 9, 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colombo, L., Franken, J., Koetje, E., van Went, J., Dons, H.J.M., Angenent, G.C., and van Tunen, A.J. (1995). The petunia MADS-box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996). Multiple interactions amongst floral homeotic MADS-box proteins. EMBO J. 15, 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro, R., Immink, R.G.H., Ferioli, V., Bernasconi, B., Byzova, M., Angenent, G.C., Kater, M.M., and Colombo, L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Gen. Genomics 268, 152–159. [DOI] [PubMed] [Google Scholar]
- Flanagan, C.A., and Ma, H. (1994). Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol. Biol. 26, 581–595. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., and Schneitz, K. (1998). The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9, 227–238. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Immink, R.G.H., Gadella, T.W.J., Ferrario, S., Busscher, M., and Angenent, G.C. (2002). Analysis of MADS-box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99, 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, D.R., and Moscovits, T. (1991). Rapid, non radioactive screening for activating ras oncogene mutations using PCR primer-introduced restriction analysis (PCR-PIRA). PCR Methods Appl. 1, 146–148. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater, M.M., Colombo, L., Franken, J., Busscher, M., Masiero, S., Van Lookeren Campagne, M.M., and Angenent, G.C. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., Kreuzaler, F., Kalman, Z., and Schell, J. (1984). A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J. 3, 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Lopez-Dee, Z.P., Wittich, P., Pe, M.E., Rigola, D., Del Buono, I., Gorla, M.S., Kater, M.M., and Colombo, L. (1999). OsMADS13, a novel rice MADS-box gene expressed during ovule development. Dev. Genet. 25, 237–244. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E.R., and Yanofsky, M.F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 6, 182–184. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Unraveling the redundant roles of MADS-box genes during carpel and fruit development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Savidge, B., Rounsley, S.D., and Yanofsky, M.F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS-box genes and the floral organ identity genes. Plant Cell 7, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R.J., Veit, B., Mandel, M.A., Mena, M., Hake, S., and Yanofsky, M.F. (1993). Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell 5, 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7, 731–749. [Google Scholar]
- Theissen, G., Strater, T., Fischer, A., and Saedler, H. (1995). Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs of AGAMOUS-like MADS-box genes from maize. Gene 156, 155–166. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto, S., van der Krol, A.R., and Chua, N.H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., and Haughn, G.W. (1999). BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J. 18, 329–336. [DOI] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]