Abstract
ARG1 (ALTERED RESPONSE TO GRAVITY) is required for normal root and hypocotyl gravitropism. Here, we show that targeting ARG1 to the gravity-perceiving cells of roots or hypocotyls is sufficient to rescue the gravitropic defects in the corresponding organs of arg1-2 null mutants. The cytosolic alkalinization of root cap columella cells that normally occurs very rapidly upon gravistimulation is lacking in arg1-2 mutants. Additionally, vertically grown arg1-2 roots appear to accumulate a greater amount of auxin in an expanded domain of the root cap compared with the wild type, and no detectable lateral auxin gradient develops across mutant root caps in response to gravistimulation. We also demonstrate that ARG1 is a peripheral membrane protein that may share some subcellular compartments in the vesicular trafficking pathway with PIN auxin efflux carriers. These data support our hypothesis that ARG1 is involved early in gravitropic signal transduction within the gravity-perceiving cells, where it influences pH changes and auxin distribution. We propose that ARG1 affects the localization and/or activity of PIN or other proteins involved in lateral auxin transport.
INTRODUCTION
Higher plant organs sense gravity primarily through the sedimentation of starch-filled amyloplasts in specialized cells called statocytes (Caspar and Pickard, 1989; Kiss et al., 1989; Kuznetsov and Hasenstein, 1996; Blancaflor et al., 1998). These cells constitute the columella of the root cap and the endodermal layer of the shoot. Amyloplast displacement and sedimentation is thought to activate signal transduction pathways that lead to the asymmetric redistribution of the plant hormone auxin across the stimulated organ (reviewed by Masson et al., 2002). A greater amount of auxin accumulates in the bottom flank of the organ, where it inhibits cell elongation in the root and promotes it in the shoot. As a result of this growth differential, the root curves downward and the shoot curves upward (reviewed by Masson et al., 2002).
In roots, the amyloplasts appear to sediment within a network of fine actin filaments that are tethered through the cortical endoplasmic reticulum (ER) and anchored at the plasma membrane (Yoder et al., 2001). Mechanical perturbation of the cytoskeleton and/or the ER was proposed to activate a gravity signal transduction pathway through the regulation of membrane channels (Sievers and Busch, 1992). Consistent with this model, transient changes in Ca2+ fluxes and alkalinization of the columella cytoplasm have been observed in response to gravistimulation (Scott and Allen, 1999; Fasano et al., 2001; Plieth and Trewavas, 2002).
Using fluorescent cytosolic pH reporters, Scott and Allen (1999) and Fasano et al. (2001) reported a transient pH increase in the S2 and S3 layers of the columella cells upon gravistimulation. Within 30 s, the cytosolic pH increased from 7.2 to 7.5. This phenomenon was accompanied by a decrease in the apoplastic pH around the columella cells (Fasano et al., 2001). Application or injection of pH-modifying agents into these cells either enhanced or reduced the gravitropic curvature (Scott and Allen, 1999; Fasano et al., 2001). Furthermore, the pgm-1 mutant of Arabidopsis, which exhibits reduced gravitropism as a result of the absence of starch in its gravity-perceiving amyloplasts, also showed a reduction and/or a delay in the onset of gravity-induced cytosolic and apoplastic pH changes (Fasano et al., 2001). Together, these observations indicate that changes in the cellular and apoplastic pH of columella cells play a vital role in root gravitropism. Possibly, they condition the root cap for the establishment of lateral polarity, such as the gradient of auxin, necessary for normal gravitropism.
Auxin influx and efflux carriers mediate the distribution of auxin in the root and the shoot. Some of these carriers show polar localization at the plasma membrane in certain tissues. The AUX1 gene encodes a transmembrane component of the influx machinery present in cells of the protophloem, the columella, the lateral root cap, and the epidermis of the root elongation zones (EZs) (Bennett et al., 1996; Marchant et al., 1999; Swarup et al., 2001). Likewise, Arabidopsis PIN genes encode putative transmembrane components of the efflux machinery (reviewed by Friml and Palme, 2002). When mutated, AUX1, AtPIN3, and AGR1/AtPIN2 (also known as EIR1 and WAV6) yield roots with altered gravitropism (Bennett et al., 1996; Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998; Utsuno et al., 1998; Friml et al., 2002b). AtPIN3 may contribute to the generation of the gravity-induced auxin gradient in the root cap, because it becomes localized at the bottom membrane of the columella cells upon gravistimulation (Friml et al., 2002b), whereas AGR1/AtPIN2 and AUX1 appear to contribute to the basipetal transport and the propagation of the auxin gradient from the root cap to the EZs. Accordingly, AGR1/AtPIN2 is associated mainly with the basal membranes of epidermal and cortical cells of the EZs (Müller et al., 1998) and with the basal side of lateral cap cells (R. Chen and P.H. Masson, unpublished data).
Auxin transport appears to be regulated by protein phosphorylation. Staurosporine, a protein kinase inhibitor, was shown to reduce auxin efflux in tobacco (Delbarre et al., 1998). Also, a mutation in Arabidopsis RCN1, which encodes a subunit of protein phosphatase 2A, resulted in increased basipetal auxin transport in the root tip and reduced gravitropic curvature (Garbers et al., 1996; Rashotte et al., 2001). Although the mechanism by which protein phosphorylation regulates auxin transport and gravitropism remains unclear, the function and/or localization of PIN efflux carriers are potential targets (Muday and Murphy, 2002).
Only a handful of genetic loci have been identified that may contribute to the early stages of gravitropic signal transduction. In roots and hypocotyls, the ARG1 (ALTERED RESPONSE TO GRAVITY; also known as RHG) and ARG1-LIKE2 (ARL2) genes of Arabidopsis have been implicated in this phase of gravitropism (Fukaki et al., 1997; Sedbrook et al., 1999; Guan et al., 2003). The arg1 and arl2 mutants exhibit slower kinetics of gravitropic curvature in these organs. Yet, they show normal growth morphology, elongation rate, growth sensitivity to plant hormones and auxin transport inhibitors, phototropic response, and accumulation of starch in their amyloplasts (Fukaki et al., 1997; Sedbrook et al., 1999; Guan et al., 2003). Thus, these genes appear to participate in an early step of gravitropism before the initiation of gravitropic curvature.
ARG1 and ARL2 encode type-II DnaJ-like proteins with the conserved J domain located at the N terminus, followed by a hydrophobic region and a C-terminal coiled-coil region (Sedbrook et al., 1999; Miernyk, 2001; Guan et al., 2003). Escherichia coli DnaJ and a number of yeast and human DnaJ-like proteins were shown to interact with the Hsp70 molecular chaperone and its homologs (reviewed by Zuber et al., 1998). The conserved tripeptide HPD present within the J domain is important for the stimulation of the intrinsic ATPase activity of Hsp70s. DnaJ-like proteins together with Hsp70s and/or other heat-shock proteins perform diverse cellular functions. For instance, E. coli DnaJ and its homologs participate in protein folding and the response to heat stress (Zuber et al., 1998). Yeast Ydj1p/Mas5p participates in protein translocation into the ER and mitochondria (McClellan and Brodsky, 2000; Artigues et al., 2002). Mammalian and yeast auxilins interact with their Hsc70 counterparts to disassemble clathrin triskelia from clathrin-coated vesicles during endocytosis (reviewed by Lemmon, 2001). Squid hsc70 and the kinesin light chain tandem repeats, which contain the J domain, regulate the movement of axonal organelles through the liberation of membrane-associated kinesin (Tsai et al., 2000).
Although DnaJ-like proteins are present in organisms from all kingdoms of life, paralogs of ARG1 have been found only in plants and nematodes to date (Sedbrook et al., 1999). These proteins may be specialized in physiological processes intrinsic to these organisms, such as gravitropism in plants. The specific mechanisms by which ARG1 and ARL2 contribute to gravitropic signaling have yet to be elucidated. Here, we examine the early events in gravitropism of arg1-2 roots as well as the localization of the ARG1 protein. Our data support a role for ARG1 in an early phase of gravity signal transduction.
RESULTS
ARG1 Expression in the Statocytes Is Sufficient for Its Function in Gravitropism
Previous studies suggested that ARG1 might function in gravity signal transduction (Sedbrook et al., 1999). This hypothesis predicts that ARG1 would be required in the statocytes of hypocotyls and roots to mediate a wild-type gravitropic response. Thus, we sought to express ARG1 in the root and shoot statocytes of arg1-2 null mutant seedlings.
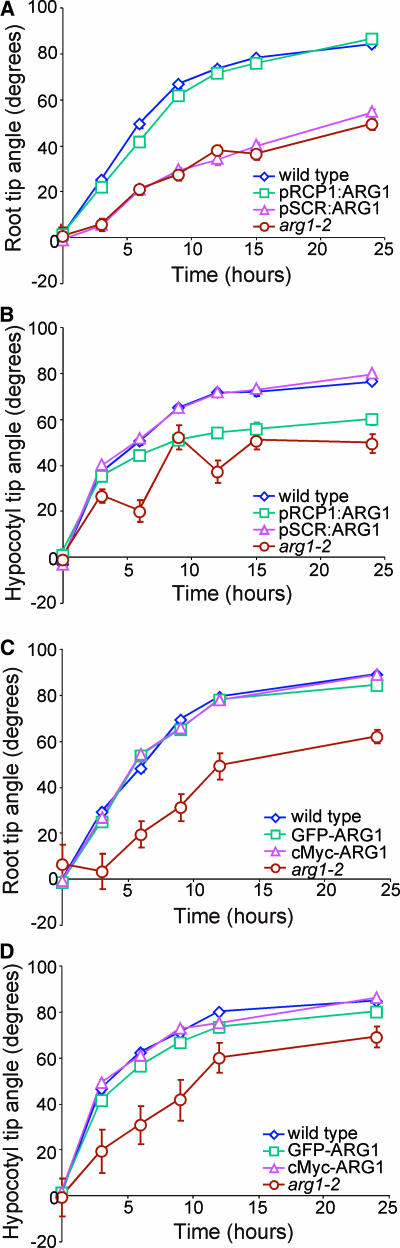
To achieve this goal, we placed the ARG1 open reading frame under the control of either the RCP1 or the SCR promoter (pRCP1:ARG1 and pSCR:ARG1, respectively). The RCP1 promoter directs gene expression specifically in the root cap (Tsugeki and Fedoroff, 1999), whereas the SCR promoter is active only in the endodermis of both shoots and roots (Malamy and Benfey, 1997a; Wysocka-Diller et al., 2000). When transformed into arg1-2, the pRCP1:ARG1 construct was able to restore a wild-type gravitropic response to mutant roots (Figure 1A). In hypocotyls, this construct allowed only partial rescue of the arg1-2 gravitropic phenotype (Figure 1B). Conversely, pSCR:ARG1 was able to restore a wild-type gravitropic response to mutant hypocotyls but not to roots (Figures 1A and 1B). These observations, especially the rescue of mutant hypocotyls by pSCR:ARG1, suggest that the primary function of ARG1 in gravitropism lies in events that occur specifically within the statocytes. This conclusion is consistent with the hypothesis that ARG1 may be involved in the signal transduction phase of gravitropism.
Figure 1.
Gravitropic Reorientation Kinetics of Transgenic arg1-2.
(A) and (B) Reorientation kinetics of root (A) and hypocotyl tips (B) of wild-type Wassilewskija (Ws), arg1-2, and representative transgenic arg1-2 carrying the pRCP1:ARG1 or pSCR:ARG1 construct (pRCP1:ARG1 and pSCR:ARG1, respectively).
(C) and (D) Reorientation kinetics of root (C) and hypocotyl tips (D) of wild-type Wassilewskija, arg1-2, and representative transgenic arg1-2 carrying the GFP-ARG1 or cMyc-ARG1 construct (GFP-ARG1 and cMyc-ARG1, respectively).
In each experiment, 90° gravistimulation was given at 0 h. Each data point represents the mean angle of organ tips. Vertical bars represent standard errors associated with the data points. For many data points, these bars were masked by the curve symbols. In all experiments, wild-type organ tips reoriented from a horizontal position at 0° toward the vertical at 90°. n = 39 to 131, 45 to 129, 22 to 80, and 22 to 80 in (A), (B), (C), and (D), respectively.
arg1-2 Mutants Show Defects in Gravity-Induced pH Changes within Root Statocytes
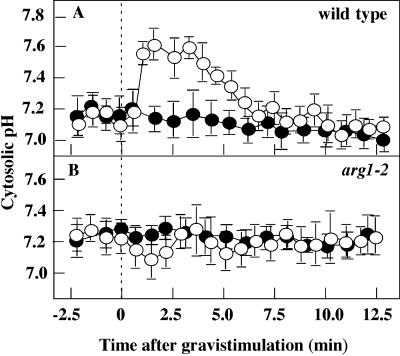
Early physiological events associated with gravitropic signal transduction have been best described in root statocytes (reviewed by Boonsirichai et al., 2002). Therefore, we focused our investigations of ARG1 function on its role in root gravitropism. One of the first manifestations of gravity signal transduction in roots is the alkalinization of their statocytes (Scott and Allen, 1999; Fasano et al., 2001). Using the pH-sensitive 2,7-bis-(2-carboxyethyl)-5(and 6)-carboxyfluorescein–dextran reporter, we analyzed this process in arg1-2 and compared the response to that of wild-type plants.
In vertically grown plants, the S3 columella cells of arg1-2 and wild-type plants showed similar, stable cytoplasmic pH levels of between 7.0 and 7.3 (Figure 2). Upon gravistimulation, wild-type S3 cells exhibited a transient increase in their cytoplasmic pH, which was not found in arg1-2 S3 cells (Figures 2A and 2B). This wild-type transient pH increase in response to gravistimulation in the Wassilewskija ecotype is similar to that reported previously for columella cells in the Columbia ecotype (Fasano et al., 2001). Similarly, the lack of a pH increase in arg1-2 is reminiscent of the reduced pH changes seen in pgm-1 root caps, which lack starch and show a severe impairment in gravity perception (Fasano et al., 2001). These observations suggest that arg1-2 disrupts a cellular event that contributes to the gravity-induced pH increase.
Figure 2.
Cytosolic Alkalinization of the S3 Columella Cells of the Wild Type and arg1-2 upon Gravistimulation.
(A) Wild type (ecotype Wassilewskija).
(B) arg1-2.
A 90° gravistimulus was given at 0 min. Closed circles represent mean cytosolic pH levels of ungravistimulated controls, and open circles represent gravistimulated roots. Data represents means ± se. n = 25 and 11 for the wild type and arg1-2, respectively.
arg1-2 Mutants Accumulate Auxin in the Root Cap
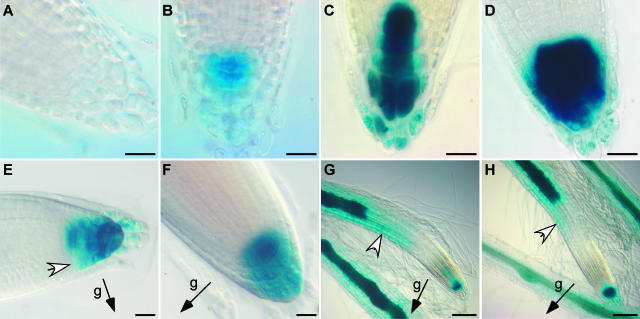
We investigated the pattern of auxin dynamics in arg1-2 seedlings by analyzing the activity of the auxin-responsive DR5 promoter (Ulmasov et al., 1997). arg1-2 plants harboring a DR5::β-glucuronidase (GUS) reporter gene construct were generated from a cross between arg1-2 and a Columbia plant carrying this construct (Ulmasov et al., 1997). Plants homozygous for both arg1-2 and the DR5::GUS insertion, or for both ARG1 and the DR5::GUS insertion, were identified among the F2 progeny. Thus, the DR5::GUS insertion is present at the same locus in both the wild-type and mutant backgrounds. In the wild-type background derived from either the parental DR5::GUS line or the F2 segregants, GUS activity was maximal in the central columella cells of the root cap (Figure 3C). The flanking columella cells stained only faintly. In arg1-2 root tips, both central and flanking columella cells stained faster and more intensely for GUS activity than in the wild-type background (Figures 3A to 3D). These observations suggest that arg1-2 seedlings accumulate a higher level of auxin in an expanded domain of the root tip compared with wild-type seedlings.
Figure 3.
DR5::GUS Activity in the Wild-Type ARG1 and Mutant arg1-2 Backgrounds.
(A) and (B) Root tips of vertically grown wild-type (A) and arg1-2 (B) plants stained with 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) buffer under the same conditions for 7 h.
(C) and (D) Root tips of vertically grown wild-type (C) and arg1-2 (D) plants stained for 15 h under the same conditions in an experiment distinct from that presented in (A) and (B).
(E) and (F) Wild-type (E) and arg1-2 (F) root tips after a 6-h gravistimulation stained with 2 mM X-Gluc buffer containing 2 mM K3Fe(CN)6 and either 0.1% (v/v) Triton X-100 for 5 h (E) or 10% (v/v) methanol for 24 h (F) at 22°C. Different staining conditions were used in (E) and (F) to allow equal staining intensity between wild-type and mutant root tips.
(G) and (H) Wild-type (G) and arg1-2 (H) root tips after 16 h of vertical growth on 1 μM NAA followed by a 7-h gravistimulation and staining overnight.
White arrowheads indicate the lateral gradient of DR5 activity in the root cap (E) and in the distal EZ ([G] and [H]). Black arrows indicate the gravity (g) vector. Ten to 25 roots were examined for each category. Bars = 20 μm for (A) through (F) and 100 μm for (G) and (H).
Upon 90° gravistimulation, the activity of the DR5 promoter expanded toward the bottom flank of the root cap in wild-type seedlings. More intense GUS staining was seen in peripheral columella cells on the bottom flank compared with the top flank (Figure 3E). In the arg1-2 background, this asymmetry was not observed (Figure 3F). DR5::GUS expression was not observed in the EZ cells of the transformed roots. However, when gravistimulated in the presence of 1 μM 1-naphthaleneacetic acid (NAA), an auxin, wild-type roots exhibited greater DR5 activity at the bottom flank of the distal EZ (Figure 3G), as reported previously (Ottenschläger et al., 2003). This gradient of DR5 activity also was observed across the distal EZ of arg1-2 roots (Figure 3H), suggesting that this region of arg1-2 was capable of displaying a lateral gradient of auxin concentration or responsiveness upon gravistimulation. Interestingly, even in the presence of 1 μM NAA, the gravity-induced auxin gradient was not observed in arg1-2 root caps (Figure 3H).
cMyc- and Green Fluorescent Protein–Tagged ARG1 Constructs Complement arg1-2
To examine the subcellular localization of ARG1, we generated the green fluorescent protein (GFP)–ARG1 and cMyc-ARG1 constructs, which carry N-terminal GFP- or cMyc-tagged ARG1, respectively. These constructs contained genomic sequences that included a 3.8-kb sequence upstream of the ARG1 initiation codon (i.e., the native promoter), the ARG1 transcribed region, and a 3.3-kb downstream sequence. Both constructs retained the full coding sequence of ARG1 and were transformed into arg1-2.
We assayed for the ability of these constructs to restore a wild-type gravitropic response to arg1-2 seedlings. Figures 1C and 1D show that roots and hypocotyls of arg1-2 carrying either construct exhibited gravitropism kinetics similar to that of the wild type. On the other hand, nontransgenic arg1-2 seedlings showed the expected alteration in their kinetics of root and hypocotyl reorientation compared with wild-type seedlings (Sedbrook et al., 1999). Thus, both constructs appeared functional and therefore should yield a fair representation of ARG1 localization. Analyses of the ARG1 expression pattern were performed on homozygous T3 plants and subsequent generations.
The ARG1 Protein Is Associated with Compartments of the Vesicular Trafficking Pathway and with Cell Plates
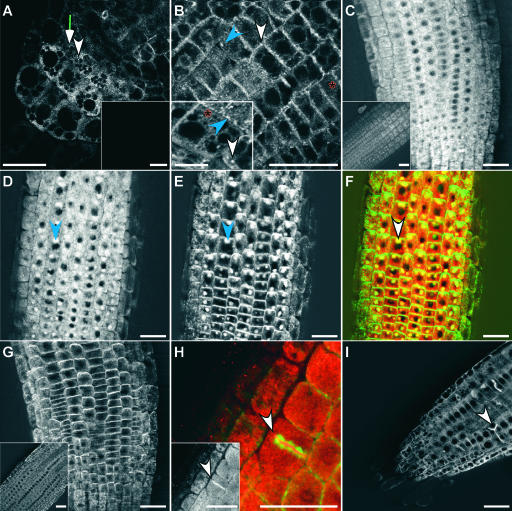
Confocal and multiphoton microscopy were used to analyze the localization of ARG1 in live samples of GFP-ARG1 transgenic roots. GFP fluorescence was highest in the root tip region, which includes the root cap, the meristematic region, and the elongation zones (Figures 4A and 4B). A lower level of GFP fluorescence also was observed in the mature part of the root (data not shown), which is consistent with previous findings of ubiquitous ARG1 expression (Sedbrook et al., 1999). In cells of the root tip, we observed a diffuse and reticulated pattern of GFP-ARG1 (Figures 4A and 4B, red asterisks), compatible with ARG1 association with the ER (Saint-Jore et al., 2002). In columella, root meristematic, and distal EZ cells, we also observed punctated signals (Figures 4A and 4B). The largest fluorescent dots were circular, oval (Figures 4A and 4B, white arrowheads), or linear in shape (Figures 4A and 4B, blue arrowheads), suggesting that they might correspond to Golgi stacks (Saint-Jore et al., 2002). Other fluorescent dots were smaller and likely correspond to vesicles (Figures 4A and 4B). In cortical cells, the majority of these structures were localized to the subcortical region close to the plasma membrane, although some were found in transvacuolar strands (Figures 4B and 4B inset). A C-terminal GFP fusion of ARG1 showed a similar localization pattern (data not shown).
Figure 4.
Localization of GFP-ARG1 and cMyc-ARG1.
(A) and (B) Multiphoton images of transgenic arg1-2 root cap (A) and distal EZ (B) carrying the GFP-ARG1 construct, showing a large abundance of small dotted signals located at the periphery of all cells, larger punctated signals that appear circular (white arrowheads) or linear (blue arrowheads) in shape, and reticulated signals (red asterisks). Insets show signals from a nontransgenic root cap (A) and a cortical EZ cell from a different root that shows the reticulate and large punctate fluorescence patterns (B).
(C) and (D) Confocal images of the distal EZ of transgenic arg1-2 carrying a cMyc-ARG1 construct immunostained with anti-cMyc antibodies. The inset in (C) shows signals from a nontransgenic root. The seedling in (D) was treated for 2 h with 50 μM BFA before fixation, showing the accumulation of the cMyc-ARG1 fusion protein in BFA compartments (blue arrowhead).
(E) The same seedling shown in (D) treated with anti-AGR1/AtPIN2 antibodies, showing the accumulation of AGR1/AtPIN2 in similar BFA compartments (blue arrowhead).
(F) A merged image of (D) and (E), showing cMyc-ARG1 in red and AGR1/AtPIN2 in green. Yellow indicates possible colocalization. Both proteins were found in the same BFA compartments (white arrowhead).
(G) Immunostaining of cMyc-ARG1 transgenic distal EZ treated with anti-AGR1/AtPIN2 without BFA treatment, showing the localization of AGR1/AtPIN2 on basal membranes. The inset shows agr1-5, an AGR1/AtPIN2 null mutant (Chen et al., 1998), treated with anti-AGR1/AtPIN2 antibodies.
(H) Immunostaining of cMyc-ARG1 transgenic EZ treated with anti-cMyc (signals in red) and anti-α-tubulin antibodies (signals in green), showing the localization of cMyc-ARG1 at the cell plate (arrowhead). The inset shows signals from anti-cMyc antibodies only (white).
(I) Confocal image of transgenic GFP-ARG1 treated for 2 h with 10 μM oryzalin followed by a 2-h wash in liquid GM medium. GFP-ARG1 is found at aberrant cell division planes (arrowhead).
Three to 15 roots were examined for each condition. Bars = 25 μm except for the inset in (B), where the bar = 10 μm.
Immunofluorescence analyses of transgenic seedlings carrying the cMyc-ARG1 construct revealed a similar localization pattern, but the signals were more diffuse (Figure 4C), possibly as a consequence of the fixation step needed in this assay. To investigate the association of ARG1 with the vesicle-trafficking pathway, we treated these seedlings before fixation with brefeldin A (BFA), a compound shown to disrupt vesicle trafficking. The cMyc-ARG1 fusion protein became associated with distinct BFA-induced compartments (Figure 4D). These structures were comparable to compartments in which specific proteins from the trans-Golgi network, the plasma membrane, and cell walls accumulate in response to BFA (Baluska et al., 2002; Nebenführ et al., 2002).
In a double immunofluorescence labeling experiment, AGR1/AtPIN2, a transmembrane component of the auxin efflux machinery, accumulated in the same subcellular compartments as cMyc-ARG1 upon BFA treatment (Figures 4E and 4F). Normally, AGR1/AtPIN2 resides at the plasma membrane of the epidermal and cortical cells of the distal EZ (Figure 4G) (Müller et al., 1998; R. Chen and P.H. Masson, unpublished data). The effect of BFA on the localization of both cMyc-ARG1 and AGR1/AtPIN2 was reversible. Both proteins assumed their normal localization if the BFA pretreatment was followed by a 2-h wash in BFA-free medium (data not shown). Together, these observations indicate that ARG1 likely resides in cellular compartments that participate in vesicular trafficking. This trafficking pathway may be the same pathway that is used by AGR1/AtPIN2 in distal EZ cells.
GFP-ARG1 and cMyc-ARG1 fusion proteins also were found to be associated with the cell plate, as indicated by double immunofluorescence staining of cMyc-ARG1 and α-tubulin (Figure 4H). Accordingly, treatment of transgenic seedlings with 10 μM oryzalin, a microtubule-destabilizing compound, resulted in GFP-ARG1 localization at aberrant cell division planes (Figure 4I).
ARG1 Is Associated with Microsomal Membranes and the Cytoskeleton
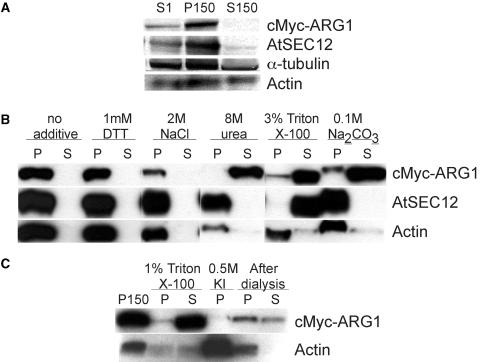
ToPred, a protein topology prediction program, predicted a single-pass transmembrane region between the J domain and the predicted coiled-coil region of ARG1 (Sedbrook et al., 1999). However, this region also is rich in Gly and Phe, suggesting that it might instead act as a linker between the J domain and the C terminus, similar to the Gly/Phe-rich region of many DnaJ-like proteins (Zuber et al., 1998; Miernyk, 2001). Furthermore, no signal peptides or potential post-translational membrane association motifs could be identified in ARG1 by sequence analysis (data not shown). Because GFP-ARG1 appeared to localize to the ER, the Golgi, and vesicles near the plasma membrane, we took a biochemical approach to investigate ARG1's association with plant membranes. We prepared microsomal membrane and soluble protein fractions from liquid cultures of cMyc-ARG1 transgenic plants, because the localization pattern of ARG1 was preserved among different root tissues (Figures 4A and 4B). Immunoblot analyses with anti-cMyc antibodies indicated that cMyc-ARG1 was associated primarily with the microsomes and was barely detectable in the soluble fraction (Figure 5A). This association was sensitive to the presence of a denaturing agent (3 and 8 M urea, data not shown and Figure 5B, respectively) and to a high-pH environment (0.1 M Na2CO3, pH 11.5) (Figure 5B). The sensitivity of ARG1's membrane association with these compounds strongly suggests that ARG1 associates peripherally with plant membranes and indicates that it is not an integral membrane protein.
Figure 5.
Immunoblot Analyses of Protein Extracts from Transgenic arg1-2 Carrying the cMyc-ARG1 Construct, Showing the Membrane and Cytoskeleton Association of ARG1.
(A) Fractionation of cMyc-ARG1, AtSEC12 (an integral membrane protein control), α-tubulin, and actin (integral/peripheral membrane protein controls) in the nucleus-free total protein extracts (S1), the microsomal membrane fraction (P150), and the soluble fraction (S150). A total of 50 μg was loaded per lane. The results shown are representative of five experiments.
(B) Fractionation of cMyc-ARG1, AtSEC12, and actin in microsomes treated with buffer containing the reagents specified at top. The pellet (P) and supernatant (S) of each treatment were derived from 90 μg of P150 starting materials, except those of the Na2CO3 treatment, which were derived from 110 μg of P150. The results shown are representative of three experiments.
(C) Fractionation of cMyc-ARG1 and actin in the microsomes, the cytoskeleton-enriched pellet, and the supernatant after treatment with 1% (v/v) Triton X-100, the pellet after depolymerization of the cytoskeleton with 0.5 M KI, and the cytoskeleton-enriched pellet and the supernatant after cytoskeleton repolymerization upon dialysis of the KI supernatant. The pellet and supernatant of Triton X-100 treatment were derived from 85 μg of P150 starting materials. A total of 30 μg of KI and postdialysis pellets and the volume equivalent of postdialysis supernatant were loaded.
The association of ARG1 with the microsomes was not affected by changes in the redox potential of its environment (up to 1 mM DTT), nor was it modified by a change in ionic interactions (up to 2 M NaCl) (Figure 5B). However, it was sensitive to the presence of Triton X-100 (Figure 5B), indicating that it interacts directly or indirectly with integral membrane proteins. Yet, a small portion of ARG1 remained in the pellet, even when AtSEC12, an integral ER membrane protein (Bar-Peled and Raikhel, 1997), was solubilized almost completely by Triton X-100. This fraction of ARG1 could have been retained in the pellet by means of its interaction with the cytoskeleton, which is resistant to Triton X-100 extraction (Tan and Boss, 1992). Sequence analyses of ARG1 also revealed the similarity of its predicted coiled-coil region to the coiled coil of cytoskeleton-interacting proteins, such as INCENP (Sedbrook et al., 1999).
To further investigate the possible ARG1-cytoskeleton association, we analyzed the fractionation of cMyc-ARG1 after a round of cytoskeleton depolymerization and repolymerization. The cytoskeleton was depolymerized partially by treatment of the cytoskeleton-enriched microsomal pellets with 0.5 M KI (Tan and Boss, 1992; Cox and Muday, 1994). A large portion of ARG1 was released from the pellet by this treatment (Figure 5C). Upon the removal of KI through dialysis, actin subunits repolymerized and were collected in the pellet (Figure 5C). Immunoblot analyses revealed that some ARG1 protein was associated with the same pellet, although some remained in the supernatant (Figure 5C). This observation suggested that a small pool of ARG1 might be associated with the cytoskeleton. Unfortunately, the low abundance of ARG1 in this pool precluded a more thorough analysis of such interactions. Thus, the association of ARG1 with microsomal membranes likely is mediated through its interaction with other membrane-associated proteins and possibly partly through its direct or indirect interaction with the cytoskeleton.
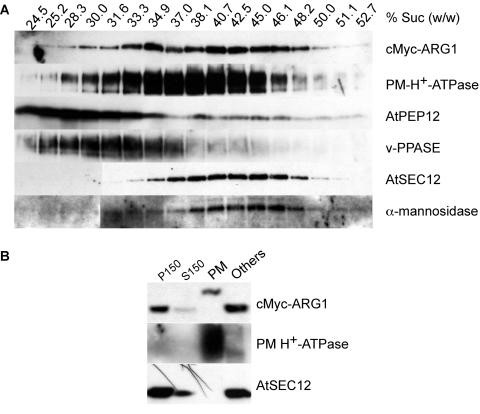
ARG1 Cofractionates with Multiple Membranes
We attempted to identify the types of membrane that ARG1 is associated with. Nucleus-free total protein extracts from 2-week-old etiolated cMyc-ARG1 transgenic plants were fractionated on a continuous 14 to 50% sucrose gradient. cMyc-ARG1 was found to fractionate in a range of the gradient at which markers from the plasma membrane (PM H+-ATPase) and other membranes, including the ER (AtSEC12) and the Golgi (α-mannosidase), can be found (DeWitt et al., 1996; Bar-Peled and Raikhel, 1997; Rancour et al., 2002). The pattern of cMyc-ARG1 fractionation also overlapped partially with that of an endosomal marker, AtPEP12 (da Silva Conceição et al., 1997) (Figure 6A). This observation is consistent with ARG1's association with cellular compartments involved in membrane trafficking and with the cell plates.
Figure 6.
Immunoblot Analyses of Protein Extracts from Transgenic arg1-2 Carrying the cMyc-ARG1 Construct, Showing the Association of ARG1 with Multiple Membranes.
(A) Fractionation of cMyc-ARG1, PM H+-ATPase (a plasma membrane marker), AtPEP12 (an endosomal marker of the vacuolar pathway), anti-vacuolar pyrophosphatase (v-PPASE; a vacuolar marker), AtSEC12 (an ER marker), and α-mannosidase (an early Golgi marker) in a 14 to 50% (w/w) sucrose gradient. The sucrose concentration of each fraction, as determined by its refractive index, is given at top. A total of 20 μL of each fraction was loaded per lane. The results shown are representative of two separate gradients.
(B) Fractionation of cMyc-ARG1, PM H+-ATPase, and AtSEC12 in the two-phase partitioning experiment. cMyc-ARG1 was found in both the plasma membrane–enriched upper phase (PM) and the plasma membrane–deprived lower phase (Others). The shift in mobility of cMyc-ARG1 in the plasma membrane fraction likely was an artifact of electrophoresis (see Results). A total of 75 μg of protein was loaded per lane. P150, microsomal membrane fraction; S150, soluble protein fraction.
Because ARG1 likely was associated with multiple membranes, we sought to determine its relative distribution in the plasma membrane versus other membranes. The plasma membrane was fractionated from the microsomal membranes using a two-phase partitioning method (Larsson et al., 1987). Immunoblot analyses showed that the PM H+-ATPase control was enriched in the upper-phase plasma membrane fraction and almost absent from the lower phase, which contains the majority of other membranes (Figure 6B). On the other hand, most of the AtSEC12 was retained in the lower phase, indicating that the plasma membrane fraction was relatively pure. cMyc-ARG1 was found in both fractions (Figure 6B), although there appeared to be less ARG1 per milligram of protein in the plasma membrane fraction than in the other fraction, in agreement with our immunofluorescence and GFP fluorescence data (Figures 4A to 4D, 4H, and 4I) and with the sucrose gradient experiment (Figure 6A). Surprisingly, the mobility of cMyc-ARG1 was altered in the plasma membrane fraction. This mobility shift probably is an electrophoresis artifact, possibly resulting from the high polyethylene glycol content within that phase. Indeed, when a more sensitive detection method was used, some AtSEC12 also was found in the plasma membrane fraction, and its mobility was shifted similarly to cMyc-ARG1 (data not shown).
DISCUSSION
We have analyzed the function and localization of the Arabidopsis ARG1 protein in roots. We showed that ARG1 expression in the statocytes is sufficient to mediate a wild-type gravitropic response. ARG1 is required for the gravity-induced cytosolic alkalinization of the columella cells. We also showed that arg1-2 root caps accumulate a higher level of auxin than do wild-type root caps and that they seem impaired in the generation of a gravity-induced lateral auxin gradient. ARG1 is a peripheral membrane protein that is associated with compartments of the secretory pathway and the cell plates. It also may interact with the cytoskeleton. These observations support a role for ARG1 in gravity signal transduction within the statocytes.
ARG1 Participates in an Early Step of Gravitropism
Analyses of GFP-ARG1 transgenic plants indicated that ARG1 is expressed in multiple root tissues. These include the root cap, the meristem, and the EZs, all of which contribute to root gravitropism (Figures 4A and 4B). However, ARG1 expression in the root cap and the endodermis was sufficient to rescue the gravitropic defects of arg1-2 roots and hypocotyls, respectively (Figures 1A and 1B). Although the expression domain of the RCP1 promoter includes the columella cells as well as other root cap tissues, the SCR promoter is active specifically in the endodermis (Malamy and Benfey, 1997a; Tsugeki and Fedoroff, 1999; Wysocka-Diller et al., 2000). Because these promoters are active in root and hypocotyl statocytes, respectively, it is likely that ARG1 function is only required in these cells for normal gravitropism. Nonetheless, we cannot exclude the possibility that ARG1 also functions in other seedling parts in which functions redundant to those of ARG1 may exist.
Partial rescue of arg1-2 hypocotyl gravitropism was observed when the pRCP1:ARG1 construct was expressed in arg1-2 mutant seedlings. Although the RCP1 promoter was reported to be root cap specific (Tsugeki and Fedoroff, 1999), it remains possible that this promoter also drives a low level of expression in hypocotyls. The generally low level of ARG1 expression driven by the RCP1 and SCR promoters in these transgenic seedlings invalidated our attempts to define expression patterns by in situ hybridization or by in situ reverse transcriptase–mediated PCR analysis (Koltai and Bird, 2000) (data not shown). However, it should be noted that the gravitropic response of arg1-2[pRCP1:ARG1] transgenic hypocotyls remained strongly affected compared with those of wild-type and arg1-2[pSCR:ARG1] hypocotyls (Figure 1B). Hence, together, these data support a role for ARG1 in gravity signal transduction within the statocytes.
arg1-2 Seedlings Lack Gravity-Related pH Changes
Upon gravistimulation, a transient increase in cytoplasmic pH, which was observed in wild-type S3 columella cells (Scott and Allen, 1999; Fasano et al., 2001), was missing from arg1-2 seedlings (Figure 2). Because cytosolic alkalinization of the statocytes is one of the earliest responses to gravistimulation and is required for normal gravitropism in roots (Scott and Allen, 1999; Fasano et al., 2001), it is reasonable to conclude that its disappearance from arg1-2 roots contributes to the arg1 mutant phenotype.
There are at least two possible mechanisms by which the columella cells could achieve cytosolic alkalinization (Scott and Allen, 1999). First, they may upregulate the transport of protons across the plasma membrane through the activation of the PM H+-ATPases or by regulating the activity of other proton antiporter/symporter systems. This mechanism would couple the cytoplasmic pH increase with a concomitant decrease in apoplastic pH (Fasano et al., 2001). Second, the cells could sequester protons into their subcellular compartments, such as the vacuole, via the tonoplast H+-ATPase (Scott and Allen, 1999). Interestingly, PM H+-ATPase accumulates in similar endosomal compartments as ARG1 and the PIN proteins upon BFA treatment (Geldner et al., 2001; Baluska et al., 2002). As discussed below, it is possible that ARG1 and PM H+-ATPase may share the same endomembrane-trafficking pathway, which may have been disturbed in arg1-2 roots.
arg1-2 Affects Auxin Transport within the Root Cap
The DR5::GUS construct, which places the uidA reporter gene under the control of an auxin-responsive promoter, has been instrumental in the analyses of auxin distribution in planta (Ulmasov et al., 1997; Sabatini et al., 1999; Rashotte et al., 2001; Tang et al., 2003). Histochemical analyses of DR5::GUS reporter activities indicated that arg1-2 seedlings might accumulate more auxin in an expanded region of the root cap than do wild-type seedlings (Figures 3A to 3D). A similar phenotype has been reported in wild-type roots upon treatment with N-1-naphthylphthalamic acid, an inhibitor of polar auxin transport (Rashotte et al., 2001). Together, these results suggest that the transport of auxin within arg1-2 root tips is disrupted in both vertically grown and gravistimulated roots.
In gravistimulated wild-type roots, we observed enhanced GUS activities in the peripheral cap cells that are located on the bottom flank of the roots (Figure 3E). Similar observations were reported by Rashotte et al. (2001) as well as by Ottenschläger et al. (2003) using a GFP reporter. This redistribution of reporter activities was not detected in arg1-2 roots (Figure 3F). Although we cannot eliminate the possibility that arg1-2 is affected in specific aspects of auxin homeostasis in the root cap, the data presented here support the hypothesis that arg1-2 might disrupt some regulatory properties of the auxin transport machinery that are needed to generate a gravity-induced gradient. In this regard, it is interesting that homozygous arg1-2 seedlings still exhibit a residual graviresponse (Sedbrook et al., 1999) (Figure 1). Thus, a narrower gravity-induced auxin gradient could have been generated in their root caps but gone undetected because of the presence of high auxin levels in mutant columella cells. This model is consistent with our observation that the arg1-2 distal EZ still was capable of generating a proper auxin gradient when gravistimulated in the presence of NAA (Figure 3H).
Because ARG1 expression in the root cap is sufficient for its function in gravitropism (Figure 1A), the primary auxin transport defects associated with arg1-2 may reside within the root cap itself. Hence, arg1-2 could affect the influx of auxin into the lateral cap cells, its efflux out of the columella cells, or both. Because the AtPIN3 and AtPIN4 auxin efflux carriers are found in columella cells but not in lateral cap cells (Friml et al., 2002a, 2002b) and AtPIN3 is redistributed rapidly within the statocytes in response to gravistimulation (Friml et al., 2002b), it seems possible that ARG1 modulates the function and/or localization of these proteins in the root cap. On the other hand, it is less likely that ARG1 would modulate the activity or localization of AUX1 in peripheral cap cells. ARG1 and AUX1 are expressed in both peripheral and columella cap cells (Figure 4A) (Swarup et al., 2001), whereas enhanced DR5::GUS expression in arg1-2 appears restricted to the columella cells (Figures 3B, 3D, and 3F). ARG1 is not likely to suppress AUX1 activity in one tissue while enhancing it or not affecting it in the other tissue.
ARG1 Cycles between the Plasma Membrane and Intracellular Compartments
Analyses of GFP-ARG1 transgenic seedlings revealed similar punctate and reticulate GFP signals, resembling Golgi stacks, vesicles, and ER, in the EZ and root cap columella cells (Figures 4A and 4B). Treatment of cMyc-ARG1 transgenic seedlings with BFA, an inhibitor of protein trafficking in the endomembrane system (Satiat-Jeunemaitre et al., 1996; Sciaky et al., 1997), caused cMyc-tagged ARG1 to accumulate in the “BFA compartment” (Figure 4D), which appears to be an aggregate that fuses the trans-most Golgi cisternae with endocytic vesicles (reviewed by Nebenführ et al., 2002). This result is compatible with the association of GFP-ARG1 with the trans-Golgi network and/or endocytic vesicles. Similarly, our biochemical analysis indicated the presence of cMyc-tagged ARG1 in the plasma membrane protein fraction (Figure 6B) as well as in other membranes, which may include the ER, the Golgi, and possibly the endosome (Figure 6A). Together, these data indicate that ARG1 is associated with cellular compartments involved in the trafficking of proteins and vesicles between intracellular compartments and the plasma membrane.
Several plasma membrane proteins, such as AtPIN1, PM H+-ATPase, and AGR1/AtPIN2, are found in the BFA compartment (Geldner et al., 2001; Baluska et al., 2002) (Figure 4E). Similarly, AtPIN3 localization within the plasma membrane of columella cells is BFA sensitive (Friml et al., 2002b). Thus, the ability to cycle between the plasma and intracellular membranes appears to be a common property of the PIN proteins (Geldner et al., 2001; Muday and Murphy, 2002; this report). Double immunolabeling experiments indicate that AGR1/AtPIN2 and cMyc-ARG1 accumulate in the same BFA compartment (Figure 4F). Because AGR1/AtPIN2, AtPIN3, and possibly PM H+-ATPase also are involved in gravitropism, it is appealing to hypothesize that ARG1 and some PIN proteins share the same membrane-trafficking pathway. Hence, these same proteins could be targets for ARG1 function.
DnaJ and its homologs encode Hsp40 molecular chaperones (Zuber et al., 1998). The presence of the J domain in ARG1 suggests that it also may participate in protein folding. The tripeptide HPD, which is required for the stimulation of Hsp70, also is conserved in ARG1 (Sedbrook et al., 1999). Because the similarity between ARG1 and DnaJ is limited to the J domain, whereas their C termini, which typically are involved in target recognition, diverge, we speculate that ARG1 recognizes and modulates the folding or activity of an exclusive set of substrates that function in specific physiological responses, such as gravitropism. In arg1-2 mutants, a portion of these proteins may not have been folded properly, leading to impaired cytosolic alkalinization, altered auxin transport in the columella cells, and reduced gravitropic response.
On the other hand, many DnaJ-like proteins have more specialized functions (Zuber et al., 1998). Like ARG1, these proteins share sequence similarity to DnaJ and its homologs only in the J domain. Our studies suggest that ARG1 may cycle between intracellular compartments and the plasma membrane. Hence, we hypothesize that ARG1 may function to regulate the trafficking of vesicles that carry specific cargo proteins such as auxin transporters or PM H+-ATPases between intracellular compartments and the plasma membrane. In arg1-2 mutants, these proteins might not be transported to their proper location at the plasma membrane or they might not be retrieved properly from the membrane when necessary.
The cycling of specific PIN proteins may be important for the regulation of auxin fluxes in the root tip. For instance, the membrane localization of AtPIN3 in the columella cells changes upon gravistimulation from a symmetrical distribution to accumulation at the membrane corresponding to the bottom flank (Friml et al., 2002b). Interestingly, we observed increased accumulation of auxin in the columella cells of nongravistimulated arg1-2 as well as a lack of auxin redistribution in its root cap upon gravistimulation. These data suggest that arg1-2 likely affects auxin fluxes in these cells and that ARG1 function might not be limited to gravitropism.
ARG1 also was present at the cell plate of dividing cells (Figures 4H and 4I). However, arg1 mutants, or their double and triple mutants with ARG1 paralogs, failed to show apparent cytokinetic defects (Sedbrook et al., 1999; Guan et al., 2003). Instead of playing a direct role in cytokinesis, ARG1 could control the localization or polar distribution of specific membrane proteins at the cell plate. Interestingly, AtPIN1 and AGR1/AtPIN2 also are localized to the cell plates (Geldner et al., 2001) (data not shown), further supporting the possible involvement of ARG1 in the localization of PIN proteins or in the regulation of their function.
In arg1-2 roots, AGR1/AtPIN2 still maintains its polar localization at the basal membrane of epidermal and cortical cells and at the outer periclinal membrane of the cortical cells of the EZ (data not shown). This observation is consistent with the ability of the mutant EZ to generate a gravity-induced auxin gradient in the presence of NAA (Figure 3H). As discussed above, ARG1 expression in the root cap was sufficient to rescue the gravitropic defect of arg1-2 roots, and arg1 mutants are not completely agravitropic, suggesting the possibility of redundant functions in regions of the root other than the cap.
Our studies have focused mainly on roots, for reasons including their amenability to whole-mount immunostaining and the spatial separation of the gravity-perceiving and organ curvature sites, allowing for more facile analyses of the early physiological events associated with gravitropism. Here, we establish that ARG1 expression within the hypocotyl statocytes is sufficient to restore normal gravitropism to arg1-2 hypocotyls (Figure 1B). We also show that ARG1 displays similar cellular localization in multiple root tissues (Figures 4A and 4B). Thus, it is likely that ARG1 functions similarly in roots and hypocotyls.
To conclude, our data strongly support a role for ARG1 in regulating the trafficking or function of specific membrane proteins needed for the early phases of gravity signal transduction. Future research involving detailed analyses of PIN protein localization in arg1-2 and the isolation of the molecular players that interact with ARG1 and its gene product should permit us to elucidate the molecular and cellular mechanisms that allow ARG1 to modulate gravity-induced cytosolic alkalinization, auxin transport, and gravity signal transduction within the statocytes.
METHODS
Plant Materials and Growth Conditions
The arg1-2 and agr1-5 mutants were isolated in the Wassilewskija and Estland backgrounds, respectively, of Arabidopsis thaliana (Chen et al., 1998; Sedbrook et al., 1999). pgm-1 (TC7) and DR5::GUS-transformed seeds in the Columbia background (Caspar and Pickard, 1989; Ulmasov et al., 1997) were obtained from T. Caspar (Dupont Central Research and Development, Wilmington, DE) and T.J. Guilfoyle (University of Missouri, Columbia, MO), respectively. arg1-2 plants carrying the DR5::GUS construct were derived from crosses between arg1-2 and the DR5::GUS-transformed plant. Plants homozygous for arg1-2 were identified based on the BamHI restriction polymorphism associated with this allele (Sedbrook et al., 1999). Plants homozygous for the DR5::GUS insertion were identified based on histochemical staining (see below).
Seeds were germinated on germination medium containing half-strength Murashige and Skoog (1962) salts with macronutrients and micronutrients and 1.5% (w/v) sucrose (premixed media obtained from Invitrogen, Carlsbad, CA, and/or Caisson Laboratories, Sugar City, ID) solidified with type-E agar (Sigma, St. Louis, MO). Plant growth conditions were as described (Rutherford and Masson, 1996). For biochemical analyses, seeds were germinated in liquid GM medium on a 100-rpm rotary platform.
Physiological Analyses
Gravitropic Reorientation Assay
Three-day-old seedlings germinated in GM medium containing 0.8% (w/v) agar were gravistimulated by 90° rotation in darkness. Plates were photographed at specified time intervals after gravistimulation with a Nikon COOLPIX800 digital camera (Tokyo, Japan). Root and hypocotyl tip angles were measured using NIH Image software version 1.62 (http://rsb.info.nih.gov/nih-image/).
Measurement of Cytoplasmic pH
Root cells were pressure-microinjected with 2,7-bis-(2-carboxyethyl)-5(and 6)-carboxyfluorescein linked to dextran (molecular weight 10,000) (Molecular Probes, Eugene, OR), and fluorescence ratio images were analyzed and calibrated as described previously (Bibikova et al., 1998). Root cells were categorized as being microinjected successfully if they maintained turgor, cytoplasmic streaming, and cytoplasmic structure during the experiment. Seedlings were examined with a Nikon Diaphot 300 epifluorescence microscope mounted on its back so that its rotatable stage was vertical with a Lambda 10-c filter wheel (Sutter, Novato, CA) in the excitation path. Each root was imaged while growing vertically and upon gravistimulation by rotating the vertical stage through 90°. Roots were observed with 440- or 480-nm excitation interference filters (20-nm half bandwidth), a 490-nm dichroic mirror, and a 520- to 560-nm emission filter (Omega Optical, Brattleboro, VT). Images were collected using a SenSys cooled charge-coupled device camera (Photometrics, Austin, TX) and processed using IPLabs Spectrum image-analysis software (Scanalytics, Fairfax, VA).
Molecular Cloning
Standard molecular techniques were used (Sambrook et al., 1989). Restriction enzymes and T4 DNA ligase were obtained from New England Biolabs (Beverly, MA). Native Pfu DNA polymerase for PCR was obtained from Stratagene (La Jolla, CA). Primers were custom-ordered from Integrated DNA Technologies (Coralville, IA).
To generate the cMyc-ARG1 and GFP-ARG1 constructs, a XhoI-SacI fragment of the genomic BAC 4N22 DNA (Sedbrook et al., 1999), containing the ARG1 promoter and part of the ARG1 open reading frame (ORF), was subcloned into pBluescript KS− (Stratagene). The ARG1 promoter region was amplified from this plasmid with a T3 promoter primer and 5′-GGGTCTAGAGGGCCCGGGCTTCTCGAAGAATTTGAA-3′. For the cMyc-ARG1 construct, part of the ARG1 coding region was amplified by PCR with the primer 5′-CCCTCTAGAATGGAGCAAAAGCTTATCAGTGAGGAAGACTTGAGCGCGAAAAAGCTTG-3′, containing a cMyc epitope (DeWitt and Sussman, 1995), and a T7 promoter primer. The promoter and the coding region were ligated using the XbaI sites that were designed into the primers. The product was used to replace the XhoI-SacI ARG1 wild-type sequence in an Asp718 fragment of BAC 4N22 that had been subcloned previously into pBIN19 (Sedbrook et al., 1999). For the GFP-ARG1 construct, the ARG1 coding region was amplified with the primers 5′-CCCTCTAGAATGAGCGCGAAAAAGC-3′ and T7. This fragment was ligated to the ARG1 promoter and subcloned into pBIN19 as described above. The mGFP5 ORF lacking the basic chitinase endoplasmic reticulum signal sequence was amplified from pBIN m-gfp5-ER (provided by Jim Haseloff) with the primers 5′-CCCCCCGGGATGAGTAAAGGAGAAGAAC-3′ and 5′-GGGTCTAGAAAGCTCATCATGTTTG-3′ and subcloned between the SmaI and XbaI sites of the same plasmid immediately upstream of the ARG1 coding region.
To generate the pSCR:ARG1 construct, the SCR promoter (Malamy and Benfey, 1997a; Wysocka-Diller et al., 2000) was amplified from Wassilewskija genomic DNA using the primer pair 5′-AGCTCTGCAGTGGTCCGGTGGTCT-3′ and 5′-ATTCGTCGACGGAGATTGAAGGGTTGT-3′. For the pRCP1:ARG1 construct, the RCP1 promoter was amplified from plasmid pKSSS (Tsugeki and Fedoroff, 1999) provided by N.V. Fedoroff using the primer pair 5′-CCCCCTGCAGGTCACTTTCTAAATGT-3′ and 5′-TATCGTCGACGTCACTTGGGTTTGGAACAG-3′. The ARG1 ORF was amplified from the cDNA (Sedbrook et al., 1999) with the primer pair 5′-AAATTAGTCGACAAGATGAGCGCGAAAAAGCT-3′ and 5′-AACAAACCCGGGCTCTGATCAACCAAGCTTC-3′. The promoters were subcloned into pCAMBIA1390 (CAMBIA, Canberra, Australia; provided by A. Bleecker) between the PstI and SalI sites. The ARG1 ORF then was subcloned into the resulting plasmids between the SalI and XmaI sites to obtain pSCR:ARG1 and pRCP1:ARG1, which carry the SCR and RCP1 promoters, respectively.
Transformations of Arabidopsis with the constructs described above were performed as described by Bent (2000).
Biochemical Analyses
Preparation of Microsomes
Total proteins were isolated from 3-week-old plants grown in liquid cultures. Plant tissues were homogenized with a Dounce homogenizer (Kontes Glass Co., Vineland, NJ) in a 2× volume of MIB [20 mM Hepes-KOH, pH 7.0, 50 mM KOAc, 1 mM Mg(OAc)2, 250 mM sorbitol, 1 mM phenylmethylsulfonyl fluoride (PMSF), a 1:200 dilution of protease inhibitor cocktail for plant extracts (PIC; Sigma), and 1 mM DTT]. A postnuclear supernatant (S1) was prepared by centrifugation at 1000g for 10 min at 4°C. The microsomes were prepared by centrifugation at 150,000g for 45 min at 5°C. The supernatant (S150) was separated from the pellet (P150), which was resuspended in MIB for further analyses. Protein concentrations were determined using the Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA) with bovine plasma γ-globulin as the standard.
Analysis of Membrane Association
The microsomes (P150) were diluted (>20 fold) in MIB or MIB lacking DTT but containing NaCl, urea, KI, Triton X-100, or Na2CO3 at the concentrations specified in Results. The dilutions were incubated at room temperature for 30 min and centrifuged at 300,000g for 30 min at 20°C. Supernatants were concentrated by precipitation as described by Aguilar et al. (1999). Pellets were rinsed and resuspended in MIB.
Analysis of Cytoskeleton Association
cMyc-ARG1 association with the cytoskeleton was analyzed using a variation of the procedures described by Tan and Boss (1992) and Cox and Muday (1994). Briefly, cytoskeleton-enriched pellets were prepared from the microsomes by treatment with 1% (v/v) Triton X-100 in CSB (5 mM Hepes-KOH, pH 7.0, 10 mM MgCl2, 2 mM EGTA, 0.1 mM sodium orthovanadate, 1 mM PMSF, and a 1:200 dilution of PIC) for 20 min at 4°C followed by a 20 min centrifugation at 50,000g. The pellets were treated for 20 min at 4°C with 0.5 M KI in 5 mM Hepes-KOH, pH 7.0, 100 mM KCl, 2 mM EGTA, 0.1 mM sodium orthovanadate, 1 mM PMSF, and a 1:200 dilution of PIC to depolymerize the cytoskeleton. Then, the samples were centrifuged at 40,000g for 20 min at 4°C. The resulting supernatant was dialyzed against 3 L of CSB (with 5 mM benzamidine hydrochloride instead of PIC) over a 24-h period at 4°C to remove KI. Repolymerized cytoskeletal elements were collected after a 5-min centrifugation at 12,000g and 4°C. The pellets were resuspended in CSB.
Sucrose Gradient Fractionation
A total of 3.5 mg of a postnuclear supernatant (S1) was applied to a 10-mL continuous gradient of 14 to 50% (w/w) sucrose in MIB with a 0.5-mL 60% (w/w) sucrose cushion. The gradient was centrifuged for 2 h at 150,000g and 4°C. Fractions (0.4 mL) were collected as described previously (Rancour et al., 2002).
Preparation of Plasma Membrane by Two-Phase Partitioning
Plants were homogenized in Serrano buffer (0.25 M Tris-HCl, pH 8.5, 25 mM EDTA, 1.15 M sucrose, 1 mM PMSF, and β-mercaptoethanol), and the microsomes were prepared as described above. P150 pellets were resuspended in 0.33 M sucrose, 4 mM KCl, and 5 mM K-phosphate, pH 7.8, and applied to the phase mixture (Larsson et al., 1987). After mixing, phases were separated by a 5-min centrifugation at 1000g and 4°C. The upper phase was purified twice more against fresh lower phase solution (obtained from the phase system; Larsson et al., 1987). The upper phase was diluted in a 2× volume of 10 mM Tris-HCl, pH 7.5, and 0.33 M sucrose. The lower phases were pooled and diluted four times in the same buffer. Membranes were pelleted at 150,000g and 5°C for 45 min.
Protein Gel Blot Analyses
Protein gel blot analyses were performed as described (Qiagen U.S.A., 2001). Anti-cMyc, clone 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-actin, clone C4 (Chemicon International, Temecula, CA), and anti-α-tubulin antibodies, clone B-5-1-2 (Sigma), were used at dilutions of 1:200, 1:1000, and 1:3000, respectively. Anti-AtSEC12, anti-AtPEP12, anti-PM H+-ATPase, anti-vacuolar pyrophosphatase, and anti-α-mannosidase antibodies were provided by S.Y. Bednarek and used at a dilution of 1:1000 (DeWitt et al., 1996; Bar-Peled and Raikhel, 1997; da Silva Conceição et al., 1997; Rancour et al., 2002; S.Y. Bednarek, personal communication). Horseradish peroxidase–conjugated goat anti-mouse IgG and goat anti-rabbit IgG antibodies (Pierce, Rockford, IL) were used at a dilution of 1:10,000.
Histochemical Staining for GUS Activity
Seedlings were incubated for up to 24 h at 37°C in 70 mM sodium phosphate, pH 7.0, containing 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, cyclohexylammonium salt (Research Organics, Cleveland, OH), 2 mM K3Fe(CN)6, and 10% (v/v) methanol, unless stated otherwise. Samples were cleared subsequently as described by Malamy and Benfey (1997b). Gravistimulated samples on medium without 1-naphthaleneacetic acid were stained as described in the legend to Figure 3 for equal intensity and cleared with chloral hydrate:water:glycerol (8:2:1, w/w/v). Samples were mounted in 50% (v/v) glycerol for examination with a Nikon Optiphot 2 microscope equipped with Nomarski optics.
Immunofluorescence Staining and Microscopy
Immunolocalization of ARG1 and PIN proteins was as described by Müller et al. (1998) with modifications. Four-day-old seedlings were fixed in 4% (w/v) p-formaldehyde in PME (50 mM Pipes-KOH, pH 6.9, 5 mM MgSO4, and 10 mM EGTA) for 1 h at 25 mm Hg. For cell wall digestion, samples were incubated for 20 min in 0.5% (w/v) macerozyme R-10 and 0.1% (w/v) pectolyase Y-23 in PME. Primary and secondary antibodies were diluted in PME containing 3% (w/v) BSA and 1% (v/v) goat serum. Colocalization with α-tubulin was as described by Goodbody and Lloyd (1994) with modifications. Samples were fixed in 4% (w/v) p-formaldehyde in 50 mM Pipes-KOH, pH 7.0, and 1 mM CaCl2 for 1 h at 25 mm Hg. Cell wall digestions and antibody incubations were as described above but using Pipes/CaCl2 buffer. In all cases, samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and imaged with a Bio-Rad MRC-1024 laser scanning confocal microscope (Keck Biological Imaging Laboratory, University of Wisconsin, Madison) with excitation lines of 488, 568, and 647 nm and emission band-path filter sets of 522, 605, and 680 nm. Anti-cMyc antibodies, clone 9E10 and A-14 (Santa Cruz Biotechnology), were used at a dilution of 1:200. Anti-α-tubulin, clone YOL1/34 (Harlan, Indianapolis, IN), was used at a dilution of 1:100. Anti-AGR1/AtPIN2 polyclonal antibodies were raised against a peptide containing amino acids 379 to 503 of the AGR1/AtPIN2 protein, affinity-purified as described (Harlow and Lane, 1988), and used at a dilution of 1:200. Rhodamine-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) and fluorescein isothiocyanate–conjugated goat anti-mouse Fab antibodies (Sigma) were used at a dilution of 1:300.
For visualization of GFP signals, excised roots from 4-day-old seedlings were mounted live in water and examined with a Bio-Rad MRC-1024 confocal microscope (Keck Biological Imaging Laboratory) with a 488-nm excitation line and a 522-nm emission band-path filter or a multiphoton optical workstation equipped with a Ti:sapphire laser tuned to 900 nm (Wokosin et al., 2003) (Laboratory for Optical and Computational Instrumentation, University of Wisconsin).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Patrick H. Masson, phmasson@wisc.edu
Acknowledgments
We thank Sebastian Bednarek and David Rancour for guidance with biochemical procedures, Lance Rodenkirch at the Keck Biological Imaging Laboratory for support with confocal microscopy, John White and Jayne Squirrell at the Laboratory for Optical and Computational Instrumentation for help with multiphoton microscopy, and Changhui Guan and Christen Yuen for helpful discussions. This research was supported by grants from the National Science Foundation (MCB 99-05675 and MCB 02-40084) and the National Aeronautics and Space Administration (NAG2-1336 and NAG2-1602) to P.H.M., by a grant from the National Science Foundation (MCB 02-12099) to S.G., and by a fellowship from the Thai government to K.B. This is article 3615 from the Laboratory of Genetics.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015560.
References
- Aguilar, R.M., Bustamante, J.J., Hernandez, P.G., Martinez, A.O., and Haro, L.S. (1999). Precipitation of dilute chromatographic samples (ng/ml) containing interfering substances for SDS-PAGE. Anal. Biochem. 267, 344–350. [DOI] [PubMed] [Google Scholar]
- Artigues, A., Iriarte, A., and Martinez-Carrion, M. (2002). Binding to chaperones allows import of a purified mitochondrial precursor into mitochondria. J. Biol. Chem. 277, 25047–25055. [DOI] [PubMed] [Google Scholar]
- Baluska, F., Hlavacka, A., Samaj, J., Palme, K., Robertson, G.D., Matoh, T., McCurdy, D.W., Menzel, D., and Volkmann, D. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells: Insights from brefeldin A-induced compartments. Plant Physiol. 130, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled, M., and Raikhel, N.V. (1997). Characterization of AtSEC12 and AtSAR1 proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 114, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273, 948–950. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (2000). Arabidopsis in planta transformation: Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 124, 1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, T.N., Jacob, T., Dahse, I., and Gilroy, S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934. [DOI] [PubMed] [Google Scholar]
- Blancaflor, E., Fasano, J., and Gilroy, S. (1998). Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 116, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsirichai, K., Guan, C., Chen, R., and Masson, P.H. (2002). Root gravitropism: An experimental tool to investigate basic cellular and molecular processes underlying mechanosensing and signal transmission in plants. Annu. Rev. Plant Biol. 53, 421–447. [DOI] [PubMed] [Google Scholar]
- Caspar, T., and Pickard, B. (1989). Gravitropism by a starchless mutant of Arabidopsis: Implications for the starch-statolith theory of gravity sensing. Planta 177, 185–197. [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95, 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D.N., and Muday, G.K. (1994). NPA binding activity is peripheral to the plasma membrane and is associated with the cytoskeleton. Plant Cell 6, 1941–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Conceição, A., Marty-Mazars, D., Bassham, D.C., Sanderfoot, A.A., Marty, F., and Raikhel, N.V. (1997). The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9, 571–582. [PMC free article] [PubMed] [Google Scholar]
- Delbarre, A., Muller, P., and Guern, J. (1998). Short-lived and phosphorylated proteins contribute to carrier-mediated efflux, but not to influx, of auxin in suspension-culture tobacco cells. Plant Physiol. 116, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt, N.D., Hong, B., Sussman, M.R., and Harper, J.F. (1996). Targeting of two Arabidopsis H+-ATPase isoforms to the plasma membrane. Plant Physiol. 112, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt, N.D., and Sussman, M.R. (1995). Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7, 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano, J.M., Swanson, S.J., Blancaflor, E.B., Dowd, P.E., Kao, T., and Gilroy, S. (2001). Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13, 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Liung, K., Woody, S., Sandberg, G., Scheres, B., Jürgens, G., and Palme, K. (2002. a). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., and Palme, K. (2002). Polar auxin transport: Old questions and new concepts? Plant Mol. Biol. 49, 273–284. [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002. b). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). The RHG gene is involved in root and hypocotyl gravitropism in Arabidopsis thaliana. Plant Cell Physiol. 38, 804–810. [DOI] [PubMed] [Google Scholar]
- Garbers, C., DeLong, A., Deruère, J., Bernasconi, P., and Soll, D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15, 2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.D., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Goodbody, K.C., and Lloyd, C.W. (1994). Immunofluorescence techniques for analysis of the cytoskeleton. In Plant Cell Biology: A Practical Approach, N. Harris and K.J. Oparka, eds (Oxford, UK: IRL Press), pp. 221–243.
- Guan, C., Rosen, E.S., Boonsirichai, K., Poff, K.L., and Masson, P.H. (2003). The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 133, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Kiss, J., Hertel, R., and Sack, F. (1989). Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177, 198–206. [PubMed] [Google Scholar]
- Koltai, H., and Bird, D.M. (2000). High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiol. 123, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, O.A., and Hasenstein, K.H. (1996). Intracellular magnetophoresis of amyloplasts and induction of root curvature. Planta 198, 87–94. [DOI] [PubMed] [Google Scholar]
- Larsson, C., Widell, S., and Kjellbom, P. (1987). Preparation of high-purity plant membranes. Methods Enzymol. 148, 558–568. [Google Scholar]
- Lemmon, S.K. (2001). Clathrin uncoating: Auxilin comes to life. Curr. Biol. 11, R49–R52. [DOI] [PubMed] [Google Scholar]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997. a). Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J. 12, 957–963. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997. b). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Marchant, A., Kargul, J., May, S.T., Muller, P., Delbarre, A., Perrot-Rechenmann, C., and Bennett, M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18, 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, P.H., Tasaka, M., Morita, M.T., Guan, C., Chen, R., and Boonsirichai, K. (2002). Arabidopsis thaliana: A model for the study of root and shoot gravitropism. In The Arabidopsis Book, E.M. Meyerowitz and C.R. Somerville, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0043, http://www.aspb.org/publications/arabidopsis. [DOI] [PMC free article] [PubMed]
- McClellan, A.J., and Brodsky, J.L. (2000). Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics 156, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk, J. (2001). The J-domain proteins of Arabidopsis thaliana: An unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday, G.K., and Murphy, A.S. (2002). An emerging model of auxin transport regulation. Plant Cell 14, 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A., Guan, C., Gälweiler, L., Tänzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nebenführ, A., Ritzenthaler, C., and Robinson, D.G. (2002). Brefeldin A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger, I., Wolff, P., Wolverton, C., Bhalerao, R.P., Sandberg, G., Ishikawa, H., Evans, M., and Palme, K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100, 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plieth, C., and Trewavas, A.J. (2002). Reorientation of seedlings in the earth's gravitational field induces cytosolic calcium transients. Plant Physiol. 129, 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiagen U.S.A. (2001). The QIAGEN guide to analytical gels. Part IX. Gel analysis: Specific antibodies-mediated detection of proteins on a membrane. Qiagen News 2, 24–28. [Google Scholar]
- Rancour, D.M., Dickey, C.E., Park, S., and Bednarek, S.Y. (2002). Characterization of AtCDC48: Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 130, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., DeLong, A., and Muday, G.K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13, 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, R., and Masson, P.H. (1996). Arabidopsis thaliana sku mutant seedlings show surface-dependent alteration in root growth vector. Plant Physiol. 111, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Saint-Jore, C.M., Evins, J., Batoko, H., Brandizzi, F., Moore, I., and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29, 661–678. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Satiat-Jeunemaitre, B., Cole, L., Bouret, T., Howard, R., and Hawes, C. (1996). Brefeldin A effects in plant and fungal cells: Something new about vesicle trafficking? J. Microsc. 181, 162–177. [DOI] [PubMed] [Google Scholar]
- Sciaky, N., Presley, J., Smith, C., Zaal, K.J.M., Cole, N., Moreira, J.E., Terasaki, M., Siggia, E., and Lippincott-Schwartz, J. (1997). Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 137, 1137–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A.C., and Allen, N.S. (1999). Changes in cytosolic pH within Arabidopsis root columella cells play a key role in the early signaling pathway for root gravitropism. Plant Physiol. 121, 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., Chen, R., and Masson, P.H. (1999). ARG1 (Altered Response to Gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc. Natl. Acad. Sci. USA 96, 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, A., and Busch, M.B. (1992). An inhibitor of the Ca2+-ATPases in the sarcoplasmic and endoplasmic reticula inhibits transduction of the gravity stimulus in cress roots. Planta 188, 619–622. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Friml, J., Marchant, A., Liung, K., Sandberg, G., Palme, K., and Bennett, M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15, 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Z., and Boss, W.F. (1992). Association of phosphatidylinositol kinase, phosphatidylinositol monophosphate kinase, and diacylglycerol kinase with the cytoskeleton and F-actin fractions of carrot (Daucus carota L.) cells grown in suspension culture. Plant Physiol. 100, 2116–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., Brady, S.R., Sun, Y., Muday, G.K., and Roux, S.J. (2003). Extracellular ATP inhibits root gravitropism at concentrations that inhibit polar auxin transport. Plant Physiol. 131, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, M.-Y., Morfini, G., Szebenyi, G., and Brady, S.T. (2000). Release of kinesin from vesicles by hsc70 and regulation of fast axonal transport. Mol. Biol. Cell 11, 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki, R., and Fedoroff, N.V. (1999). Genetic ablation of root cap cells in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 12941–12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuno, K., Shikanai, T., Yamada, Y., and Hashimoto, T. (1998). AGR, an Agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 39, 1111–1118. [DOI] [PubMed] [Google Scholar]
- Wokosin, D.L., Squirrell, J.M., Eliceiri, K.W., and White, J.G. (2003). Optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities. Rev. Sci. Instrum. 74, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller, J.W., Helariutta, Y., Fukaki, H., Malamy, J.E., and Benfey, P.N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595–603. [DOI] [PubMed] [Google Scholar]
- Yoder, T.L., Zheng, H., Todd, P., and Staehelin, L.A. (2001). Amyloplast sedimentation dynamics in maize columella cells support a new model for the gravity-signal apparatus of roots. Plant Physiol. 125, 1045–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber, U., Buchberger, A., Laufen, T., and Bukau, B. (1998). DnaJ proteins. In Molecular Chaperones in the Life Cycle of Proteins: Structure, Function and Mode of Action, A.L. Fink and Y. Goto, eds (New York: Marcel Dekker), pp. 241–273.