Abstract
Plants have evolved sophisticated defense mechanisms against pathogen infections, during which resistance (R) genes play central roles in recognizing pathogens and initiating defense cascades. Most of the cloned R genes share two common domains: the central domain, which encodes a nucleotide binding adaptor shared by APAF-1, certain R proteins, and CED-4 (NB-ARC), plus a C-terminal region that encodes Leu-rich repeats (LRR). In Arabidopsis, a dominant mutant, suppressor of npr1-1, constitutive 1 (snc1), was identified previously that constitutively expresses pathogenesis-related (PR) genes and resistance against both Pseudomonas syringae pv maculicola ES4326 and Peronospora parasitica Noco2. The snc1 mutation was mapped to the RPP4 cluster. In snc1, one of the TIR-NB-LRR–type R genes contains a point mutation that results in a single amino acid change from Glu to Lys in the region between NB-ARC and LRR. Deletions of this R gene in snc1 reverted the plants to wild-type morphology and completely abolished constitutive PR gene expression and disease resistance. The constitutive activation of the defense responses was not the result of the overexpression of the R gene, because its expression level was not altered in snc1. Our data suggest that the point mutation in snc1 renders the R gene constitutively active without interaction with pathogens. To analyze signal transduction pathways downstream of snc1, epistasis analyses between snc1 and pad4-1 or eds5-3 were performed. Although the resistance signaling in snc1 was fully dependent on PAD4, it was only partially affected by blocking salicylic acid (SA) synthesis, suggesting that snc1 activates both SA-dependent and SA-independent resistance pathways.
INTRODUCTION
Plants use different mechanisms to fight against microbial pathogen infections. One of the main mechanisms is disease resistance mediated by plant resistance (R) genes (Staskawicz et al., 1995). Recognition of pathogens carrying Avr genes by the cognate R genes often leads to a localized hypersensitive response (HR) and restriction of pathogen spread (Hammond-Kosack and Jones, 1996). A number of R genes have been cloned from different species, and most of them encode proteins that contain nucleotide binding site (NB) and Leu-rich repeat (LRR) domains (Bent, 1996; Hammond-Kosack and Jones, 1997; Ellis et al., 2000). Some R proteins also contain a conserved ARC domain located between the NB and the LRR (van der Biezen and Jones, 1998a).
Because of the specificity of the genetic interactions between R genes and their cognate Avr genes, NB-LRR proteins have long been suggested to be the receptors of pathogen-derived ligands encoded by the Avr genes, and binding of the ligands to the NB-LRR proteins was believed to activate the downstream signal transduction cascade (Staskawicz et al., 1995; Baker et al., 1997). Recently, it was proposed that the interaction of the NB-LRR and Avr proteins is indirect and may require a third host protein as a mediator (van der Biezen and Jones, 1998b; Dangl and Jones, 2001; Bonas and Lahaye, 2002). Activation of NB-LRR proteins results from the modification of host proteins by pathogen effectors rather than from direct interactions between NB-LRR and Avr proteins. This model is strongly supported by studies of RIN4, a protein that was identified in a yeast two-hybrid screen for plant proteins that interact with AvrB (Mackey et al., 2002), and its interaction with RPS2, an NB-LRR protein that recognizes bacterial pathogens that express AvrRpt2 (Innes et al., 1993; Bent et al., 1994; Mindrinos et al., 1994; Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). AvrRpt2 causes the elimination of RIN4 during the activation of the RPS2 pathway. Suppression of RIN4 by gene silencing activates defense responses constitutively. On the other hand, overexpression of RIN4 inhibits RPS2 function. The constitutive activation of defense responses caused by the loss of RIN4 function requires a functional RPS2. These data suggest that RPS2 senses the disappearance of RIN4 induced by AvrRpt2 and that the dissociation of RPS2 from RIN4 results in the activation of the downstream signal transduction cascade.
The localized HR that results from the recognition of pathogen Avr genes by the cognate plant R genes also triggers a secondary defense response termed systemic acquired resistance (SAR) in uninfected leaves (Ryals et al., 1996). Arabidopsis NPR1 is an essential regulator of SAR (Cao et al., 1997; Ryals et al., 1997). Mutations in NPR1 abolish both pathogenesis-related (PR) gene expression and resistance induced by avirulent pathogens or SAR-inducing agents such as salicylic acid (SA) (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). snc1 (suppressor of npr1-1, constitutive1) is an Arabidopsis mutant isolated from a screen for suppressors of npr1-1 (Li et al., 2001). The snc1 mutation results in constitutive PR gene expression and resistance to Pseudomonas syringae pv maculicola (P.s.m.) ES4326 and Peronospora parasitica Noco2 in the npr1-1 background. snc1 plants are smaller than wild-type plants, accumulate high levels of SA, and often have curly leaves. Although snc1/SNC1 plants constitutively express PR genes, their morphology is indistinguishable from that of wild-type plants. It appears that both copies of SNC1 must be mutated for the mutant morphological phenotypes to be visible.
Previously, the snc1 mutation was mapped to a 120-kb region on chromosome 4. This region contains a cluster of RPP5 orthologs, including the recently cloned RPP4 (Parker et al., 1997; van der Biezen et al., 2002). The snc1 mutant phenotypes are suppressed completely by eds1, but not by ndr1, suggesting that SNC1 is located upstream of EDS1 and that one of the RPP5 homologs is activated constitutively in snc1 (Li et al., 2001). Here, we report the identification of snc1 and discuss the molecular mechanism by which this mutation affects R gene–mediated defense.
RESULTS
snc1 Contains a Mutation in At4g16890
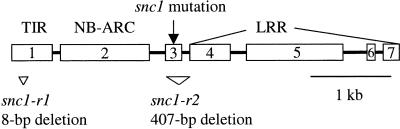
To identify the molecular lesion in snc1, PCR fragments covering the entire 120-kb region to which snc1 was mapped were amplified from snc1 DNA and sequenced. The sequence then was compared with that of the wild type, and a single G-to-A mutation was found in the coding region of At4g16890 (Figure 1), suggesting that SNC1 is At4g16890. The cDNA sequence of the gene was obtained by sequencing reverse transcriptase–mediated (RT) PCR fragments and found to be consistent with the annotation of At4g16890. At4g16890 encodes a putative protein of 1468 amino acids that is highly similar to RPP4 (62% identical) and RPP5 (68% identical), two closely related R genes of the Toll Interleukin1 Receptor (TIR)-NB-LRR class. As shown in Figure 1, the TIR and NB-ARC domains are encoded by exon 1 and 2, respectively. A stretch of 80 to 90 amino acids is encoded by exon 3. The snc1 mutation is located in this region, which recently was named NL linker (Meyers et al., 2003). The remaining exons towards the 3′ end encode the LRRs. The snc1 mutation converts the Glu at position 552 to a Lys in the NL linker.
Figure 1.
Gene Structure of SNC1 (At4g16890).
Exons (rectangles) and introns (lines) are drawn in proportion to their lengths. Exons encoding the TIR (exon 1), NB-ARC (exon 2), and LRR (exons 4 to 7) domains are indicated. The position of the G-to-A mutation in snc1 is indicated by the arrow. The two revertant mutations (snc1-r1 and snc1-r2) are shown as triangles.
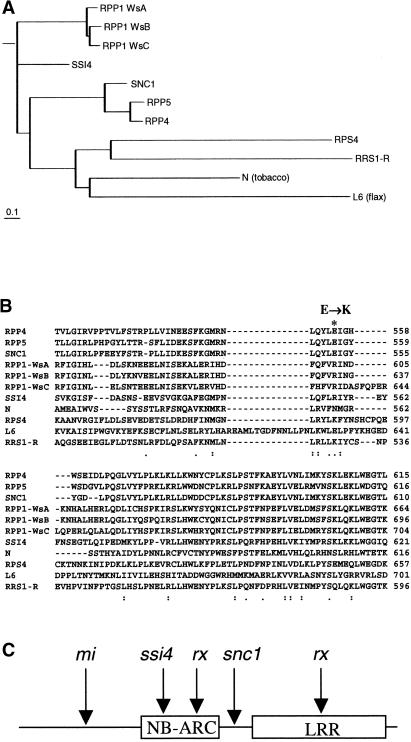
To investigate the relationship between SNC1 and other TIR-NB-LRR–type R proteins, full-length amino acid sequences of R proteins that were shown previously to be functional were used to generate a phylogenetic tree (Figure 2A). Alignment of the amino acid sequences encoded by exon 3 of SNC1 and the corresponding sequences from other TIR-NB-LRR R proteins also was performed (Figure 2B). Although Glu-552 is conserved among SNC1, RPP4, and RPP5, other TIR-NB-LRR–type R proteins contain different residues at this position. However, closely related R proteins tend to have the same corresponding residues. Among the aligned R proteins, all except one have a charged amino acid at this position, suggesting potential functional conservation of these residues.
Figure 2.
Sequence Analysis of SNC1 and the snc1 Mutation.
(A) Phylogenetic relationship of SNC1 and other TIR-NBS-LRR–type R proteins. The neighbor-joining tree was constructed with BIONJ (Gascuel, 1997) using proteins aligned with CLUSTAL W. Distances were calculated with TREE-PUZZLE 5.0 (Strimmer and von Haeseler, 1996) using the WAG substitution matrix.
(B) Amino acid sequence comparison of SNC1 exon 3 with other TIR-NBS-LRR proteins. The alignment was performed using CLUSTAL W. The predicted Glu-to-Lys (E→K) change in snc1 is highlighted (asterisk). Colons indicate high amino acid identity, and dots indicate low identity. RPP4, Resistance to Peronospora parasitica 4 (van der Biezen et al., 2002); RPP5, Resistance to P. parasitica 5 (Parker et al., 1997); RPP1-WsA to RPP1-WsC, Resistance to P. parasitica 1, Arabidopsis thaliana accession Wassilewskija A to C (Botella et al., 1998); SSI4, suppressor of salicylic acid insensitivity of npr1-5, 4 (Shirano et al., 2002); N, Nicotiana glutinosa virus resistance gene (Whitham et al., 1994); RPS4, Resistance to Pseudomonas syringae 4 (Gassmann et al., 1999); L6, Linum usitatissimum rust resistance gene (Lawrence et al., 1995); RRS1-R, Resistance to Ralstonia solanacearum 1-recessive (Deslandes et al., 2002).
(C) Locations of the snc1 mutation and amino acid changes in confirmed gain-of-function R protein variants that cause constitutive HR.
To compare the mutation in the snc1 protein with other gain-of-function R protein variants, the location of these mutations is summarized in Figure 2C. In the tomato Mi protein, there is an unusually long N-terminal extension. Replacing the N-terminal 161 amino acids with the corresponding region of a nonfunctional homolog causes localized HR when expressed transiently in Nicotiana benthamiana leaves (Hwang et al., 2000). Several rx mutations also were found to constitutively activate HR in transient assays (Bendahmane et al., 2002). These mutations are located in either the NB or the LRR region. Both Mi and Rx encode coiled-coil (CC)-NB-LRR–type R proteins. The lesion mimic Rp1-D–21 mutant also contains a rearrangement in the LRR-encoding region of a Rp1-D haplotype, but it is unclear whether the lesion mimic phenotype is actually caused by this mutant haplotype (Sun et al., 2001). Recently, a mutation in the NB region of a TIR-NB-LRR–type R protein was found to cause spontaneous lesions in ssi4 (Shirano et al., 2002). The mutation in snc1 is the only one located in the NL linker region, and it is the only confirmed R gene mutation that causes constitutive disease resistance in the absence of cell death.
Deletions in At4g16890 Revert the snc1 Mutant to Wild Type
To confirm that the mutation in At4g16890 is responsible for the snc1 phenotypes, we performed a genetic screen to identify revertant mutations of the snc1 gene that restored wild-type morphology. Seeds of snc1 npr1-1 were mutagenized by fast neutron bombardment, and M2 plants were screened for mutants with wild-type morphology. M3 seeds from the candidate mutants were planted again to test for segregation of the wild-type and snc1 morphologies. Because snc1 is a dominant mutation and snc1/SNC1 heterozygous plants have wild-type morphology, mutants that produce segregating progeny are likely to have defects in one copy of the snc1 mutant gene.
Indeed, when nine such mutants were analyzed by PCR for mutations in At4g16890, eight had either large deletions or rearrangements detectable by agarose gel electrophoresis. All eight mutations affected at least part of the At4g16890 coding region, with some also affecting neighboring genes. In one case, the entire exon 3 that contains the original snc1 mutation was deleted (snc1-r2; Figure 1). In the only mutant with no large deletion detected, a small deletion of 8 bp (snc1-r1; Figure 1) was identified in the first exon by direct sequencing of snc1-r1.
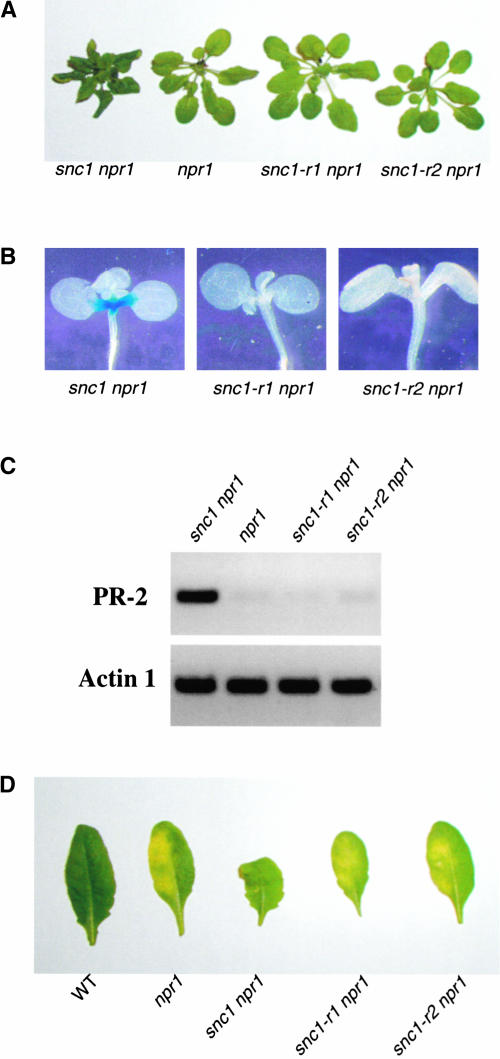
We selected snc1-r1 npr1-1 and snc1-r2 npr1-1 for further characterization because the deletions in these two mutants were within At4g16890. As shown in Figure 3A, snc1-r1 npr1-1 and snc1-r2 npr1-1 plants were larger than snc1 npr1-1 plants and no longer exhibited curly leaves. Both snc1-r1 npr1-1 and snc1-r2 npr1-1 had completely lost the constitutive expression of the pBGL2-β-glucuronidase (GUS) reporter gene (Figure 3B). RT-PCR analysis revealed that constitutive expression of the endogenous BGL2 (PR-2) gene also was abolished in these plants (Figure 3C). In addition, both mutants exhibited enhanced susceptibility to P.s.m. ES4326, a phenotype similar to that of npr1-1 plants (Figure 3D). Overall, these data indicate that deletions in the mutated At4g16890 reversed the phenotypes of the snc1 npr1-1 mutant to those of npr1-1.
Figure 3.
Phenotypic Analysis of snc1 Revertants.
(A) Morphological phenotypes of the snc1 revertants. All plants were grown in parallel on soil and photographed when they were 4 weeks old.
(B) Suppression of snc1-induced pBGL2-GUS reporter gene expression in the revertants. Three-week-old seedlings grown on MS medium were stained for GUS activity from pBGL2-GUS.
(C) PR-2 (BGL2) gene expression in snc1 revertants. RNAs were prepared from 20-day-old plants grown on MS medium and reverse transcribed to obtain total cDNA. The cDNA samples were normalized by real-time PCR using an Actin1 probe. PR-2 and Actin1 in different mutant plants then were amplified by 35 cycles of PCR using equal amounts of total cDNA, and the products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
(D) Enhanced disease susceptibility of snc1 revertants to P.s.m. ES4326. Leaves from 4-week-old plants were infiltrated with a P.s.m. ES4326 suspension in 10 mM MgCl2 (OD600 = 0.0001). Photographs were taken of representative leaves 4 days after infection. WT, wild type.
SNC1/snc1 heterozygous plants were shown previously to constitutively express the pBGL2-GUS reporter gene (Li et al., 2001). To determine whether snc1 plants containing deletions in one copy of the snc1 gene (referred to hereafter as snc1/−) still express pBGL2-GUS, seeds from heterozygous revertants were plated on MS medium (Murashige and Skoog, 1962) and the seedlings were stained for expression of the GUS reporter gene. We found that 36 of 47 plants from seeds of snc1/snc1-r1 did not stain and 23 of 28 plants from seeds of snc1/snc1-r2 did not stain, suggesting that snc1/− plants do not constitutively express pBGL2-GUS. This is consistent with the finding that no pBGL2-GUS expression was observed in the leaves of heterozygous snc1 revertants (data not shown).
The snc1 Phenotype Is Not Attributable to Overexpression of At4g16890
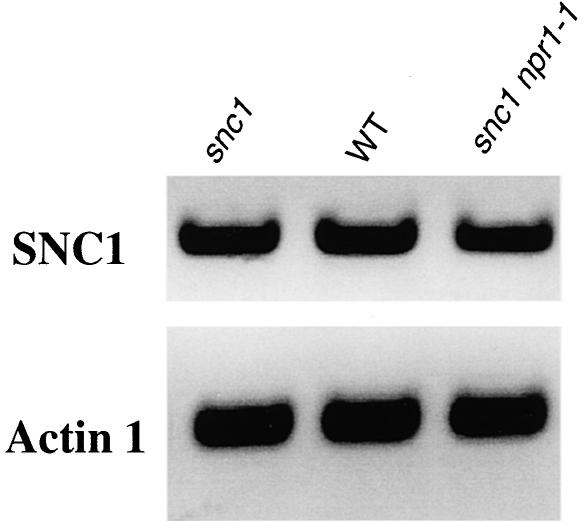
Previously, bal plants, as well as Arabidopsis transgenic plants overexpressing At4g16890, were shown to constitutively express PR genes (Stokes et al., 2002). Given this finding, we analyzed the expression level of At4g16890 in snc1 npr1-1, snc1, and wild-type plants to determine whether the snc1 mutant phenotype is caused by the overexpression of At4g16890. RT-PCR analysis showed that the expression level of At4g16890 was not altered in the mutant plants (Figure 4). Similar results also were obtained using real-time quantitative RT-PCR (data not shown). Thus, the snc1 phenotype is caused by the point mutation in At4g16890 rather than by overexpression of the gene. It is unclear whether the snc1 mutant protein accumulates to a higher level than the wild-type protein.
Figure 4.
Analysis of SNC1 Expression Levels in Wild-Type and Mutant Plants.
RNAs were prepared from 20-day-old plants grown on MS medium and reverse transcribed to obtain cDNA. The cDNA samples were first normalized by real-time PCR using an Actin1 probe. SNC1 and Actin1 in wild-type (WT) and mutant plants then were amplified by 30 cycles of PCR using equal amounts of total cDNA, and the products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
To test whether the expression of At4g16890 carrying the snc1 mutation would confer the snc1-like phenotype to wild-type plants, genomic clones containing either wild-type At4g16890 or a mutant At4g16890 containing the snc1 point mutation were transformed into wild-type plants. The snc1-like morphology was observed in 15 of 21 plants (>70%) transformed with the mutant At4g16890. On the other hand, only 5 of 23 plants (<25%) transformed with the wild-type gene developed snc1-like morphology, which could be the result of the overexpression of the At4g16890 transgene in these plants. Based on these data, we conclude that SNC1 is At4g16890 and that the Glu-552–to–Lys-552 mutation causes the constitutive activation of this R protein homolog.
The snc1 Phenotype Is Fully Dependent on PAD4
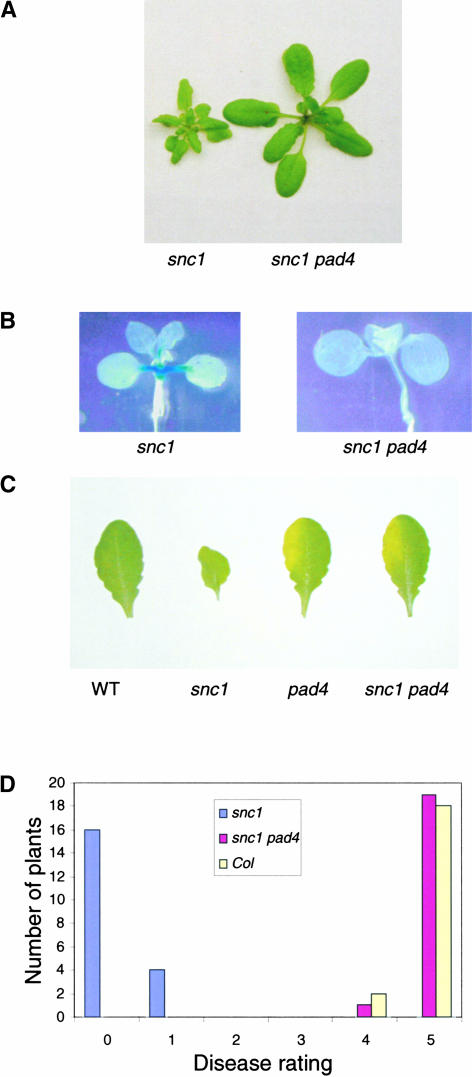
Previously, we showed that the snc1 mutant phenotype is suppressed completely by eds1. This finding is consistent with the fact that SNC1 belongs to the TIR-NB-LRR class of R genes, which usually require EDS1 as a downstream signaling component (Aarts et al., 1998). PAD4 interacts with EDS1 in vivo (Feys et al., 2001). To determine whether PAD4 also is required for the manifestation of the snc1 phenotype, a double mutant was constructed between snc1 and pad4-1 (Glazebrook et al., 1996; Jirage et al., 1999). As shown in Figure 5A, pad4-1 completely suppressed the morphological phenotypes of snc1. The double mutant also lost constitutive pBGL2-GUS expression (Figure 5B) and resistance to both P.s.m. ES4326 and P. parasitica Noco2 (Figures 5C and 5D). These data suggest that PAD4 is fully required for downstream signaling in snc1.
Figure 5.
Analysis of snc1 pad4-1.
(A) Morphology of snc1 pad4-1 plants. The photograph shows 4-week-old plants grown on soil.
(B) Suppression of constitutive pBGL2-GUS reporter gene expression in snc1 by pad4-1. Twenty-day-old seedlings grown on MS medium were stained for GUS activity.
(C) Enhanced susceptibility of the snc1 pad4-1 double mutant to P.s.m. ES4326. The leaves of 4-week-old soil-grown plants were infiltrated with a suspension of P.s.m. ES4326 in 10 mM MgCl2 (OD600 = 0.00005). The photograph was taken 3 days after infection. At this dose, wild-type (WT) Arabidopsis normally is resistant to the bacterium.
(D) Susceptibility of snc1 pad4-1 to P. parasitica Noco2. Two-week-old seedlings were sprayed with Noco2 spores at a conidiospore suspension concentration of 5 × 103 spores per milliliter of water. The infection was rated as follows on 20 plants at 6 days after infection by counting the number of conidiophores per infected leaf: 0, no conidiophores on the plants; 1, no more than 5 conidiophores per infected leaf; 2, 6 to 20 conidiophores on a few of the infected leaves; 3, 6 to 20 conidiophores on most of the infected leaves; 4, 5 or more conidiophores on all infected leaves; 5, 20 or more conidiophores on all infected leaves. Col, Columbia wild type.
snc1 Activates Both SA-Dependent and SA-Independent Defense Pathways
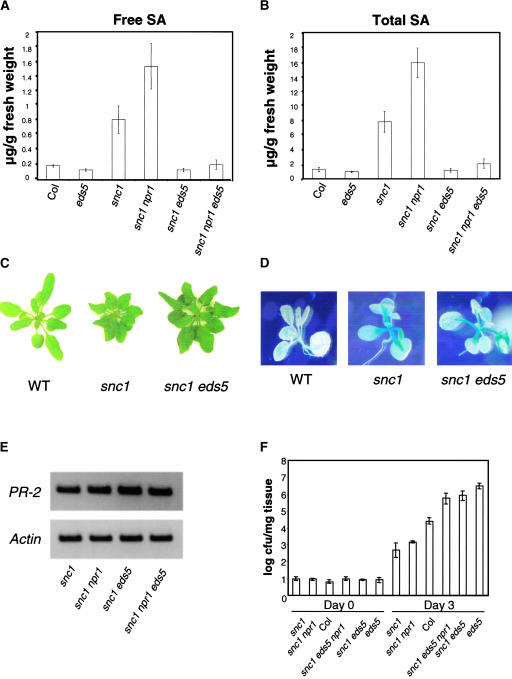
To test whether the increased SA level in snc1 is required for the activation of downstream defense pathways, a double mutant was constructed between snc1 and eds5-3, a mutant defective in pathogen-induced SA synthesis (Nawrath and Métraux, 1999; Nawrath et al., 2002). As shown in Figures 6A and 6B, snc1 eds5-3 had a similar amount of SA as eds5-3. snc1 eds5-3 plants were slightly smaller than wild-type plants, and the leaves of snc1 eds5-3 were still curly (Figure 6C). Constitutive expression of pBGL2-GUS in snc1 was not affected by eds5-3 (Figure 6D). RT-PCR analysis of PR-2 (BGL2) expression confirmed that eds5-3 did not affect the expression of PR-2 in the double mutant (Figure 6E). To determine whether the resistance of snc1 was affected by the decreased SA level, growth of the virulent bacterial pathogen P.s.m. ES4326 on snc1 eds5-3 also was determined (Figure 6F). Although snc1 npr1 was resistant to P.s.m. ES4326, snc1 eds5-3 was more susceptible than wild-type plants, suggesting that SA is required for the resistance of snc1 to P.s.m. ES4326. Compared with eds5-3, snc1 eds5-3 was slightly less susceptible, indicating that the SA-independent pathway also may contribute to resistance to the bacterial pathogen.
Figure 6.
Epistasis Analysis of snc1 and eds5-3.
(A) and (B) Free SA (A) and total SA (B) levels in the mutants. Leaf tissue was harvested 4 weeks after germination and used for SA extraction. Each treatment had four replicates. Col, Columbia wild type.
(C) Morphology of snc1 eds5 compared with snc1 and the wild type (WT).
(D) GUS staining of the pBGL2-GUS reporter gene in snc1 and snc1 eds5. Staining was performed on 20-day-old plants grown on MS medium.
(E) PR-2 expression in the mutants. RT-PCR was used to analyze the expression of PR-2 as described for Figure 3.
(F) Bacterial growth of P.s.m. ES4326 in the mutants. Two leaves of each plant were infiltrated with the bacteria (OD600 = 0.00005). Leaf discs within the inoculated areas were taken after 0 and 3 days of infiltration. Four replicates were taken for each treatment. Error bars represent 95% confidence limits of log-transformed data. cfu, colony-forming units.
DISCUSSION
To identify the mutation that causes constitutive PR gene expression and pathogen resistance in snc1 plants, we sequenced the complete region to which the snc1 mutation was mapped and identified a single mutation in At4g16890, an NB-LRR–class R gene that is highly similar to RPP4 and RPP5. We showed that deletions of this mutated R gene revert the mutant plants to the wild-type phenotype. In addition, we observed the snc1-like phenotype in >70% of wild-type plants transformed with a genomic clone containing the snc1 mutation. These data indicate that At4g16890 encodes SNC1 and that the point mutation we identified in SNC1 is a gain-of-function mutation that renders this R protein constitutively active. Unlike bal plants, snc1 plants do not overexpress At4g16890. In addition, snc1 is genetically stable but bal is metastable (Stokes et al., 2002). In our effort to identify revertants of snc1, we obtained nine revertants as well as a small number of recessive mutants (Y. Zhang and X. Li, unpublished data) by screening M2 plants from ∼4000 M1 families. This is quite different from the results of the genetic screen that Stokes et al. (2002) performed to identify revertants of bal, in which a high frequency of revertants (∼7% of the M2 plants) was observed in the progeny of mutagenized bal plants.
Mutational analysis of the NB-LRR class of R genes has suggested that both the NB and LRR domains are essential for the functions of these R proteins. A large number of mutations in both the NB and LRR domains have been found to inactivate the R proteins (Warren et al., 1998; Dinesh-Kumar et al., 2000; Tao et al., 2000, Axtell et al., 2001; Tornero et al., 2002), implying that the mutated residues are important for R protein function. Although negative regulation of this group of R proteins also may play a crucial role in R gene–mediated resistance, as suggested by the data that RPS2 can be activated constitutively by the elimination of its negative regulator RIN4 (Axtell and Staskawicz, 2003; Mackey et al., 2003), it is not known which regions of the R genes are important for this negative regulation. Here, we report that a single amino acid change in SNC1, located in the NL linker region, constitutively activates this R protein and that the NL linker region in SNC1 probably is involved in its negative regulation.
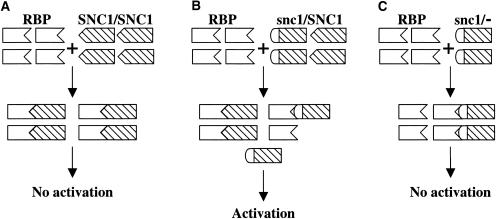
A model is proposed to explain how the mutation in snc1 results in the constitutive activation of defense responses (Figure 7). In wild-type plants, SNC1 interacts with an R PROTEIN BINDING PROTEIN (RBP) equivalent to RIN4. This interaction involves the NL linker region in SNC1, and amino acid Glu-552 is one of the critical residues for this interaction. Changing the negatively charged Glu-552 to the positively charged Lys-552 results in reduced binding affinity between snc1 and the negative regulator and dissociation of snc1 from the complex, which then leads to the constitutive activation of the downstream resistance pathways.
Figure 7.
Model for the Activation of SNC1 by the snc1 Mutation.
(A) In wild-type plants, SNC1 is sequestered by the proposed negative regulator RBP and is inactive.
(B) In snc1/SNC1 plants, some snc1 protein dissociates from its negative regulator and activates downstream defense pathways.
(C) In heterozygous revertant snc1/− plants, all snc1 protein is sequestered as a result of the excess amount of the negative regulator.
Our model explains the phenotypic differences between snc1/SNC1, snc1/−, and snc1/snc1 plants. In snc1/SNC1 plants, both the wild-type and mutant proteins bind to the negative regulator of SNC1. As a result of reduced binding affinity between snc1 and RBP, a portion of the snc1 protein dissociates from the negative regulator and partially activates the downstream signal transduction pathways. Because there is no wild-type protein in snc1/− plants, the excess amount of RBP sequesters all of the snc1 protein and blocks the activation of downstream defense responses. Because snc1/snc1 plants most likely contain more snc1 protein than snc1/SNC1 plants, the phenotype is more dramatic in snc1/snc1 plants. The constitutive PR gene expression in transgenic plants overexpressing SNC1 (At4g16890) also can be explained by the excess amount of SNC1 protein unbound to the negative regulator of SNC1. Furthermore, the existence of a negative regulator of SNC1 equivalent to RIN4 is supported by the finding that a loss-of-function mutation in BON1/CPN1 (Hua et al., 2001; Jambunathan et al., 2001) leads to the constitutive activation of SNC1-dependent resistance pathways (J. Hua, personal communication).
How downstream signaling pathways are activated by snc1 protein dissociated from its negative regulator is unclear. Recently, different domains of Rx have been shown to interact with each other in vivo (Moffett et al., 2002). It was proposed that disruption of these intramolecular interactions leads to the activation of downstream pathways. It is possible that dissociation of the snc1 protein from its negative regulator also induces conformational changes of the protein, which subsequently triggers either the release or recruitment of active effectors, as suggested previously (Moffett et al., 2002).
Analysis of residues that correspond to Glu-552 in SNC1 revealed significant divergence of the residues among different TIR-NB-LRR–type R proteins (Figure 2B). RPS4 and RRS1-R actually contain a Lys at this position, indicating that the Glu-to-Lys difference at this position does not necessarily render other R genes constitutively active. The effects of this change may depend strictly on the nature of the interactions between the R proteins and their interacting partners. Compared with mutations in other constitutively active R gene variants, the mutation in snc1 is unique. The region in which the snc1 mutation is located is different from the regions of other R proteins in which HR-inducing mutations were found (Figure 2C). The chimeric Mi proteins that induce localized HR contain replacements in the N-terminal extension (Hwang et al., 2000), whereas mutations in Rx that lead to constitutive HR are localized either in the NB or the LRR (Bendahmane et al., 2002). In addition, both Mi and Rx are CC-NB-LRR–type R genes. The only reported TIR-NB-LRR–type R gene mutation that causes spontaneous lesion formation is ssi4, which also is found in the NB region (Shirano et al., 2002).
Unlike ssi4, snc1 plants do not develop spontaneous lesions. snc1 is the first confirmed R gene mutant that activates downstream resistance pathways in the absence of HR, suggesting that R gene–mediated resistance can be uncoupled from cell death. Transgenic plants overexpressing the snc1 mutant gene do not have spontaneous lesions either (data not shown). The lack of HR-like lesions in snc1 plants and the unique location of the snc1 mutation suggest that the mutation may activate the R protein by a mechanism different from those in the constitutive HR-inducing mutants. We hypothesize that the activation of downstream defense pathways in snc1 is attributable to the disruption of intermolecular interactions, whereas the HR induction by constitutively active Rx, Mi, and SSI4 variants is caused by the disruption of intramolecular interactions by the mutations, as suggested previously (Hwang et al., 2000; Moffett et al., 2002).
The absence of spontaneous lesions in snc1 also makes it a very useful tool for studying resistance pathways downstream of R genes without the interference of cell death. This is demonstrated by the analysis of the snc1 pad4-1 and snc1 eds5-3 double mutants. Because SNC1 encodes an R protein belonging to the TIR-NB-LRR class and EDS1 is required for resistance conferred by this group of R genes, it is not surprising that eds1 completely suppressed the snc1 mutant phenotypes. Resistance conferred by the TIR-NB-LRR class of R genes often is only partially dependent on PAD4 (Glazebrook et al., 1996; Feys et al., 2001; van der Biezen et al., 2002). It is surprising that pad4 completely suppressed the snc1 mutant phenotype. One of the main differences between resistance in snc1 and resistance conferred by other TIR-NB-LRR R genes such as RPP4 and RPP5 is that HR is not involved in the resistance in snc1 because snc1 plants do not form spontaneous lesions. As suggested previously by Feys et al. (2001), most likely, EDS1 and PAD4 both are fully required for the HR-independent resistance responses, whereas EDS1 encodes additional functions for the generation of HR. Because the morphological phenotype of snc1 is not nearly as dramatic as that of the transgenic plants overexpressing this gene (Stokes et al., 2002), SNC1 probably is only partially activated by snc1. This effect also may contribute to the full dependence of the snc1 phenotype on PAD4.
Another interesting observation is that eds5-3 had no effect on constitutive PR-2 expression in snc1, whereas the expression of the SA-degrading enzyme NahG in snc1 suppressed the pBGL2-GUS reporter gene (Li et al., 2001). Because catechol, the product of NahG, induces disease susceptibility in plants (van Wees and Glazebrook, 2003), the suppression of pBGL2-GUS by NahG might be caused by the accumulation of catechol rather than by the reduction of SA. Alternatively, SA synthesized independent of EDS5 might be required for constitutive PR-2 expression, and the lack of PR-2 expression in snc1 NahG might be the result of the hydrolysis of SA synthesized independent of EDS5.
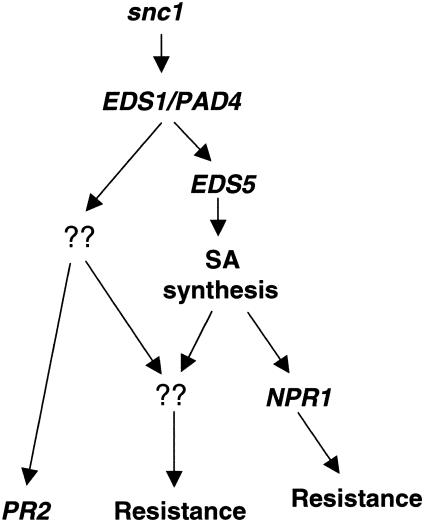
A simplified model is proposed in Figure 8 to describe the resistance pathways activated by snc1. The activation of downstream pathways appears to require both EDS1 and PAD4, because mutations in EDS1 and PAD4 completely suppressed the snc1 phenotype. On the other hand, increased SA levels were only partially responsible for the snc1 phenotypes, because snc1 eds5-3 double mutants with wild-type levels of SA had intermediate sizes, curly leaves, and constitutive PR-2 expression. Thus, both SA-dependent and SA-independent pathways exist downstream of snc1. Previously, it was shown that the induction of PR-2 by avirulent pathogens was not affected in the inoculated leaves of eds5 mutants (Nawrath and Métraux, 1999). The expression levels of PR-2 in cpr1 and cpr5 also were not affected by eds5 (Clarke et al., 2000). Thus, the constitutive expression of PR-2 appears to be a hallmark of the SA-independent resistance pathway.
Figure 8.
Model for Pathways Activated in snc1 Plants.
Although snc1 eds5-3 plants are susceptible, snc1 npr1 plants are resistant to the bacterial pathogen P.s.m. ES4326, indicating that both NPR1-dependent and NPR1-independent pathways are activated downstream of SA. The SA-dependent and NPR1-independent pathway appears to be the major contributor to the resistance to P.s.m. ES4326 in snc1 npr1 plants. On the other hand, SA alone cannot induce resistance to P.s.m. ES4326 in the npr1 background (Cao et al., 1994), suggesting that the SA-independent pathway(s) also is required for resistance to the bacterial pathogen in snc1 npr1 plants.
METHODS
Screening for snc1 Revertants
The snc1 npr1-1 mutant seeds of Arabidopsis thaliana were treated by fast-neutron bombardment at a dose of 60 Gray by Andrea Kodym (Agriculture and Biotechnology Laboratory, International Atomic Energy Agency, Vienna, Austria). M1 plants were grown on soil and allowed to self-pollinate. M2 seeds from 10 to 20 plants were pooled upon harvesting. M2 plants were grown on soil at 22°C under 16-h-light/8-h-dark cycles. Approximately 40,000 M2 plants from ∼4000 M1 families were screened for those with wild-type size and morphology. Seeds from putative mutants were collected and planted again. Lines producing progeny with both wild-type (approximately three-fourths) and snc1 (approximately one-fourth) morphology were analyzed further for the presence of deletions in At4g16890 by PCR and sequence analysis.
Mutant Characterization
To test pBGL2-GUS reporter gene expression, seeds were surface-sterilized and plated on MS medium (Murashige and Skoog, 1962). The plates were incubated in a TC16 plant growth chamber from Conviron (Winnipeg, Canada) at 22°C under 16-h-light/8-h-dark cycles. After 20 days, GUS staining was performed using a protocol described previously (Bowling et al., 1994). Infection of the plants with Pseudomonas syringae pv maculicola ES4326 and Peronospora parasitica Noco2 was performed as described previously (Li et al., 2001). Salicylic acid (SA) was extracted from fresh leaf tissue of 4-week-old plants and measured as described previously (Li et al., 1999).
Expression Analysis
To analyze the level of gene expression by reverse transcriptase–mediated PCR, total RNA samples were prepared from 20-day-old plants grown on MS medium using the Totally RNA kit from Ambion (Austin, TX). Reverse transcription was performed using the RT-for-PCR kit from Clontech (Palo Alto, CA). Real-time PCR was performed using the QuantiTect SYBR Green PCR kit from Qiagen (Valencia, CA). The primers used to amplify Actin1 were 5′-CGATGAAGCTCAATCCAAACGA-3′ and 5′-CAGAGTCGAGCACAATACCG-3′. The primers used to amplify PR-2 were 5′-GCTTCCTTCTTCAACCACACAGC-3′ and 5′-CGTTGATGTACCGGAATCTGAC-3′. The primers used to amplify SNC1 were 5′-AGATGTCCCCGATGTCATCC-3′ and 5′-CCAAACATTTTCAGACTTACAAGACTTG-3′. In each primer pair combination, one of the primers was designed to be cDNA specific according to the instructions of the QuantiTect SYBR Green PCR kit to avoid amplification of genomic DNA.
Reverse Complementation
A 7.2-kb PstI-BamHI genomic fragment containing the wild-type At4g16890 gene was first cloned from BAC clone F5D3 to pGEM3Z (from Perkin-Elmer). The same fragment then was cloned from pGEM3Z to pGreen229 (Hellens et al., 2000) to obtain pG229-SNC1. A 1.2-kb KpnII-AvrII genomic fragment containing the snc1 mutation was amplified by PCR from the genomic DNA of snc1 plants and used to replace the wild-type fragment in pG229-SNC1. The resulting clone was named pG229-snc1. Both pG229-SNC1 and pG229-snc1 then were used to transform the wild-type plants by the floral-dip method (Clough and Bent, 1998).
Creating the snc1 pad4-1 Double Mutant
To create the snc1 pad4-1 double mutant, the original snc1 npr1-1 (as female) was crossed with pad4-1. The resulting F1 plants had wild-type morphology and were grown to set seeds. In the F2 population, 36 seedlings were grown on soil and 7 of the 36 plants showed the distinct snc1 morphology of small stature and curly leaves. The F3 seeds of these seven snc1-like plants were collected and replanted. Among them, three had approximately one-quarter large wild-type plants segregating out. The plants with wild-type morphology were potential snc1 pad4-1 double mutants. These lines were plated on MS plates with 0.2 mM SA to check for NPR1 homozygosity, because npr1 mutants bleach to death on high concentrations of SA that wild-type plants can survive (Cao et al., 1997). These lines also were plated on MS medium containing 50 μg/mL kanamycin to check for the presence of the pBGL2-GUS reporter gene. The lines that were both NPR1 and pBGL2-GUS homozygous were used for further characterization. The resulting lines were backcrossed with snc1 to confirm the presence of the snc1 mutation, and as expected, the F1 plants had snc1 morphology. The presence of pad4-1 in the final mutant was confirmed by sequencing analysis of the pad4-1 locus.
Creating the snc1 eds5-3 Double Mutant
The strategy for obtaining the snc1 eds5-3 double mutant was similar to that used to isolate snc1 pad4-1. The original snc1 npr1-1 (as female) was crossed with eds5-3, and the resulting F1 plants had wild-type morphology and were grown to set seeds. In the F2 population, 72 seedlings were grown on soil and 19 of the 72 plants showed the distinct curly leaves of snc1. Among these 19 plants, 3 were larger, whereas the other 16 remained small. The F3 seeds of four small snc1-like plants were collected and replanted. Among them, two had approximately one-quarter large plants with curly leaves. The large plants with curly leaves were potential snc1 eds5-3 double mutants. These lines were plated on MS plates with 0.2 mM SA to check for NPR1 homozygosity. Lines that were NPR1 or npr1-1 homozygous were used for further characterization. The resulting lines were backcrossed with snc1 to confirm the presence of the snc1 mutation, and as expected, the F1 plants had snc1 morphology. The presence of eds5-3 in the final mutant lines was confirmed by sequencing analysis of the eds5-3 locus.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Xin Li, xinli@interchange.ubc.ca.
Acknowledgments
We thank Mark Tessaro, Patricia Lam, Sarah Westelmajer, Joyce Wu, Yu-ti Cheng, and Victor Ho for their technical assistance; Jane Glazebrook for pad4-1 seeds; Christiane Nawrath for eds5-3 seeds; Jane Parker for Peronospora strains; Phil Mullineaux for pGreen229; Jian Hua for sharing unpublished results; Patrick Keeling and his laboratory members for help with the phylogenetic analysis; and Kristoffer Palma, Wendy Durrant, and Jim Kronstad for careful reading of the manuscript. We are grateful for financial support to X.L. from the Natural Sciences and Engineering Research Council of Canada and the Canadian Foundation for Innovation and for financial support to X.D. from the National Science Foundation. S.G. is supported by the doctoral scholarship program of the Austrian Academy of Sciences.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015842.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., McNellis, T.W., Mudgett, M.B., Hsu, C.S., and Staskawicz, B.J. (2001). Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding Pseudomonas syringae avrRpt2 avirulence gene. Mol. Plant-Microbe Interact. 14, 181–188. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Farnham, G., Moffett, P., and Baulcombe, D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (1996). Plant disease resistance genes: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bonas, U., and Lahaye, T. (2002). Plant disease resistance triggered by pathogen-derived molecules: Refined models of specific recognition. Curr. Opin. Microbiol. 5, 44–50. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Theulieres, F., Hirsch, J., Feng, D.X., Bittner-Eddy, P., Beynon, J., and Marco, Y. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., Tham, W.H., and Baker, B.J. (2000). Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. USA 97, 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3, 278–284. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.-A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signalling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel, O. (1997). BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14, 685–695. [DOI] [PubMed] [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hua, J., Grisafi, P., Cheng, S.H., and Fink, G.R. (2001). Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 15, 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, C.F., Bhakta, A.V., Truesdell, G.M., Pudlo, W.M., and Williamson, V.M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes, R.W., Bent, A.F., Kunkel, B.N., Bisgrove, S.R., and Staskawicz, B.J. (1993). Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J. Bacteriol. 175, 4859–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N., Siani, J.M., and McNellis, T.W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13, 2225–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Clarke, J.D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant-Microbe Interact. 14, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., III, Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated disease resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.-L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Moffett, P., Farnham, G., Peart, J., and Baulcombe, D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Métraux, J.-P. (2002). EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Métraux, J.-P. (1999). Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabò, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 Receptor–Nucleotide Binding Site–Leucine-Rich Repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Stokes, T.L., Kunkel, B.N., and Richards, E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer, K., and von Haeseler, A. (1996). Quartet puzzling: A quartet maximum-likelihood method for reconstruction tree topologies. Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- Sun, Q., Collins, N.C., Ayliffe, M., Smith, S.M., Drake, J., Pryor, T., and Hulbert, S.H. (2001). Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Yuan, F., Leister, R.T., Ausubel, F.M., and Katagiri, F. (2000). Mutational analysis of the Arabidopsis nucleotide binding site–leucine-rich repeat resistance gene RPS2. Plant Cell 12, 2541–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Chao, R.A., Luthin, W.N., Goff, S.A., and Dangl, J.L. (2002). Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D.G. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D. (1998. a). The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 26, R226–R227. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D.G. (1998. b). Plant disease resistance proteins and the “gene-for-gene” concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33, 733–742. [DOI] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R.W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 9, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the Interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]