Abstract
Background
Previous studies have established a correlation between electrophoretic polymorphism of esterase B, and virulence and phylogeny of Escherichia coli. Strains belonging to the phylogenetic group B2 are more frequently implicated in extraintestinal infections and include esterase B2 variants, whereas phylogenetic groups A, B1 and D contain less virulent strains and include esterase B1 variants. We investigated esterase B as a marker of phylogeny and/or virulence, in a thorough analysis of the esterase B-encoding gene.
Results
We identified the gene encoding esterase B as the acetyl-esterase gene (aes) using gene disruption. The analysis of aes nucleotide sequences in a panel of 78 reference strains, including the E. coli reference (ECOR) strains, demonstrated that the gene is under purifying selection. The phylogenetic tree reconstructed from aes sequences showed a strong correlation with the species phylogenetic history, based on multi-locus sequence typing using six housekeeping genes. The unambiguous distinction between variants B1 and B2 by electrophoresis was consistent with Aes amino-acid sequence analysis and protein modelling, which showed that substituted amino acids in the two esterase B variants occurred mostly at different sites on the protein surface. Studies in an experimental mouse model of septicaemia using mutant strains did not reveal a direct link between aes and extraintestinal virulence. Moreover, we did not find any genes in the chromosomal region of aes to be associated with virulence.
Conclusion
Our findings suggest that aes does not play a direct role in the virulence of E. coli extraintestinal infection. However, this gene acts as a powerful marker of phylogeny, illustrating the extensive divergence of B2 phylogenetic group strains from the rest of the species.
Background
In humans, Escherichia coli strains can be commensal (part of the normal intestinal microbiota) and/or the cause of various infectious diseases (intestinal and extraintestinal infections) [1]. The extent of commensal or virulent properties displayed by a strain is determined by a complex balance between the status of the host and the production of virulence factors in the bacteria. The role of the intrinsic virulence of the isolates needs to be clarified and molecular markers of virulence are required to predict the invasiveness of clinical strains isolated during the course of extraintestinal infection or patient colonization.
E. coli has a clonal genetic structure and exhibits a low level of recombination [2]. E. coli strains can be categorised into four main phylogenetic groups, A, B1, B2, and D. These groups have been defined based on proteic (multi-locus enzyme electrophoresis including the electrophoresis of esterases [3]) and genetic markers (restriction fragment length polymorphism [4], random amplified polymorphic DNA [4] and multi-locus sequence typing (MLST) [5,6]). Seven types of esterases (A, B, C, D, I, F and S), differing in their ability to hydrolyse synthetic substrates and their sensitivity to di-isopropyl fluorophosphate, have been identified by separation on polyacrylamide agarose gels [7-9]. The most frequently observed type in this group of enzymes corresponds to esterase B (EC 3.1.1.1). This protein shows two types of electrophoretic mobility: B1 from Mf = 74 to Mf = 66 and B2 from Mf = 63 to Mf = 57 [9]. Strains with type B2 esterase belong to the phylogenetic group B2, whereas those with type B1 esterase belong to the non-B2 phylogenetic groups [10]. Several studies have shown a correlation between long-term evolutionary history (strain phylogeny) and virulence in E. coli, with most extraintestinal E. coli pathogens (including urinary tract infection strains) belonging to just one of the four main E. coli phylogenetic groups, the phylogenetic group B2 [11-13]. This correlation suggests a possible link between esterase polymorphism and extraintestinal virulence in an asexual species with a low level of recombination. Esterase B allozymes therefore appear to act as efficient molecular markers of virulence dividing E. coli strains into two genetically distinct groups, which differ significantly in their pathogenicity. However, the direct role of esterase B, or of its B1 and/or B2 allozymes, in the virulence process remains unknown.
The aims of this study were (i) to identify the gene encoding esterase B, (ii) to analyse its polymorphic counterparts in relation to E. coli clonal structure, (iii) to identify a potential physical link between this genetic locus and regions known to be associated with pathogenicity in the E. coli genome, and (iv) to test a potential direct role of esterase B in virulence in a mouse model of extraintestinal infection.
Results and Discussion
The acetyl esterase gene (aes) encodes esterase B
Seven candidate genes encoding proteins with predicted esterase activity were identified, based on their respective PM and pI values, using the MaGe system [14] (aes [15], yddV, glpQ, ndk, yzzH and cpdA). Of these, Aes exhibited several characteristics particularly reminiscent of esterase B: i) a major esterase domain, ii) a theoretical pI of 4.72 for the K-12 strain protein (esterase B1, pI ranging from 4.5 to 4.8) and 5.18 for CFT073 protein (esterase B2, pI ranging from 4.85 to 5.0), and iii) the presence of a serine in the active site [9]. The inactivation of aes by gene disruption in K-12 MG1655 and CFT073 strains and complementation of the mutant strains with the aes gene confirmed that Aes was esterase B (Additional file 1: Fig. S1 and data not shown).
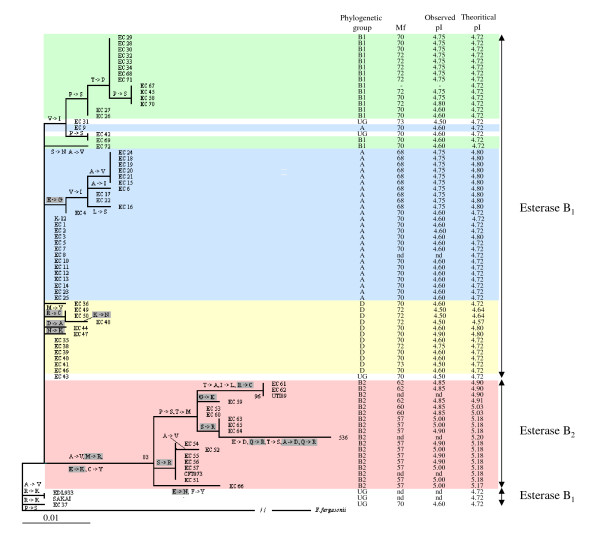
We then studied the correlation between Aes sequences and esterase B electrophoretic polymorphism. The comparison of the Aes phylogenetic tree with the theoretical and observed pI values and the esterase B electrophoretic mobilities (Mf values) for the 72 ECOR strains [10] is shown in Fig. 1. Overall analysis of the tree confirmed separation of esterase B into two variants: esterase B1 and esterase B2. Indeed, the Aes tree showed a clear distinction between Aes from the phylogenetic group B2 strains and Aes proteins from other strains, separated by a long branch, well supported by bootstrap (83%). Moreover, the characterisation of the phylogenetic group B2, based on Aes polymorphism, was consistent with the pI and Mf values of esterase B2 (pI: 4.85 to 5.0 and Mf 57 to Mf 62), which were previously demonstrated to be specific to the phylogenetic group B2. Likewise, the characterisation of the phylogenetic groups A, B1 and D, based on Aes polymorphism, correlated with the pI and Mf values of esterase B1 (pI: 4.60 to 4.80 and Mf 68 to Mf 72) [10]. Amino-acid substitutions detected from the branches of the Aes tree were analysed taking into account variation in esterase B mobility and pI values [16] (Fig. 1). In most cases, for the Aes phylogenetic group B2 strains, substitutions of acidic to neutral, neutral to basic or acidic to basic amino acids corresponded to increases in pI (from 4.85 to 5.00) and decreases in Mf values (from 62 to 57) among esterase B variants. However, there were some discrepancies. For example, the substitution of a basic amino acid in the ECOR 53 and 60 strains by a neutral amino acid in the ECOR 61 and 62 strains (R?C) corresponded to a faster migration in the ECOR 61 and 62 strains (Mf values 62 versus 60), with no effect on pI (4.85) (Fig. 1).
Figure 1.
Phylogenetic tree of Aes sequences from the 72 ECOR strains and 6 E. coli reference strains. The tree was reconstructed with PHYML [50]. E. fergusonii was used as an outgroup. Bootstraps are shown for values higher than 70%. Differences in amino acids are indicated on the branches. Differences for each branch were derived from comparison of consensus amino-acid sequences of the ancestors and descendants. Boxed amino-acid substitutions correspond to substitutions that change the overall pI of the protein. The phylogenetic groups A (blue box), B1 (green box), B2 (red box), D (yellow box) and ungrouped strains (UG) (white box), electrophoretic mobilities (Mf) obtained by polyacrylamide agarose gel electrophoresis [10] and the observed [10] and theoretical pI of Aes are indicated. nd: non determined. -: non significant results.
A more complex pattern of polymorphism was found among the A, B1 and D phylogenetic group strains. Taking the most frequent esterase B electrophoretic variant (pI: 4.60 and Mf 70) detected in the phylogenetic group A and D strains, an acidic to neutral amino-acid change (E?G) led to an increase in pI (from 4.60 to 4.75) and a decrease of Mf (from 70 to 68) of the esterase B variant, as expected. This amino-acid change was detected in 11 strains in the phylogenetic group A (Fig. 1). In contrast, several discrepancies were found among strains belonging to the phylogenetic B1 group: Aes polymorphism included several substitutions of neutral to neutral amino acids but with increased pI values (from 4.60 to 4.75) and in some cases paradoxical increases of Mf values (from 70 to 72) was observed (Fig. 1). These apparent discrepancies may be due to the effects of conformational or post-translational modifications of the protein.
The phylogenetic history of aes reflects the species phylogeny
To determine the evolutionary history of aes, we tested for selection using the aes sequence from 78 studied strains. First, we used a one-ratio model (M0) to estimate the average ratio ω (dN/dS) for all sites and all lineages at 0.18. The likelihood ratio test suggested that aes was under strong global purifying selection (compared to the neutral hypothesis which is ω = 0). The M1a, M2a, M7 and M8 models, estimating the selection on specific codons, confirmed that the vast majority (91%) of the sites are under negative selection. Finally, the branch-site model A did not detect positive selection along the branch separating group B2 from group non-B2 strains. Thus, the use of several tests for selection has shown that the evolution of aes has been driven essentially by purifying selection, as observed for the housekeeping genes used in the MLST (data not shown); no positively selected site was identified for the branch separating esterase B2 from esterase B1 strains. Previous studies based on whole genome sequencing data using PAML have not identified aes to be under positive selection [17,18].
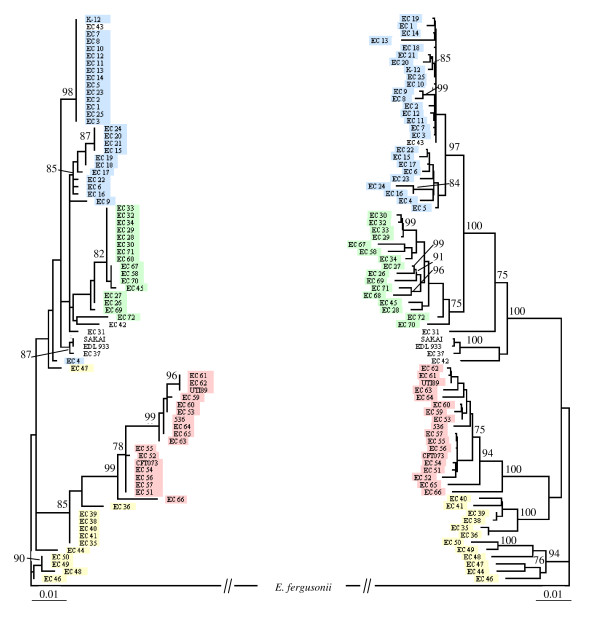
Visual comparison of the phylogenetic history of aes with that of the six concatenated housekeeping genes, reflecting the species phylogeny, revealed a similar topology with four main phylogenetic groups (Fig. 2). Indeed, all strains belonging to the B2 phylogenetic group were clustered in a monophyletic group (bootstrap 99%) with ECOR 66 at its base, as observed in the MLST tree. Likewise, two sub-groups were observed for phylogenetic group D, one of which was associated with the phylogenetic group B2 (ECOR 35, 36, 38, 39, 40, 41) (bootstrap 85%), also observed in the MLST tree. Phylogenetic group A also constituted two sub-groups, although these were not sister groups. By contrast, the B1 phylogenetic group was monophyletic overall, with only two strains (ECOR 4 and ECOR 47) clearly misclassified (Fig. 2).
Figure 2.
Phylogenetic trees for the 72 ECOR strains and six E. coli reference strains. The trees were constructed from (A) aes sequences and (B) multi-locus sequence typing of six housekeeping genes representing the species phylogeny (trpA, trpB, pabB, putP, icd and polB) [5], obtained using PHYML procedure [50]. E. fergusonii was used as an outgroup. Bootstraps are shown for values higher than 70%. Strains studied and belonging to phylogenetic groups A (blue boxed), B1 (green boxed), B2 (red boxed), D (yellow boxed) and UG (white boxed) are indicated.
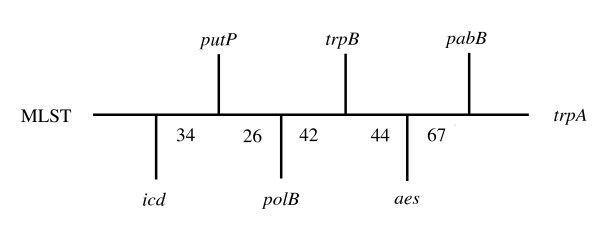
We used a recently developed technique ("TreeOfTree") allowing the level of congruence between phylogenetic trees to be tested [19]. We tested each individual housekeeping gene tree, the MLST tree, and the aes tree. All the bootstraps are low enough (less than 67%) to suggest that all the gene trees can be view as not incongruent, the aes gene tree itself clustering with pabB and trpA gene trees with very low bootstrap (44%) (Fig. 3). Thus, aes tree topology showed that aes is a powerful marker of the species phylogeny, as observed for each housekeeping gene used in the MLST scheme.
Figure 3.
Tree representing the distance matrix generated from comparisons between gene tree structures. Gene tree structure comparisons were between trees based on aes sequences, six individual housekeeping genes (trpA, trpB, pabB, putP, icd and polB) and multi-locus sequence typing (concatenation of the six housekeeping genes), with distances derived from path-length difference. Numbers are bootstraps.
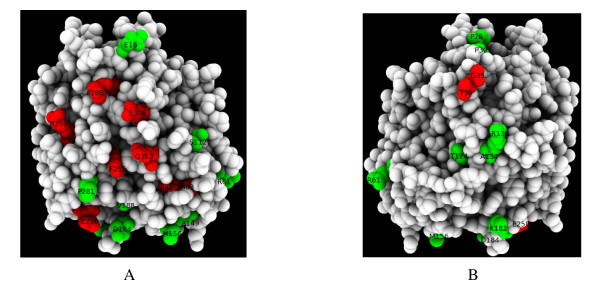
Aes B1 and B2 protein variants were then compared by protein modelling. We found that residues S 157, D 254 and H 284 had a geometry similar to that of the esterase catalytic site. Thirty-eight polymorphic sites were identified by comparing the 319 Aes amino-acid sequences obtained for the 72 ECOR strains and six reference strains. For four of these sites, variation has become fixed in both B1 and B2 types, with the identified residues differing between the two types at each site. These polymorphisms could thus be used to distinguish between the types: the B1 conserved amino acids A 53, M 64, E 73 and C 78 correspond to the B2 conserved amino acids V, R, K and Y, respectively. These four polymorphic sites were found on the long B2/non B2 branch in the proteic tree, explaining the observed high bootstrap (83%) (Fig. 1). Fig. 4 shows the location of 24 additional sites at the protein surface with observed amino-acid variants for either type B1 (green) or type B2 (red). No one site was polymorphic for both B1 and B2 types. But for all the polymorphic sites within types B1 and B2, some of the amino-acid variants are shared by the two types. Consequently, these sites cannot be considered to be specific to either one type or the other and cannot be used to distinguish between the two types of protein. Polymorphic sites were clustered, localised at the surface and were not found in the active site, consistent with previous observations of similarity in the catalytic activity of B1 and B2 esterases with synthetic substrates [7,9]. These differences in location of the polymorphic sites between the two variants support the divergence of the B2 phylogenetic group strains from the A, B1 and D phylogenetic groups strains within this species.
Figure 4.
Models of the Aes protein variants. Of the 38 polymorphic sites identified, only the 24 sites at the protein surface are represented. Polymorphic sites are in green for carboxylesterase type B1 and red for type B2. The views A and B correspond to two opposite faces of the structure obtained by a rotation of 180° around the Y axis. Images were generated using PMG [57].
Is Aes involved in virulence?
The previously observed correlation between electrophoretic esterase B polymorphism and the distinction between B2 and non-B2 phylogenetic group strains [10] - and thus with the extraintestinal virulence of the strains - suggested a putative role for the enzyme, or certain variants, as a virulence factor. The esterase B hydrolase function may have a direct role in the colonization or invasion of the eukaryotic cells as it was observed for esterases in other bacteria [20,21]. Indeed, esterase B2 variants belonging to phylogenetic group B2 may confer higher levels of virulence to the strain during extraintestinal infection. There are several examples of proteins with variants playing different roles in extraintestinal infections: the adhesins FimH [22], PapG [23] and the somatic antigen O [24,25].
Previous studies of Aes have not demonstrated a role of the protein in virulence. Firstly, experimental studies characterising Aes as an enzyme with esterase activity have demonstrated the inhibitory interaction of Aes with MalT, a transcriptional regulator of the maltose regulon. These findings suggested a role for Aes in the regulation of maltose metabolism [15,26]. Aes may also play a role in the regulation of raffinose metabolism by inhibiting α-galactosidase [27]. However, these data were obtained from overexpression of aes from plasmids, thus raising the question of their relevance in vivo. An illustration of aes overexpression from the plasmid pACS2 [28] is shown in Additional file 1: Fig. S1. Secondly, a previous study of aes expression in the K-12 strain in vitro did not find significant effects on expression under the various metabolic, stress or environmental conditions tested http://genexpdb.ou.edu/, with the exception of aes overexpression observed in strains cultured in the presence of acetate [29]. Interestingly, esterase B exhibits Michaelis-Menten kinetics for the hydrolysis of 1-naphtyl acetate [9]. Finally, aes expression was found to be homogeneous across 10 representative strains of E. coli/Shigella cultured in 869 medium [30].
Our previous findings from the study of the genetic sequence surrounding aes did not suggest a role for the encoded protein in virulence. Indeed, comparisons, using the MaGe system, of 75 kbp of sequence upstream and downstream from aes in the 20 strains of E. coli [31] showed that aes is not located in/or adjacent to any regions linked to extraintestinal pathogenicity specific to B2 strains (Additional file 2: Table S1).
To gain insight into Aes function we tested the mutants under different conditions. Firstly, we studied the in vitro growth of parent-type strains and their respective mutants on several carbon sources. We did not observe any difference between parent-type strains K-12 or CFT073 and their respective mutants K-12 Δaes and CFT073 Δaes in competition studies with LB and gluconate minimum media (data not shown). Additionally, growth of the strains CFT073, K-12, CFT073 Δaes and K-12 Δaes, in the presence of different carbon sources, was the same for parent and mutant strains. These results suggested that Aes does not play a role in regulation of the growth of the strains in these conditions. Secondly, we studied whether Aes is involved in the virulence of E. coli in vivo using a septicaemia mouse model. Kaplan-Meyer curves obtained for CFT073 and its mutants CFT073 Δaes and CFT073 Δaes:Cm were similar, suggesting that Aes is not involved in the virulence process (p = 0.87) (Additional file 1: Fig. S2).
Conclusion
Selection tests and phylogenetic analyses indicate that aes is under purifying selection, showing a similar evolutionary history to that of the species. The differences in electrophoretic properties between the variant types B1 and B2 were consistent with analyses of the amino-acid sequence tree for Aes and protein structure models obtained for these variants. These findings illustrated the marked divergence of the B2 phylogenetic group from the A, B1 and D phylogenetic groups in this species. This confirms the classical characteristics of esterases as excellent markers in the study of population genetics for prokaryotes, particularly Enterobacteriaceae [32], and eukaryotes such as Drosophila spp [33]. Findings from an in vivo experimental model of septicaemia did not show direct involvement of Aes in extraintestinal virulence. Moreover, we did not find any virulence-associated genes in the chromosomal region surrounding aes. Thus, esterase B does not appear to play a direct role as a virulence factor in E. coli extraintestinal infection, but may serve as an informative marker of phylogeny.
Methods
Bacterial strains
We used E. coli K-12 MG1655 (phylogenetic group A) and CFT073 (phylogenetic group B2) reference strains, their mutants, K-12 Δaes (obtained from the KEIO collection [34]) and CFT073 Δaes (obtained during the course of this study) and the aes complemented mutant strains K-12 Δaes pACS2 [28] andCFT073 Δaes pACS2 for the identification of the esterase B-encoding gene. The strains K-12 MG1655, CFT073 and their aes mutants were also used for the investigation of the putative role of esterase B. We used the 72 strains from the E. coli reference (ECOR) collection, encompassing commensal and pathogenic strains representative of the genetic diversity of the species [35], and four additional pathogenic reference strains (536, UTI89, Sakaï and EDL 933) for the sequencing of aes. The E. fergusonii strain ATCC 35469T, the most closely related species to E. coli [36], was used as an outgroup.
Candidate gene selection using bioinformatic tools
The MaGe (Magnifying Genome) software program [14] was used for candidate gene selection and comparative analysis of genetic sequences surrounding aes. The MaGe software program allows gene annotation and comparative analysis of available E. coli and closely related genomes, with visualisation of E. coli genome annotations enhanced by a synchronized display of synteny groups in the other genomes chosen for comparison [14]. Protein motifs and domains can be identified using the InterPro databank [37]. Candidate genes were obtained after the selection of proteins showing esterase motifs and compatible molecular weights (from 15,000 to 60,000 Da) and pI values (from 4.0 to 5.5) [9].
Inactivation of the aes gene and control experiments
Inactivation was carried out as previously described [38], using a PCR product obtained with primers aesW1 (5'-TTTCATGGCAGTGGTTCCTTACAATGACGTAATTTG AAAGGAGTTTTTGCGTTAGGCTGGAGCTGCTTC-3') and aesW2 (5'-GCCACGCCG GAACATATCGAAATGATGGCTAATCTTGTTGCCGCGTATCGCATATGAAATATCCTCCTTAG-3'). The PCR product contained (i) the FRT-flanked chloramphenicol acetyltransferase (cat) gene responsible for chloramphenicol resistance and (ii) the adjacent sequences homologous to the 5' and 3' flanking regions of aes. Inactivation of the aes gene was confirmed by PCR using the following primers: aes1 (5'-ACTGAGCGGCGAATGTTAACA-3') and aes2 (5'-ATTGTCTGGAGACGCTGGAA-3'), targeting sequences upstream and downstream from the aes gene, respectively; and c1 (5'-TTATACGCAAGGCGACAAGG-3') and c2 (5'-GATCTTCCGTCACAGGTAGG-3'), targeting sequences within cat gene. The antibiotic resistance gene was removed using the pCP20 plasmid [38].
Complementation analysis of the mutant strains was carried out by electroporation of the multicopy plasmid pACS2 [28] containing the aes gene under its native promoter.
The esterase B phenotype was investigated by vertical slab polyacrylamide gel electrophoresis of crude extracts of parent type, mutant and complemented mutant strains, using 12% (w/v) acrylamide and discontinous Tris/glycine buffer, pH 8.7. Esterase activity was detected by testing for the hydrolysis of 1-naphtyl acetate, as previously described [39].
Nucleotide sequencing, sequence alignments and selection tests
The aes gene was amplified by PCR, using the primers aes1 and aes2 (see above). The resulting 1250 bp PCR product was then sequenced by the Sanger method [40]. We compared aes sequences of 894 bp by sequence alignment using the ClustalW program [41]. The 72 aes sequences of the ECOR strains have GenBank accession numbers GQ167069 to GQ167140.
Amino-acid sequences deduced from the nucleotide sequences of aes were also analysed. After the generation of the maximum likelihood tree (see below), amino-acid substitutions for each branch of the Aes tree were identified by comparison of consensus sequences between different branches using the SEAVIEW program [42].
We tested for selection with code ML, implemented in PAML [43,44]. Using a maximum likelihood algorithm, PAML assigns likelihood scores to the data according to the various models of selection. Assignment of a higher likelihood score to a model incorporating selection than to a null model without selection and a significative likelihood ratio test are indicative of selection. The overall Ka/Ks ratio (or ω, dN/dS), reflecting selective pressure on a protein-encoding gene, was estimated using the M0 model (one-ratio) [45] for all isolate sequences, with the E. fergusonii sequence as an outgroup. We also used the M1a (null) and M2a (positive selection) models [46,47] and the more powerful M7 and M8 models [46,48] to detect positive selection on specific codons (sites). We used the branch-site model A [47,49] for the B2/non-B2 partition. This model is based on the hypothesis that positive selection occurs only in certain branches/lineages.
Tree reconstruction
Maximum-likelihood phylogenetic trees were all reconstructed using the PHYML program [50] and the GTR+G+I model. This general model is not necessarily the most parsimonious one. However, we also wanted to obtain the bootstrap support values for each partition. Given that (i) the most parsimonious model may differ from one bootstrap resampling to another, and (ii) a very long computer processing time would be required to choose the best model among the 88 possible models for each of the 500 resamplings, we chose a less time-consuming strategy, simply selecting the most general model (GTR+G+I) for all resamplings. We checked that the trees remain the same for the different models, whether the most parsimonious or the most general model is used. Additional file 2: Table S2 gives the different parsimonious models, and their estimated parameters, selected by the Akaike criterion (jMODELTEST version 0.1.1, written by Posada [51], available at http://darwin.uvigo.es/software/jmodeltest.html).
Tree comparisons
We compared the phylogenetic history of aes to the phylogenetic history of the strains, based on the concatenated nucleotide sequences of six housekeeping genes (trpA, trpB, pabB, putP, icd and polB) and individual gene sequences, as described elsewhere [19]. Briefly, each phylogenetic tree Ti is firstly transformed into a tree-distance matrix Di, the distance between two strains being the number of branches with positive length connecting them along the tree. The resulting tree distance matrix Di allows the initial tree structure Ti to be recovered, independently of branch length. Two tree distance matrices (Di and Dj) (corresponding to two gene trees i and j) can be compared by calculating the Euclidian distance between them (δij) [52]. A low δij value means that the similarity between the two tree distance matrices Di and Dj is high, and, consequently, that their tree structures Ti and Tj are close. As several gene tree structures are compared through this Euclidian distance metric, a new distance matrix Δ can be built with the δij elements. This Δ matrix can then be transformed into a "tree of gene trees" using a neighbour-joining algorithm [53].
To obtain a support value for each partition of this tree, we applied this same procedure to 500 bootstrapped sets of data, obtaining 500 Δ matrices and finally, a bootstrapped consensus "tree of gene trees". A high bootstrap support value separating two sets of gene trees allows incongruent sets of gene trees to be identified; however, a low bootstrap value suggests that the two sets of trees are not incongruent or that there is insufficient phylogenetic information to reject the hypothesis of incongruence. The "TreeOfTree" package is available from the website http://bioinformatics.lif.univ-mrs.fr.
Protein structure modelling and analysis
Modelling of the Aes protein structure was based on comparison of the available models from MODBASE [54] with models previously obtained using the Tasser-Lite homology modelling server [55,56]. Although some differences were observed between the models obtained by these two independent approaches, in particular in the N terminus region, the best models proposed by Tasser-Lite and MODBASE were similar overall. Given that our aim was to determine only the approximate location of the Aes polymorphism within the protein structure, the MODBASE model was used for further analysis. The model was finally tested to ensure that it contains an active site consistent with esterase activity. This was carried out using the 3D MSS-Sites program http://bioserv.rpbs.jussieu.fr/[57] and the Catalytic Site Atlas [58]. Polymorphic sites were identified by sequence alignment using ClustalW [41] for B1 and B2 variants separately.
Theoritical pIs of Aes were calculated using the program compute pI of the ExPASY home page http://www.expasy.ch/tools/pi_tool.html.
In vitro growth studies
Competition studies of parent strains K-12 and CFT073, with their respective mutants K-12 Δaes:Kan and CFT073 Δaes:Cm (1/1 ratio), were performed in Luria Bertani (LB) and gluconate minimum liquid media. Gluconate minimal medium mimics the intestinal environment [59]. For each medium and for each competition experiment, bacteria were plated on media with or without the appropriate antibiotic and counted after 2 h (exponential phase) and 18 h (stationary phase). Each experiment was repeated twice. Biolog GN2 (Biolog, Inc., Hayward, CA) plates were used to detect carbon utilisation of 95 substrates. Utilisation of various C sources is coupled to the reduction of a tetrazolium dye and generation of a purple colour [60]. Each strain was grown in LB medium, washed and resuspended to an optical density of 0.01 at 600 nm in mineral medium [60]. Plates were incubated at 37°C and colour changes were measured by changes in optical density (measured on a Tecan microplate reader) at a wavelength of 600 nm. The cut-off for positive results was an optical density of 0.2.
Septicaemia mouse model
A mouse model of systemic infection was used to assess the intrinsic virulence of the strains [11]. For each strain, 10 outbred female swiss OF1 mice (3-4 weeks old, 14-16 g) were challenged with a standardized subcutaneous bacterial inoculum (2 × 108 CFU of E. coli). Mortality was assessed over seven days following the challenge. Assays were performed using the CFT073 strain as a positive control (killing 10/10 mice), the K-12 strain as a negative control (killing 0/10 mice) [61] and the CFT073 Δaes and CFT073 Δaes:Cm mutant strains. Data were analysed using the StatView software to obtain Kaplan-Meyer curves; statistical analysis was carried out using the logrank test, with p values < 0.05 considered as significant.
Authors' contributions
ML carried out the in silico and in vitro studies of Aes, participated to the in vivo studies of Aes and wrote the manuscript. CH contributed to the in silico studies. OC contributed to the sequencing. LG carried out in vivo studies. PD carried out tree comparisons. PT carried out the structural analysis of the protein. ED participated in the design of the study and wrote the manuscript. BP was involved in the initial design of the study and wrote the manuscript. All authors read and approved the final manuscript.
Authors' Information
ML and CH are PhD students, OC is a research engineer, LG is a technician. PD, PT, ED and BP are researchers.
Supplementary Material
Supplemental figures. A figure showing the electrophoretic patterns of esterases from various E. coli strains. Fig. S1: Polyacrylamide gel electrophoresis of Aes. Gels were stained using 1-naphtyl acetate hydrolysis to detect esterase activity. Esterases B was detected in strains. K-12 (lane 1) and K-12 Δaes pACS2 (lane 3), but not in strain K-12 Δaes (lane 2), thus confirming that aes encodes esterase B. The dilution factor used for the crude extract of the complemented strain K-12 Δaes pACS2 was 40 times greater than that of the parent and mutant strains due to overexpression of the aes gene on the plasmid. This did not allow us to detect esterase A in the complemented strain, whereas it was clearly visible for the K-12 and K-12 Δaes strains. Fig. S2: Kaplan-Meyer curves showing the comparative scores of virulence in the mouse model of septicaemia as a function of the presence or absence of Aes in the K-12 strain (blue line), CFT073 strain (green line and squares), CFT073 Δaes:Cm strain (red line and circles) and CFT073 Δaes strain (violet line and triangles). Mice inoculated with K-12 strain were still alive at day 7.
Supplemental Tables. A table describing the genes surrounding the aes gene. Table S1: List of genes of the strain CFT073 and their characteristics within a total region of 150 kbp surrounding the aes gene. The aes gene and its characteristics are highlighted in red. Table S2: Parsimonious models, and their estimated parameters, selected by the Akaike criterion (jMODELTEST version 0.1.1, written by Posada, 2008, available at http://darwin.uvigo.es/software/jmodeltest.html) used for each tree reconstruction.
Contributor Information
Mathilde Lescat, Email: mathilde.lescat@inserm.fr.
Claire Hoede, Email: claire_hoede@yahoo.fr.
Olivier Clermont, Email: olivier.clermont@inserm.fr.
Louis Garry, Email: louis.garry@inserm.fr.
Pierre Darlu, Email: pierre.darlu@inserm.fr.
Pierre Tuffery, Email: pierre.tuffery@univ-paris-diderot.fr.
Erick Denamur, Email: erick.denamur@inserm.fr.
Bertrand Picard, Email: bertrand.picard@avc.aphp.fr.
Acknowledgements
ML was supported by the "Fondation pour la Recherche Médicale". We are grateful to Olivier Tenaillon for advice throughout this study, to Odile Bouvet for metabolic studies and Olivier Meilhac for protein electrophoresis. We acknowledge Evelyne Richet for providing the plasmid bearing the aes gene (pACS2).
References
- Donnenberg M. Escherichia coli virulence mechanisms of a versatile pathogen. San Diego, California; 2002. [Google Scholar]
- Selander RK, Levin BR. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- Herzer PJ, Inouye S, Inouye M, Whittam TS. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172(11):6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41(4):440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- Escobar-Paramo P, Sabbagh A, Darlu P, Pradillon O, Vaury C, Denamur E, Lecointre G. Decreasing the effects of horizontal gene transfer on bacterial phylogeny: the Escherichia coli case study. Mol Phylogenet Evol. 2004;30(1):243–250. doi: 10.1016/S1055-7903(03)00181-7. [DOI] [PubMed] [Google Scholar]
- Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goullet P. An esterase zymogram of Escherichia coli. J Gen Microbiol. 1973;77(1):27–35. doi: 10.1099/00221287-77-1-27. [DOI] [PubMed] [Google Scholar]
- Goullet P. Esterase electrophoretic pattern relatedness between Shigella species and Escherichia coli. J Gen Microbiol. 1980;117(2):493–500. doi: 10.1099/00221287-117-2-493. [DOI] [PubMed] [Google Scholar]
- Goullet P, Picard B, Laget PF. Purification and properties of carboxylesterase B of Escherichia coli. Ann Microbiol (Paris) 1984;135A(3):375–387. doi: 10.1016/s0769-2609(84)80079-4. [DOI] [PubMed] [Google Scholar]
- Goullet P, Picard B. Comparative electrophoretic polymorphism of esterases and other enzymes in Escherichia coli. J Gen Microbiol. 1989;135(1):135–143. doi: 10.1099/00221287-135-1-135. [DOI] [PubMed] [Google Scholar]
- Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67(2):546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Delavari P, Kuskowski M, Stell AL. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. 2001;183(1):78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177(3):642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Medigue C. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34(1):53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peist R, Koch A, Bolek P, Sewitz S, Kolbus T, Boos W. Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J Bacteriol. 1997;179(24):7679–7686. doi: 10.1128/jb.179.24.7679-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B, Goullet P, Krishnamoorthy R. A novel approach to study of the structural basis of enzyme polymorphism. Analysis of carboxylesterase B of Escherichia coli as model. Biochem J. 1987;241(3):877–881. doi: 10.1042/bj2410877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L, Bollback JP, Dimmic M, Hubisz M, Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17(9):1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci USA. 2006;103(15):5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, Weinert K, Tenaillon O, Matic I, Denamur E. Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog. 2009;5(1):e1000257. doi: 10.1371/journal.ppat.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter AJ, Kidd SP, Edwards JL, Falsetta ML, Apicella MA, Jennings MP, McEwan AG. Esterase D is essential for protection of Neisseria gonorrhoeae against nitrosative stress and for bacterial growth during interaction with cervical epithelial cells. J Infect Dis. 2009;200(2):273–278. doi: 10.1086/599987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garau G, Lemaire D, Vernet T, Dideberg O, Di Guilmi AM. Crystal structure of phosphorylcholine esterase domain of the virulence factor choline-binding protein e from Streptococcus pneumoniae: new structural features among the metallo-beta-lactamase superfamily. J Biol Chem. 2005;280(31):28591–28600. doi: 10.1074/jbc.M502744200. [DOI] [PubMed] [Google Scholar]
- Hommais F, Gouriou S, Amorin C, Bui H, Rahimy MC, Picard B, Denamur E. The FimH A27V mutation is pathoadaptive for urovirulence in Escherichia coli B2 phylogenetic group isolates. Infect Immun. 2003;71(6):3619–3622. doi: 10.1128/IAI.71.6.3619-3622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MC, Mobley HL. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 2007;72(1):19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- Plainvert C, Bidet P, Peigne C, Barbe V, Medigue C, Denamur E, Bingen E, Bonacorsi S. A new O-antigen gene cluster has a key role in the virulence of the Escherichia coli meningitis clone O45:K1:H7. J Bacteriol. 2007;189(23):8528–8536. doi: 10.1128/JB.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M, Heuzenroeder M, Kusecek B, Ochman H, Caugant D, Selander RK, Vaisanen-Rhen V, Korhonen TK, Stuart S, Orskov F. Clonal analysis of Escherichia coli O2:K1 isolated from diseased humans and animals. Infect Immun. 1986;51(1):268–276. doi: 10.1128/iai.51.1.268-276.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N, Danot O, Schlegel A, Boos W, Richet E. The Aes protein directly controls the activity of MalT, the central transcriptional activator of the Escherichia coli maltose regulon. J Biol Chem. 2002;277(19):16606–16613. doi: 10.1074/jbc.M200991200. [DOI] [PubMed] [Google Scholar]
- Mandrich L, Caputo E, Martin BM, Rossi M, Manco G. The Aes protein and the monomeric alpha-galactosidase from Escherichia coli form a non-covalent complex. Implications for the regulation of carbohydrate metabolism. J Biol Chem. 2002;277(50):48241–48247. doi: 10.1074/jbc.M207398200. [DOI] [PubMed] [Google Scholar]
- Schlegel A, Danot O, Richet E, Ferenci T, Boos W. The N terminus of the Escherichia coli transcription activator MalT is the domain of interaction with MalY. J Bacteriol. 2002;184(11):3069–3077. doi: 10.1128/JB.184.11.3069-3077.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem. 2005;280(16):15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- Le Gall T, Darlu P, Escobar-Paramo P, Picard B, Denamur E. Selection-driven transcriptome polymorphism in Escherichia coli/Shigella species. Genome Res. 2005;15(2):260–268. doi: 10.1101/gr.2405905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1):e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goullet P, Picard B. The electrophoretic polymorphism of bacterial esterases. FEMS Microbiol Rev. 1995;16(1):7–31. doi: 10.1111/j.1574-6976.1995.tb00153.x. [DOI] [Google Scholar]
- Babcock CS, Anderson WW. Molecular evolution of the Sex-Ratio inversion complex in Drosophila pseudoobscura: analysis of the Esterase-5 gene region. Mol Biol Evol. 1996;13(2):297–308. doi: 10.1093/oxfordjournals.molbev.a025589. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H, Hartl DL. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137(8):1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29(1):37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz WW, Williams PP. Characterization of esterases produced by a ruminal bacterium identified as Butyrivibrio fibrisolvens. J Bacteriol. 1973;113(3):1170–1176. doi: 10.1128/jb.113.3.1170-1176.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12(6):543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13(5):555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 1998;15(12):1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168(2):1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20(1):18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Penny DWE, Steel MA. Trees from languages and genes are very similar. Syst Biol. 1993;42:382–384. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006. pp. D291–295. [DOI] [PMC free article] [PubMed]
- Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR_3 threading algorithm. Proteins. 2004;56(3):502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci USA. 2004;101(20):7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alland C, Moreews F, Boens D, Carpentier M, Chiusa S, Lonquety M, Renault N, Wong Y, Cantalloube H, Chomilier J. RPBS: a web resource for structural bioinformatics. Nucleic Acids Res. 2005. pp. W44–49. [DOI] [PMC free article] [PubMed]
- Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004. pp. D129–133. [DOI] [PMC free article] [PubMed]
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101(19):7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11(7):1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J Infect Dis. 2006;194(8):1141–1150. doi: 10.1086/507305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures. A figure showing the electrophoretic patterns of esterases from various E. coli strains. Fig. S1: Polyacrylamide gel electrophoresis of Aes. Gels were stained using 1-naphtyl acetate hydrolysis to detect esterase activity. Esterases B was detected in strains. K-12 (lane 1) and K-12 Δaes pACS2 (lane 3), but not in strain K-12 Δaes (lane 2), thus confirming that aes encodes esterase B. The dilution factor used for the crude extract of the complemented strain K-12 Δaes pACS2 was 40 times greater than that of the parent and mutant strains due to overexpression of the aes gene on the plasmid. This did not allow us to detect esterase A in the complemented strain, whereas it was clearly visible for the K-12 and K-12 Δaes strains. Fig. S2: Kaplan-Meyer curves showing the comparative scores of virulence in the mouse model of septicaemia as a function of the presence or absence of Aes in the K-12 strain (blue line), CFT073 strain (green line and squares), CFT073 Δaes:Cm strain (red line and circles) and CFT073 Δaes strain (violet line and triangles). Mice inoculated with K-12 strain were still alive at day 7.
Supplemental Tables. A table describing the genes surrounding the aes gene. Table S1: List of genes of the strain CFT073 and their characteristics within a total region of 150 kbp surrounding the aes gene. The aes gene and its characteristics are highlighted in red. Table S2: Parsimonious models, and their estimated parameters, selected by the Akaike criterion (jMODELTEST version 0.1.1, written by Posada, 2008, available at http://darwin.uvigo.es/software/jmodeltest.html) used for each tree reconstruction.