Abstract
Nuclear LIM domains interact with a family of coregulators referred to as Clim/Ldb/Nli. Although one family member, Clim-2/Ldb-1/Nli, is highly expressed in epidermal keratinocytes, no nuclear LIM domain factor is known to be expressed in epidermis. Therefore, we used the conserved LIM-interaction domain of Clim coregulators to screen for LIM domain factors in adult and embryonic mouse skin expression libraries and isolated a factor that is highly homologous to the previously described LIM-only proteins LMO-1, -2, and -3. This factor, referred to as LMO-4, is expressed in overlapping manner with Clim-2 in epidermis and in several other regions, including epithelial cells of the gastrointestinal, respiratory and genitourinary tracts, developing cartilage, pituitary gland, and discrete regions of the central and peripheral nervous system. Like LMO-2, LMO-4 interacts strongly with Clim factors via its LIM domain. Because LMO/Clim complexes are thought to regulate gene expression by associating with DNA-binding proteins, we used LMO-4 as a bait to screen for such DNA-binding proteins in epidermis and isolated the mouse homologue of Drosophila Deformed epidermal autoregulatory factor 1 (DEAF-1), a DNA-binding protein that interacts with regulatory sequences first described in the Deformed epidermal autoregulatory element. The interaction between LMO-4 and mouse DEAF-1 maps to a proline-rich C-terminal domain of mouse DEAF-1, distinct from the helix–loop–helix and GATA domains previously shown to interact with LMOs, thus defining an additional LIM-interacting domain.
The LIM motif, a cysteine-rich zinc-coordinating domain, originally was discovered adjacent to homeodomains in three transcription factors, lin-11, Isl-1, and mec-3 (1, 2). Several additional LIM homeodomain factors have been discovered, and these, as well as the original three members of this gene family, have been established to have critical functions in lineage specification and differentiation in diverse cellular systems (1, 2).
Other nuclear LIM domain-containing factors have been identified, including the LIM-only (LMO) proteins, so named because they are composed almost entirely of two tandem LIM domains (1, 2). This subgroup of LIM proteins has three members: LMO-1 (RBTN1/TTG1) and LMO-2 (RBTN2) were isolated at sites of chromosomal translocations in acute T-cell leukemia, and the third, LMO-3 (RBTN3) was isolated based on sequence similarity (3, 4). Both LMO-1 and LMO-2 have been shown to act as oncoproteins in lymphocytes (3). In addition to its role in human cancers, gene deletion of LMO-2 showed that this gene is essential for normal blood cell formation in mice (5, 6), and thus has roles in cellular determination and differentiation. Instead of directly binding DNA, LMO factors are thought to regulate gene transcription by associating with DNA-binding proteins. In addition to the nuclear LIM domain factors, LIM domains have been found on several cytoplasmic proteins, some of which also contain kinase domains (1).
Prevailing evidence supports the notion that the LIM domain acts as a protein–protein interaction domain. Recently, several laboratories isolated cofactors that interact strongly with the LIM domains of LIM homeodomain and LMO proteins (7–10). These factors, referred to as Clim-1/Ldb2 and Clim-2/Ldb1/NLI, have been shown to promote (8) or inhibit (11) transcriptional synergism. Because these LIM-associating factors have no direct DNA interaction, it has been suggested that they may act as adapter molecules facilitating assembly of large complexes (12, 13), or that they may contribute directly to activation and/or repression. A recently isolated Drosophila homologue of the Clim family, Chip, has been implicated as playing a role in altering chromatin structure to facilitate remote enhancer-promoter interactions (14).
Prompted by the observation that Clim-2 is prominently expressed in epidermis (8), we isolated and characterized an epidermally expressed LMO factor, referred to as LMO-4. In addition to high affinity interactions with Clims, we show that LMO-4 interacts with a proline-rich C-terminal domain of the mouse homologue of the Drosophila DNA-binding protein, Deformed epidermal autoregulatory factor 1 (DEAF-1), suggesting that a complex of Clims, LMO-4, and mouse DEAF-1 (mDEAF-1) may be involved in transcriptional regulation.
MATERIALS AND METHODS
In Situ Hybridization and Immunohistochemistry.
In situ hybridization studies with 35S-labeled cRNA probes were performed on paraffin-embedded and frozen sections as described (15). For whole-mount in situ hybridization studies, embryos were briefly fixed in 4% paraformaldehyde followed by methanol and hydrogen peroxide washes and proteinase K treatment. Embryos were hybridized with 1 μg/ml of digoxigenin-labeled probe overnight at 55°C. Transcripts were detected with alkaline phosphatase-conjugated antibody and stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate tablets (Boehringer Mannheim). For immunohistochemistry, transfected CV-1 cells were fixed in 10% buffered formalin for 15 min, incubated with a monoclonal myc antibody, and stained with peroxidase.
Protein–Protein Interaction Cloning.
The C terminus of Clim-1 was expressed as a fusion with several protein kinase A phosphorylation sites and glutathione S-transferase (GST). Lambda gt11 libraries from neonatal and embryonic mouse skin were screened with a 32P-labeled protein as described (8).
Yeast Two-Hybrid Screening.
Baits were expressed as fusions with the GAL4 DNA-binding domain, and library and target plasmids were expressed as fusions with the GAL4 activation domain. β-galactosidase units were calculated according to standard methods (CLONTECH, Matchmaker two-hybrid system).
Protein–Protein Interaction Assays.
Coimmunoprecipitations were performed as described, by using 35S-labeled proteins and either a monoclonal myc antibody or a polyclonal rabbit antisera recognizing Clim-1 and Clim-2 (8). GST-interaction assays have been described (8).
Mammalian Two-Hybrid System.
LMO-4 was cloned as a fusion with the GAL4 DNA-binding domain and domains of mDEAF-1 were cloned in-frame with the VP-16 activation domain, both under control of the simian virus 40 enhancer promoter. Interaction was tested by transfecting these plasmids into HeLa cells along with a luciferase reporter under the control of GAL4 binding sites and a minimal promoter. Transfections and luciferase assays were performed as described (15).
RESULTS
Cloning of LMO-4 from Epidermis.
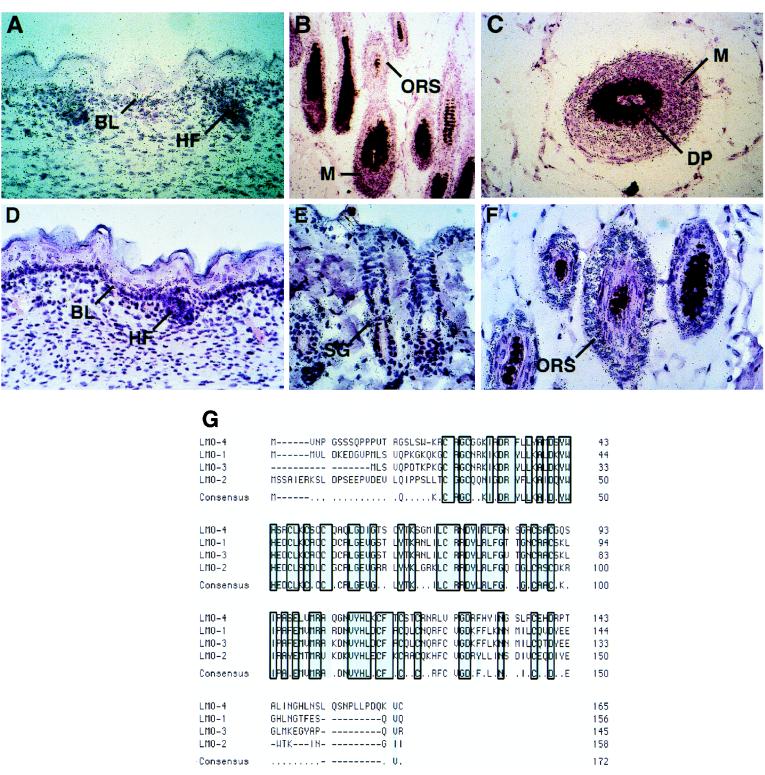
Previously, we showed by RNase protection assays that Clim-2 is highly expressed in skin (8). To investigate the precise expression pattern of Clim-2 in skin, we performed in situ hybridization studies and found that Clim-2 was expressed at its highest levels in, but not restricted to, the basal compartment of interfollicular epidermis (Fig. 1A). In addition, strong expression was observed in developing hair follicles during embryogenesis (Fig. 1A). In adult hair, Clim-2 was expressed at its highest levels in matrix cells and the outer root sheath (Fig. 1 B and C).
Figure 1.
Clim-2 and LMO-4 exhibit overlapping expression in epidermis. In situ hybridization of Clim-2 (A–C) and LMO-4 (D–F) in skin. (A) Clim-2 expression at e17.5. (B and C) Clim-2 expression in adult hair follicles. (D) LMO-4 expression at e17.5. (E and F) LMO-4 expression in adult hair follicles. (G) Comparison of the amino acid sequence of mouse LMO-4 with mouse LMO-1, -2, and -3. Conserved residues in the two LIM domains are shaded. BL, basal layer; HF, hair follicle; ORS, outer root sheath; M, matrix cells; DP, dermal papilla; SG, sebaceous gland.
Because Clim factors are known to associate with nuclear LIM proteins, these studies suggested the possibility that a novel LIM domain gene might be expressed in epidermis. To search for epidermal LIM factors, we cloned the C-terminal 100 amino acids of Clim-1, encompassing the conserved region responsible for LIM domain interactions, into a GST bacterial expression vector containing several protein kinase A phosphorylation sites, thus allowing labeling of the interaction domain with 32P in vitro. This radiolabeled protein was used to screen mouse λgt11 expression libraries from neonatal and embryonic day (e) 14.5 embryonic skin. Several interacting clones were obtained from the neonatal skin library, and one interacting clone was isolated from the embryonic skin library, all of which represented mRNAs transcribed from the same gene. Sequence analyses predicted that these cDNAs encoded a protein with two tandem LIM domains, but no other sequence homology, similar to the LIM-only factors LMO-1, -2, and -3 (Fig. 1G), and thus we refer to this factor as LMO-4. A search of GenBank databases showed that a homologous human gene, referred to as Human Breast Tumor Autoantigen, had been deposited in the GenBank database (accession no. U24576) and that an expressed sequence tag encompassing this sequence had been mapped to human chromosome 1, reference interval D1S203-D1S2865. While this manuscript was in preparation, another group reported the isolation of mouse LMO-4 (16).
In situ hybridization experiments showed that LMO-4 was expressed in epidermis with the highest levels in hair follicles, especially in the outer root sheath, sebaceous glands, and matrix cells (Fig. 1 E and F). While LMO-4 transcripts were less abundant in interfollicular epidermis than in hair follicles, they were clearly detectable in, although not limited to, the basal layer (Fig. 1D). The expression pattern of LMO-4 overlaps with that of Clim-2 (compare with Fig. 1 A–C) both in hair follicles and interfollicular epidermis, consistent with the notion that these proteins could be components of the same transcriptional complex in epidermis.
Expression of LMO-4 During Mouse Development and in the Adult Nervous System.
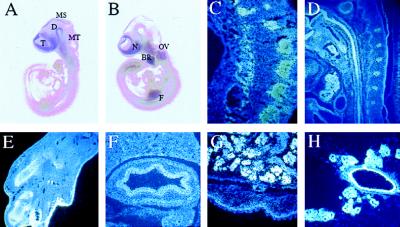
By using in situ hybridization we found that LMO-4 was expressed at least as early as e7.0. At this stage, expression was detected in the three major embryonic layers but absent from extraembryonic tissues with the exception of a weak signal in the amnion (data not shown). At e9.5 (Fig. 2A) and e10.5 (Fig. 2B) LMO-4 expression was prominent in neuroepithelium of the telencephalon, diencephalon, mesencephalon, metencephalon, and in the olfactory epithelium. High expression also was found in the first branchial arch, oral ectoderm, Rathke’s pouch, otic pit, dorsal aorta, dermatomyotome of somites, hindlimb, forelimb, and the tail bud (Fig. 2 A and B). Cross sections revealed heavy labeling in developing motor neurons of the spinal cord and dorsal root ganglia (Fig. 2C). At e11.5 LMO-4 expression pattern was similar but also included second and third branchial arches, right atrium, and urogenital ridge (data not shown).
Figure 2.
LMO-4 expression pattern during mouse development by in situ hybridization. (A) Whole-mount in situ hybridization of e9.5, lateral view. (B) Whole-mount in situ hybridization of e10.5, lateral view. (C) Cross section at e10.5 displaying strong signal in dorsal root ganglia. (D) Sagittal section of the neck area at e14.5 showing strong expression in esophagus, vertebrae and thymus. (E) Section of a hindlimb showing expression in cartilage and developing epidermis. (F) Section from e17.5 showing expression in stomach. (G) Section from e17.5 showing expression in submandibular gland and in basal layer of the epidermis. (H) Section from a breast gland showing expression in epithelial cells in a 14.5-day pregnant mother. T, telencephalon; D, diencephalon; MS, mesencephalon; MT, metencephalon; N, nasal epithelium, OV, otic vesicle; BR, first brancial arch; F, forelimb.
At e14.5, LMO-4 transcripts were prominent in the developing teeth, Meckel’s cartilage, epithelial lining of the gut from the mouth and to the intestinum (Fig. 2D), thymus (Fig. 2D), restricted regions of the atria, endothelium of large vessels, developing epidermis, cartilage of developing bone (Fig. 2 D and E), genital tubercle, and in specific regions of the nervous system, such as cortex (Fig. 3A), restricted regions of the midbrain and hindbrain, developing cerebellum, and spinal cord.
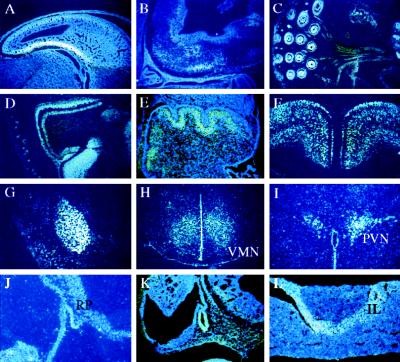
Figure 3.
Expression of LMO-4 in the nervous system and the pituitary gland by in situ hybridization. (A) Developing neocortex at e14.5. (B) Developing medulla and pituitary at e14.5. (C) Whisker pad at e17.5 showing strong signal in hair follicles and maxillary branches of the trigeminal nerve. (D) Olfactory bulb at P0. (E) Developing cerebellum at P0 showing expression in Purkinje cells. (F) Transverse section showing expression in neocortex of adult brain. (G) Transverse section showing expression in adult amygdala. (H) Transverse section showing expression in the adult ventromedial hypothalamic nucleus. (I) Transverse section showing expression in the adult paraventricular hypothalamic nucleus. (J) Sagittal section showing expression in Rathke’s pouch at e10.5. (K) Sagittal section showing expression in developing pituitary at e11.5. (L) Section from an adult pituitary gland. VMN, ventromedial nucleus; PVN, paraventricular nucleus; R, Rathke’s pouch; IL, intermediate lobe of the pituitary.
At e17.5, LMO-4 was strongly expressed in epithelial cells of the gastrointestinal tract (Fig. 2F), kidney and lung, the outer root sheath and matrix cells of whiskers (Fig. 3C), basal cells of the developing skin (Fig. 2G), submandibular gland (Fig. 2G), and developing teeth. The developing neocortex displayed strong signal in neopallidum and much weaker signal in intermediary and ventricular zones. Strong expression also was seen in selective regions of the thalamus and the olfactory bulb with strong signal in the external and internal plexiform layers, whereas weaker and absent labeling was detected in the granular layer and mitral cell layers, respectively. Robust expression was detected in restricted regions of the midbrain, pons, and medulla, with especially high signal in the pontine gray nucleus, the developing Purkinje cell layer of the cerebellum, spinal motor neurons, sensory ganglia, and pia and dura mater. LMO-4 mRNA also was detected in peripheral nerves consistent with glial expression (Fig. 3C).
LMO-4 mRNA in the nervous system at P0 paralleled the expression seen at e17.5 in most regions, such as in the olfactory bulb (Fig. 3D) and the developing Purkinje cell layer of the cerebellum (Fig. 3E). A prominent signal also was seen in neocortex with the highest levels in infragranular and supragranular layers but with absent or low levels in the molecular layer. LMO-4 expression was abundant in basal cells of the epidermis, submandibular glands, hair follicles, and at the periphery of ossification in vertebrae.
Transverse sections of the adult brain revealed that LMO-4 was strongly expressed in cortical layers 2–6 with slightly weaker signal in layer 4 (Fig. 3F), dorsal hippocampus, amygdala (Fig. 3G), striatum, and restricted regions of the thalamus, the lateral hypothalamus as well as in ventromedial (Fig. 3H) and paraventricular (Fig. 3I) hypothalamic nuclei.
During pituitary development, LMO-4 transcripts were observed as early as Rathke’s pouch stage at e9.5 and e10.5 (Fig. 3 J and K). At e14.5, labeling was detected throughout the developing pituitary gland, but in the adult, expression was highest in the intermediate lobe (Fig. 3L). In addition, expression was prominent in epithelial cells of the breast, especially during acinar formation (Fig. 2H).
LMO-4 Interacts with Clim-1 and Clim-2.
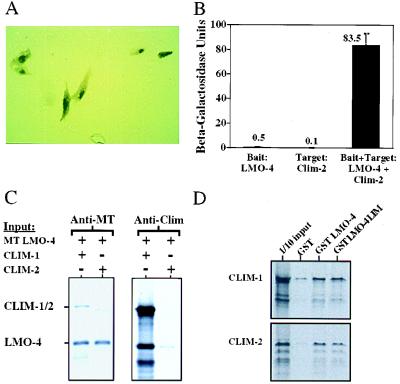
We tested whether LMO-4 was localized to the nucleus by transfecting a Myc-tagged cDNA into CV-1 cells. Immunohistochemical analyses showed that while there was some cytoplasmic staining, the protein was mainly nuclear (Fig. 4A), and this staining pattern was independent of whether Clim-1 or -2 was cotransfected with LMO-4 (data not shown). These results indicate that LMO-4 belongs to the class of nuclear LMO proteins, which previously have been shown to interact with the nuclear localized Clims (1, 2). The interaction between LMO-4 and Clim-2 appears to be strong as evidenced by the yeast two-hybrid interaction assays where Clim-2 fused to an activation domain led to strong LMO-4-dependent activation (Fig. 4B). In addition, 35S-labeled in vitro-translated LMO-4 and Clim proteins could be coimmunoprecipitated (Fig. 4C). By using GST pulldown assays we showed that the LMO-4 LIM domain interacted as strongly or better than the holoprotein with both Clim-1 and Clim-2, indicating that the interaction was mediated by the LIM domain (Fig. 4D), in accordance with findings previously described for the interaction of LMO-2 with Clim-2 (10). Collectively, these results suggest that LMO-4 and Clim proteins form a stable nuclear complex in cells where they are coexpressed.
Figure 4.
LMO-4 is a nuclear protein and interacts with Clim-1 and Clim-2. (A) Myc-tagged LMO-4 was transfected into CV-1 cells and detected with a monoclonal Myc antibody, by using peroxidase staining. (B) Interaction between LMO-4 and Clim-2 in the yeast two-hybrid system. (C) Coimmunoprecipitation of LMO-4 with Clim-1 and Clim-2. (D) A GST-interaction assay with the indicated GST proteins and 35S-labeled Clim-1 and Clim-2.
LMO-4 Interacts with the Mammalian Homologue of Drosophila DEAF-1.
While LIM homeodomain proteins exert their biological activity through DNA binding, there is no evidence that LMOs directly bind DNA (1, 2). Instead, LMOs and Clims are believed to interact with DNA-binding proteins, leading us to hypothesize that a complex containing LMO-4 and Clim coregulators might control gene activity by associating with unknown DNA-binding protein(s) in cell types where these factors are coexpressed.
To search for interacting DNA-binding proteins, we used LMO-4 as a bait in the yeast two-hybrid system with cDNA libraries from mouse skin and pituitary and human breast. From all three libraries, we isolated several LMO-4-interacting clones that included Clim-2, as well as cDNAs encoding the mouse homologue of Drosophila DEAF-1 (Fig. 5A) (17). Homologous genes, referred to as nuclear deformed epidermal autoregulatory factor-1-related (NUDR) transcriptional regulator protein (18) and Suppressin (19), have been isolated from other mammals.
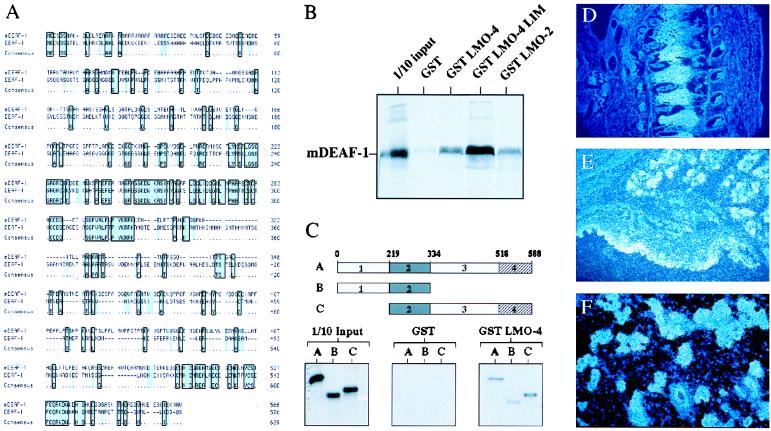
Figure 5.
Isolation and characterization of an LMO-4-interacting protein, mDEAF-1. (A) The amino acid sequence of mDEAF-1 compared with Drosophila DEAF-1. (B) A GST-interaction assay with the indicated GST proteins and 35S-labeled mDEAF-1. (C) A GST-interaction assay with the indicated GST proteins and the following 35S-labeled mDEAF-1 proteins shown schematically on top: A, full length; B, residues 1–344; C, residues 219–588. (D) In situ hybridization showing mDEAF-1 expression in dorsal root ganglia at e15.5, sagittal cut of neck region. (E) In situ hybridization showing mDEAF-1 expression in submandibular gland and epidermis at e17.5. (F) In situ hybridization showing expression of mDEAF-1 in breast epithelial cells from a 12.5-day pregnant mother.
The interaction between mDEAF-1 and LMO-4 was tested in GST-interaction assays (Fig. 5 B and C). In vitro-translated mDEAF-1 bound to the LMO-4 LIM domain, analogous with the observation that the LIM domain of LMO-2 mediates interactions with the transcription factors Tal-1 and GATA-1 (20). Furthermore, LMO-2 interacted with mDEAF-1, suggesting that the mDEAF-1-LIM interaction is not specific for LMO-4 (Fig. 5B). We used deletion mutants of mDEAF-1 to map and demonstrate specificity of the in vitro interaction. While in vitro-translated mDEAF-1 encompassing amino acids 1–334 interacted weakly with GST-LMO-4, an N-terminal deletion mutant encompassing amino acids 219–588 interacted as strongly as the holoprotein, suggesting that the LMO-4-interaction domain maps to the C-terminal region between amino acids 334 and 588 (Fig. 5C). Collectively, these in vitro experiments suggest that there is a direct interaction between LMO-4 and mDEAF-1.
In situ hybridization studies showed that mDEAF-1 had almost ubiquitous expression pattern during mouse embryogenesis, with higher expression in several tissues where both Clims and LMO-4 are expressed. These include regions of the central nervous system (data not shown), dorsal root ganglia (Fig. 5D), submandibular gland and epidermis (Fig. 5E), and breast (Fig. 5F). The observation that Clims, LMO-4, and mDEAF-1 are coexpressed in several tissues is consistent with the idea that they may form complexes in vivo.
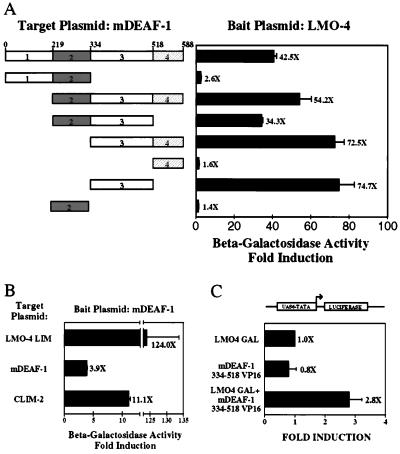
To characterize further the in vitro interactions and to test their in vivo relevance, we have used yeast two-hybrid interaction assays (Fig. 6A). In the yeast β-galactosidase reporter strain the LMO-4 bait plasmid alone gave low or nonexistent β-galactosidase activity that was increased 40-fold when a target plasmid encoding full-length mDEAF-1 was introduced with the bait. Introduction of several N- and C-terminal deletion mDEAF-1 mutants indicated that the region responsible for the LIM domain interaction mapped to the proline-rich domain between amino acids 334 and 518 (region 3; Fig. 6A), consistent with the in vitro mapping data. A bait plasmid encoding DEAF holoprotein also resulted in negligible β-galactosidase background in the yeast reporter strain. When a target plasmid encoding the LIM domain of LMO-4 was cointroduced, a more than 100-fold increase in β-galactosidase activity was observed, further confirming the in vivo interaction between mDEAF-1 and LMO-4 and mapping it to the LIM domain of LMO-4 (Fig. 6B). Cointroduction of Clim-2 activation plasmid also significantly increased β-galactosidase activity, supporting the possibility that there may be a direct mDEAF-1-Clim interaction (Fig. 6B), which was consistent with GST pulldown assays testing interactions between mDEAF-1 and Clims (data not shown).
Figure 6.
LMO-4 and mDEAF-1 interact in yeast and mammalian cells. (A) Yeast two-hybrid interaction assay using LMO-4 as a bait with the indicated mDEAF-1 target plasmids. Results are expressed as fold induction over bait alone. (B) Yeast two-hybrid interaction assay using mDEAF-1 as a bait with the indicated target plasmids. (C) Mammalian two-hybrid interaction assay using LMO-4 as a bait and the interaction domain of mDEAF-1 as a target plasmid. These plasmids were introduced with a UAS6-TATA reporter into CV-1 cells.
To test whether the interaction between LMO-4 and mDEAF-1 also occurred in mammalian cells, we linked LMO-4 to a GAL4 DNA-binding domain and the LIM-interaction domain of mDEAF-1 to the VP16 activation domain. These expression plasmids were cotransfected with a GAL-site dependent luciferase reporter gene into HeLa cells. A clear LMO-4-dependent activation was observed with the LIM interaction domain of mDEAF-1 (Fig. 6C) but not with a noninteracting domain of mDEAF-1 (data not shown). Together, our results suggest that mDEAF-1, LMO-4, and Clims may form transcriptionally active complexes in cells where they are coexpressed, such as in keratinocytes, and neural and breast epithelial cells. Thus these data support the hypothesis that LMO-4 exerts its function by interacting with specific DNA-binding transcription factors, including mDEAF-1.
DISCUSSION
Three mammalian LMO factors (LMO-1, -2, and -3) previously have been described. It has been shown that LMO-2 interacts strongly with the basic helix–loop–helix domain of the transcription factor TAL1 and that these proteins, as well as Clim-2, exist in a complex in erythroid cells (10, 20–24). Interestingly, mutations of TAL1 resulted in a phenotype in erythroid cells identical to that of LMO-2 mutated mice (25). Furthermore, mice harboring a mutation in the gene encoding the DNA-binding protein, GATA-1, exhibit a similar phenotype (26), and it has been shown that LMO-2 can interact with GATA factors (22, 24). These findings suggest a model in which LMO factors can tether to DNA by associating with DNA-binding proteins, thus allowing the Clim coregulators to interact with transactivators that do not contain a covalently linked LIM domain. A second model for LMO action has been proposed based on genetic experiments in Drosophila (27, 28). Misexpression of Drosophila LMO (dLMO) in the developing wing imaginal disc leads to a phenotype that is similar to that seen with inactivation of the LIM homeodomain factor Apterous, suggesting that dLMO may sequester the Clim homologue CHIP, thus inactivating Apterous. It has been suggested that a similar mechanism may explain the oncogenic action of LMO-2 in lymphocytes (27).
Because of the widespread, yet selective, expression of LMO-4 throughout development (16) and in the adult, it is likely that LMO-4 has multiple roles. Thus, while LMO-4 often is expressed in proliferative cells, such as in the epidermis, it is also clearly expressed in postmitotic cells such as in the adult nervous system. Considering the molecular organization of LMOs, two tandem LIM domains with little other sequence, it is tempting to speculate that in addition to regulating the levels of Clims available for interaction with LIM homeodomain factors, LMO-4 may be used as an adapter molecule in many different DNA-binding molecular complexes. Consistent with this idea are the present findings that LMO-2 and LMO-4 can interact with a domain found in mDEAF-1, and preliminary results suggesting that LMO-4 specifically interacts with additional transcription factors (B.A., unpublished observations).
mDEAF-1 appears to be ubiquitously expressed (18, 19) whereas LMO-4 expression is more restricted. Thus, LMO-4 may selectively modulate the activity of mDEAF-1 during development and in distinct tissues, particularly in the nervous system and many epithelial cell types. In all assays, the interactions between LMO-4 and Clims appear to be of very high affinity, consistent with previous observations for the interactions between LIM homeodomain factors and Clims and between LMO-2 and Clims (7–10). In contrast to the binding to Clims, the interaction of LMO-4 with mDEAF-1 appears to be weaker, analogous to interactions of LMO-2 with GATA and helix–loop–helix factors (10), thus possibly allowing dynamically regulated interactions between LMO/Clim complexes and transcription factors.
Previous studies in Drosophila established that the homeodomain gene Deformed contained a complex autoregulatory element where the Deformed binding site was necessary but not sufficient for autoregulation in transgenic flies; additional sequences adjacent to the Deformed binding site were required (17). These sequences are bound by DEAF-1, which therefore acts as a cofactor for homeodomain function. Indeed, binding sites for DEAF-1 were identified in several genes adjacent to Deformed binding sites, suggesting that it might have a general role as a cofactor for enhancers targeted by Deformed (17). While it remains to be determined whether the mammalian homologue of DEAF-1 has similar roles, our results suggest the possibility that an LMO/Clim complex may be involved in gene control by mammalian DEAF-1, such as in the regulation of proenkephalin transcription (18).
In summary, we have identified an additional member of the LIM-only protein family, referred to as LMO-4. This factor associates with the LIM coregulators Clim-1 and -2, and with the DNA-binding protein mDEAF-1, all of which are expressed in an overlapping manner in several tissues, including the nervous system and epidermis. Our results suggest that LMO-4 may coordinate the assembly of regulatory complexes on potential target genes. Furthermore, it is plausible to propose a function for LMOs as critical adapter molecules for several distinct classes of transcription factors in pattern formation and cell-type determination during embryogenesis.
Acknowledgments
We thank Catherine Carrière, Ola Hermanson, Kristen Jepsen, and Bill McGinnis for advice, discussions, and comments, and Shawn O’Connell for help with in situ hybridization. This work was supported by the Irving Weinstein Foundation and National Institutes of Health Grant AR044882 (to B.A.). M.G.R. is an Investigator with the Howard Hughes Medical Institute.
ABBREVIATIONS
- DEAF-1
Deformed epidermal autoregulatory factor 1
- mDEAF-1
mouse DEAF-1
- GST
glutathione S-transferase
- e(n)
embryonic day
Footnotes
References
- 1.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 2.Jurata L W, Gill G N. Curr Top Microbiol Immunol. 1998;228:75–113. doi: 10.1007/978-3-642-80481-6_4. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Garcia I, Rabbitts T H. Semin Cancer Biol. 1993;4:349–358. [PubMed] [Google Scholar]
- 4.Foroni L, Boehm T, White L, Forster A, Sherrington P, Liao X B, Brannan C I, Jenkins N A, Copeland N G, Rabbitts T H. J Mol Biol. 1992;226:747–761. doi: 10.1016/0022-2836(92)90630-3. [DOI] [PubMed] [Google Scholar]
- 5.Yamada Y, Warren A J, Dobson C, Forster A, Pannell R, Rabbitts T H. Proc Natl Acad Sci USA. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren A J, Colledge W H, Carlton M B, Evans M J, Smith A J, Rabbitts T H. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 7.Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Nature (London) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 8.Bach I, Carriere C, Ostendorff H P, Andersen B, Rosenfeld M G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 9.Jurata L W, Kenny D A, Gill G N. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visvader J E, Mao X, Fujiwara Y, Hahm K, Orkin S H. Proc Natl Acad Sci USA. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurata L W, Gill G N. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breen J J, Agulnick A D, Westphal H, Dawid I B. J Biol Chem. 1998;273:4712–4717. doi: 10.1074/jbc.273.8.4712. [DOI] [PubMed] [Google Scholar]
- 13.Jurata L W, Pfaff S L, Gill G N. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- 14.Morcillo P, Rosen C, Baylies M K, Dorsett D. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen B, Weinberg W C, Rennekampff O, McEvilly R J, Bermingham J R, Jr, Hooshmand F, Vasilyev V, Hansbrough J F, Pittelkow M R, Yuspa S H, Rosenfeld M G. Genes Dev. 1997;11:1873–1884. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- 16.Kenny D A, Jurata L W, Saga Y, Gill G N. Proc Natl Acad Sci USA. 1998;95:11257–11262. doi: 10.1073/pnas.95.19.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross C T, McGinnis W. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 18.Huggenvik J I, Michelson R J, Collard M W, Ziemba A J, Gurley P, Mowen K A. Mol Endocrinol. 1998;12:1619–1639. doi: 10.1210/mend.12.10.0181. [DOI] [PubMed] [Google Scholar]
- 19.LeBoeuf R D, Ban E M, Green M M, Stone A S, Propst S M, Blalock J E, Tauber J D. J Biol Chem. 1998;273:361–368. doi: 10.1074/jbc.273.1.361. [DOI] [PubMed] [Google Scholar]
- 20.Wadman I, Li J, Bash R O, Forster A, Osada H, Rabbitts T H, Baer R. EMBO J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson R C, Lavenir I, Larson T A, Baer R, Warren A J, Wadman I, Nottage K, Rabbitts T H. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 22.Osada H, Grutz G, Axelson H, Forster A, Rabbitts T H. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valge-Archer V E, Osada H, Warren A J, Forster A, Li J, Baer R, Rabbitts T H. Proc Natl Acad Sci USA. 1994;91:8617–8621. doi: 10.1073/pnas.91.18.8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt F W, Orkin S H. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 26.Fujiwara Y, Browne C P, Cunniff K, Goff S C, Orkin S H. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milan M, Diaz-Benjumea F J, Cohen S M. Genes Dev. 1998;12:2912–2920. doi: 10.1101/gad.12.18.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng C, Justice N J, Abdelilah S, Chan Y M, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1998;95:10637–10642. doi: 10.1073/pnas.95.18.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]