Abstract
A phospholipase D (PLD) was shown recently to decorate microtubules in plant cells. Therefore, we used tobacco BY-2 cells expressing the microtubule reporter GFP-MAP4 to test whether PLD activation affects the organization of plant microtubules. Within 30 min of adding n-butanol, a potent activator of PLD, cortical microtubules were released from the plasma membrane and partially depolymerized, as visualized with four-dimensional confocal imaging. The isomers sec- and tert-butanol, which did not activate PLD, did not affect microtubule organization. The effect of treatment on PLD activation was monitored by the in vivo formation of phosphatidylbutanol, a specific reporter of PLD activity. Tobacco cells also were treated with mastoparan, xylanase, NaCl, and hypoosmotic stress as reported activators of PLD. We confirmed the reports and found that all treatments induced microtubule reorganization and PLD activation within the same time frame. PLD still was activated in microtubule-stabilized (taxol) and microtubule-depolymerized (oryzalin) situations, suggesting that PLD activation triggers microtubular reorganization and not vice versa. Exogenously applied water-soluble synthetic phosphatidic acid did not affect the microtubular cytoskeleton. Cell cycle studies revealed that n-butanol influenced not just interphase cortical microtubules but also those in the preprophase band and phragmoplast, but not those in the spindle structure. Cell growth and division were inhibited in the presence of n-butanol, whereas sec- and tert-butanol had no such effects. Using these novel insights, we propose a model for the mechanism by which PLD activation triggers microtubule reorganization in plant cells.
INTRODUCTION
Microtubules are dynamic tubular structures formed by 13 protofilaments, each of which is formed by a linear polymerization of α- and β-tubulin heterodimers (Nogales et al., 1999). They are 25 nm in diameter and range up to 20 μm in length (Nogales, 2000). The ability of microtubules to grow by polymerization and shrink by depolymerization, together with a tendency to shift between these phases, make them highly dynamic cellular entities (Mitchison and Kirschner, 1984; Dhonukshe and Gadella, 2003). Cells use them to produce different microtubular conformations that are tailored to fit growth, division, and stimulus-response requirements (Desai and Mitchison, 1997). Cell wall–confined and vacuolated plant cells have evolved specialized microtubular arrays that perform important tasks (Newcomb, 1969; Goddard et al., 1994; Kost and Chua, 2002; Wasteneys, 2002). For example, transversely arranged interphase microtubules assist cellulose deposition in expanding cells (Green, 1962; Ledbetter and Porter, 1963; Cyr, 1994; Burk and Ye, 2002). At the onset of mitosis, cortical microtubules form a compact preprophase band (PPB) encircling the nucleus that marks the plane of the future cell wall (Mineyuki, 1999), whereas the mitotic noncentrosomal spindle microtubules divide the duplicated chromosomes (Baskin and Cande, 1990). Finally, during cytokinesis, the phragmoplast microtubules form tracks for vesicles that deposit material for the new cell wall that physically separates the daughter cells (Samuels et al., 1995; Smith, 2001; Verma, 2001).
During interphase, microtubules can be seen as spirals that run the cell length (Lloyd and Chan, 2002) and remain very close to the plasma membrane (Ledbetter and Porter, 1963; Newcomb, 1969; Seagull and Heath, 1980; McClinton and Sung, 1997). They are believed to bind the membrane and assist cellulose microfibril deposition (Green, 1962; Ledbetter and Porter, 1963; Mueller and Brown, 1982a, 1982b) by regulating the movement of transmembrane cellulose synthase complexes (Heath, 1974; Baskin, 2001) that can be seen as rosettes in freeze-fracture preparations (Brown and Montezinos, 1976; Mueller and Brown, 1982a, 1982b). When these enzymes polymerize glucose and form cellulose microfibrils, the rosette is continuously pushed away from the nascent polymer, driving it through the fluid lipid membrane (Heath, 1974; Giddings et al., 1980; Mueller and Brown, 1982a). The rosette is free to move but is restricted between the microtubule/membrane corridors (Herth, 1985; Giddings and Staehelin, 1988; Hasezawa and Nozaki, 1999). The resulting deposition of cellulose microfibrils determines cell shape by favoring expansion along the long axis of the cell (Giddings and Staehelin, 1988; Williamson, 1991). This cell shape can be influenced by many factors, from physical pressure (Fisher and Cyr, 2000), gravity (Himmelspach et al., 1999), electric field (Hush and Overall, 1991), and pathogen attack (Kobayashi et al., 1994) to treatment with hormones (Shibaoka, 1994). Such treatments reorient the microtubules (Nick, 1998), and in many cases, this change is reflected in the orientation of cellulose microfibrils in the cell wall, which in turn dictates cell expansion and shape (Pickett-Heaps, 1967; Robinson and Quader, 1982; Quader, 1986; Baskin, 2001).
How are microtubules linked to the membrane? In electron micrographs, direct cross-bridges are visible (Hardham and Gunning, 1978; Giddings and Staehelin, 1988; Sonobe and Takahashi, 1994; Vesk et al., 1996; Sonobe et al., 2001). Transmembrane proteins probably are involved, because some extracted tubulin behaves as if it is bound to hydrophobic proteins (Sonesson et al., 1997). Also, the intracellular microtubules are affected by factors outside of the protoplast. For example, they detach from the plasma membrane when trypsin is added outside of the cell, and microtubules in protoplasts depolymerize in the cold but become more stable when a wall is formed on the protoplast surface (Akashi and Shibaoka, 1991). External application of wall protein extensin, or charged polymers such as poly-l-Lys, to the outside of protoplasts results in the stabilization of cortical microtubules (Akashi et al., 1990). Additionally, membrane ghosts bind microtubules polymerized from extracts of the same cells when the preexisting microtubules are removed by treatment (Sonobe and Takahashi, 1994). These findings gave rise to the concept of microtubule–plasma membrane attachments.
Various plant microtubule-associated proteins (MAPs) have been characterized (Lloyd and Hussey, 2001), including motor proteins (Reddy, 2001). None of them have proven to be membrane linkers, except for a recently isolated 90-kD polypeptide from tobacco membranes (Marc et al., 1996). Moreover, this p90 was identified recently as a phospholipase D (PLD) and was shown to associate strongly with microtubules and membranes in both immunofluorescence and cosedimentation assays (Gardiner et al., 2001).
PLDs are enzymes that can hydrolyze structural phospholipids such as phosphatidylcholine to produce phosphatidic acid (PA) and free choline. Some PLDs are involved in basic phospholipid metabolism, whereas others were discovered to play a role in cell signaling by producing the second messenger PA (Munnik, 2001; Wang, 2001; Wang et al., 2002). In the Arabidopsis genome, 12 PLD genes can be identified, of which 6 have been cloned and characterized by Wang and colleagues (Elias et al., 2002; Qin and Wang, 2002). The PLDs fall into two groups, referred to as PX/PH- and C2-PLDs, based on the presence of lipid binding domains (PX and PH or C2) in their N-terminal sequences (Elias et al., 2002). Ten of them are C2-PLDs that are interesting because they seem to be plant specific. They have been divided further into PLDα, -β, -γ, and -δ subclasses based on sequence homology and their in vitro dependence on Ca2+ and phosphoinositides (Qin et al., 2002; Qin and Wang, 2002).
We do not know which genes are involved in signaling as opposed to metabolism, but we do know that several stress treatments rapidly stimulate PLD activity (reviewed by Wang, 2001, 2002; Meijer and Munnik, 2003). They include wounding (Wang et al., 2000; Zien et al., 2001), water stress (Frank et al., 2000; Munnik et al., 2000; Katagiri et al., 2001; Sang et al., 2001), cold stress (Ruelland et al., 2002; Welti et al., 2002), treatment with the stress hormone abscisic acid (Ritchie and Gilroy, 1998; Jacob et al., 1999; Hallouin et al., 2002), and pathogen/symbiont infection (Young et al., 1996; van der Luit et al., 2000; den Hartog et al., 2001; Laxalt et al., 2001; Laxalt and Munnik, 2002). In some cases, evidence for PA stimulating the downstream responses also was provided (Munnik, 2001). In general, PA is thought to work by binding target proteins downstream in the signaling cascade. Binding may activate these components directly or indirectly by concentrating them at membrane loci where they activate each other (Laxalt and Munnik, 2002).
Apart from the stimuli already mentioned, two groups of compounds are used as general PLD stimulators in plant cells. They are mastoparans, which are derivatives of tetradecapeptides present in wasp venom, and alcohols such as butanol (Munnik et al., 1995; van Himbergen et al., 1999). The mechanism by which these compounds activate PLD remains unknown, but they both have the practical advantage that active and inactive analogs are known. For example, Mas7 and n-butanol are active, whereas Mas17 and tert-butanol are inactive (Munnik et al., 1995; van Himbergen et al., 1999). Importantly, PLD activity can be measured in vivo, because it can transfer the phosphatidyl group of its substrate not just to water, forming PA, but also to a primary alcohol such as n-butanol, forming phosphatidylbutanol (PBut). Thus, after n-butanol is added to living cells, the amount of PBut formed provides a quantitative measure of PLD activity (Munnik et al., 1995; Munnik, 2001).
Immunofluorescence labeling has shown that the p90 PLD decorates microtubules and, in their absence, binds to the plasma membrane (Gardiner et al., 2001). Still, no association between the activation status of PLD and microtubule reorganization events has been observed directly in live plant cells. If PLD links microtubules to the plasma membrane, what would be the response to stimulating PLD activity? A hypothesis involving PLD holding microtubules to the membranes has been proposed (Munnik and Musgrave, 2001). It is based on the fact that when PLD hydrolyzes a phospholipid, it forms a covalent link with the phosphatidyl group in the membrane before transferring it to water. If this intermediate is stable for some time in vivo, then PLD and the microtubules will remain attached to the membrane. However, on activation, attachment will become ephemeral, because the phosphatidyl group is transferred immediately to water. Using the green fluorescent protein (GFP)–MAP4 plant microtubule reporter (Marc et al., 1998; Granger and Cyr, 2000; Dhonukshe and Gadella, 2003) in stably transformed tobacco Bright Yellow-2 (BY-2) cells, we demonstrate here that PLD activation triggers the reorganization of plant microtubules.
RESULTS
n-Butanol Releases Microtubules from the Plasma Membrane in a Time-Dependent Manner
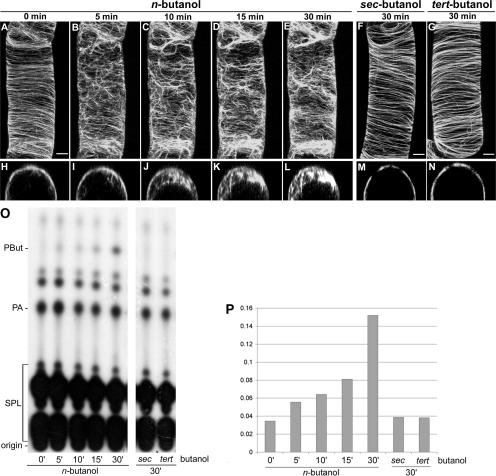
To test the effect of PLD activation on BY-2 cells expressing GFP-MAP4, we used 0.5% butanol, because we have shown previously that n-butanol, but not sec- or tert-butanol, is active at this concentration (Munnik et al., 1995). Although cortical microtubules in interphase cells normally lie in spirals very close to the plasma membrane (Figure 1A), 0.5% n-butanol dramatically induced their release into the cytosol (Figures 1B to 1I). Dissociation started almost immediately and progressed throughout the entire cell, as visualized by four-dimensional confocal imaging (Figures 1C to 1E and 1J to 1L). After 30 min, most microtubules were dissociated from the membrane. When equal concentrations of sec- or tert-butanol were used, no such effects were observed (Figures 1F, 1G, 1M, and 1N). Light microscopy analysis of control and n-, sec-, or tert-butanol–treated cells revealed no obvious differences in terms of cell shape and cytoplasmic streaming (data not shown). The effect of n-butanol was not specific to cells expressing GFP-MAP4, because cell lines expressing other microtubule reporters (i.e., GFP-TUA6 and YFP-CLIP1701-1240) (Dhonukshe and Gadella, 2003) responded very similarly (data not shown).
Figure 1.
Effect of n-Butanol on Plant Microtubules and Phospholipid Metabolism.
(A) to (E) Confocal laser scanning micrographs of an interphase BY-2 cell, stably expressing GFP-MAP4, after treatment with 0.5% n-butanol.
(F) and (G) Confocal laser scanning micrographs of GFP-MAP4–transformed BY-2 cells 30 min after the addition of 0.5% sec-butanol and 0.5% tert-butanol.
(H) to (N) Confocal laser scanning micrographs of single xz cross-sections of the cells shown in (A) to (G).
(O) TLC analysis of 32P-labeled lipids extracted from GFP-MAP4–transformed BY-2 cells after treatment with 0.5% n-, sec-, or tert-butanol. Times are shown in minutes. SPL, structural phospholipids.
(P) Quantification of the in vivo PLD activity obtained from the autoradiograph shown in (O). 32P-PBut levels were calculated as a percentage of total radioactive structural phospholipids. The PBut level at time 0 represents background radioactivity.
(A) to (G) show maximum projections of 40 confocal slices covering a depth of 15 μm and denoting approximately a hemicylinder of each cell. Bars = 5 μm in the xy plane in (A) to (N).
To check the ability of PLD to use these butanol isomers as transphosphatidylation substrates, in vivo PLD activity assays were performed in parallel. GFP-MAP4–transformed BY-2 cells were prelabeled with 32Pi and then treated with the different alcohols for the times indicated in Figure 1. Lipids were extracted subsequently, separated by thin layer chromatography (TLC), and visualized by autoradiography. As shown in Figures 1O and 1P, cells readily produced 32P-PBut from n-butanol but not from sec- or tert-butanol, in correlation with each alcohol's ability to induce the microtubular detachments from the plasma membranes. No significant change in the overall lipid composition was observed.
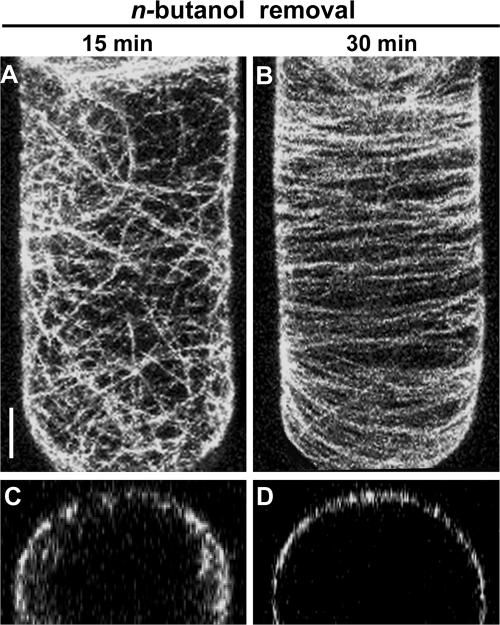
To determine whether the microtubular dissociation was reversible, cells were washed after 60 min of n-butanol treatment and reanalyzed. As shown in Figure 2, within 15 min, microtubules started reappearing at the plasma membrane (Figures 2A and 2C), whereas after 30 min, cells resembled controls that were washed but not treated with alcohol (Figures 2B and 2D). This finding indicates that the effect of n-butanol was dynamic, reversible, and nontoxic.
Figure 2.
Effect of n-Butanol on Microtubular Structures in BY-2 Cells Is Reversible.
(A) and (B) Confocal laser scanning micrographs of a GFP-MAP4–transformed interphase cell 15 and 30 min after the removal of n-butanol (by washing four times with BY-2 medium). These images are maximum projections of 40 confocal slices covering a depth of 15 μm and denoting approximately a hemicylinder of each cell. Bar = 5 μm.
(C) and (D) Confocal laser scanning micrographs of cross-sections (xz plane) of GFP-MAP4–transformed BY-2 cells after n-butanol removal.
Mastoparan, a PLD Activator, Triggers Plant Microtubule Reorganization
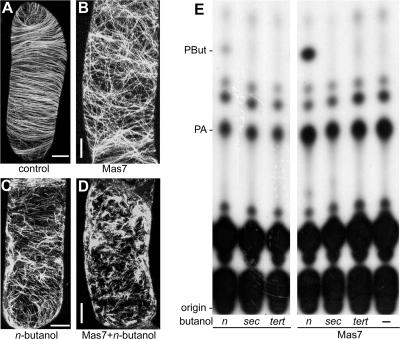
Mastoparan is a polypeptide of 14 amino acids that can activate G-proteins (Law and Northrop, 1994). It is one of the most potent in vivo activators of plant PLD, as witnessed by responses in the green alga Chlamydomonas moewusii (Munnik et al., 1995; van Himbergen et al., 1999), in suspension-cultured tomato cells (van der Luit et al., 2000; Laxalt et al., 2001), in carnation flower petals (de Vrije and Munnik, 1997), in Craterostigma plantagineum leaves (Frank et al., 2000), and in roots of Vicia sativa seedlings (den Hartog et al., 2001). Treatment of our BY-2 cells with Mas7 (5 μM), an active synthetic analog of mastoparan, rapidly reorganized the cortical microtubules by changing their orientation from transverse to random (Figures 3A and 3B). The Mas7 effect was even stronger when cells were treated in the presence of 0.5% n-butanol (Figure 3D) and compared with cells treated with n-butanol alone (Figure 3C). This synergy between n-butanol and Mas7 was not observed with sec- or tert-butanol (data not shown). Treatment of cells with the control peptide Mas17 (5 μM), an inactive mastoparan analog, had no effect on the microtubules (data not shown).
Figure 3.
The PLD Activator Mastoparan Induces the Reorganization of Microtubules in BY-2 Cells.
(A) to (D) Confocal laser scanning micrographs of GFP-MAP4–transformed interphase BY-2 cells that were treated with or without the synthetic mastoparan analog Mas7 (5 μM) in the presence or absence of 0.5% n-butanol. These images are maximum projections of 40 confocal slices covering a depth of 15 μm and denoting approximately a hemicylinder of each cell. Bars = 5 μm.
(A) Control.
(B) Mas7.
(C) n-Butanol.
(D) Mas7 and n-butanol.
(E) TLC analyses of 32P-prelabeled and mastoparan-treated or untreated cells in the presence or absence of n-, sec-, or tert-butanol. PBut and PA are indicated.
To confirm whether Mas7 activated PLD in transgenic BY-2 cells, 32Pi-labeling experiments were performed to monitor its activity in vivo. As shown in Figure 3E, Mas7 stimulated the formation of PA, the natural product of PLD activity. However, PA is not a specific reporter of PLD activity, because it also is produced by the activation of PLC in conjunction with diacylglycerol kinase (Munnik et al., 1998b; Munnik, 2001). That PLD nonetheless was activated was found when n-butanol was included as a PLD reporter. As shown in Figure 3E, Mas7 strongly stimulated the production of PBut, which did not occur in the presence of sec- or tert-butanol, because these alcohols cannot be transphosphatidylated.
Effects of Other Reported PLD Activators on the Microtubular Cytoskeleton
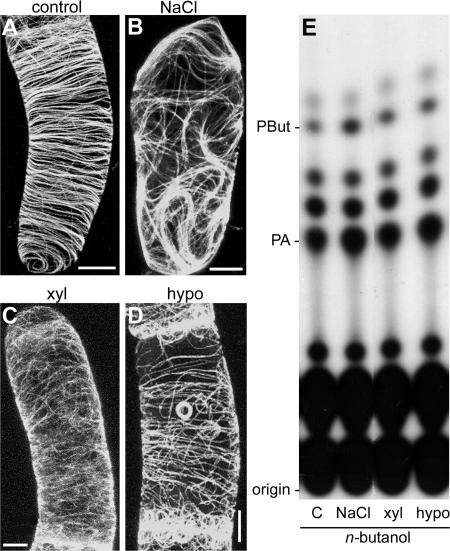
Previously, we found for suspension-cultured tomato cells that PLD activity is stimulated rapidly by hyperosmotic stress (Munnik et al., 2000), the fungal elicitor xylanase (van der Luit et al., 2000; Laxalt et al., 2001), and hypoosmotic stress (T. Munnik, unpublished results). To investigate what effects they have on the microtubular cytoskeleton, GFP-MAP4–transformed BY-2 cells were treated for 15 min with NaCl (150 mM) or xylanase (200 μg/mL) or diluted 1:1 with distilled water (hypoosmotic stress). As shown in Figures 4A to 4D, all three treatments induced rearrangements in microtubule organization. Hypoosmotic stress had the least effect but resulted in the formation of peculiar ring-like structures (Figure 4D). To correlate these results with the activity of PLD in vivo, lipid analyses of 32P-labeled cells were performed in parallel, using PBut as readout. As shown in Figure 4E, all three treatments increased the formation of PBut, although with varying intensities. These findings again show that there is a correlation between PLD activation and microtubular rearrangements, irrespective of the nature of the activator.
Figure 4.
Effect of Some Other PLD Activators on Microtubules in BY-2 Cells.
(A) to (D) Confocal laser scanning micrographs of GFP-MAP4–transformed interphase cells after treatment with some known PLD activators. These images are maximum projections of 40 confocal slices covering a depth of 15 μm and denoting approximately a hemicylinder of each cell. Bars = 5 μm.
(A) Control.
(B) Osmotic stress (150 mM NaCl).
(C) Fungal elicitor xylanase (100 μg/mL).
(D) Hypoosmotic stress (1:1 dilution with distilled water).
(E) TLC analyses of lipid extracts from 32P-prelabeled cells treated with or without the agonists noted above in the presence of 0.5% n-butanol.
Effects of Microtubule Drugs and PLC Inhibitors
To determine whether PLD activation is the cause or a consequence of plant microtubular reorganization, we pretreated BY-2 cells with the microtubule-stabilizing drug taxol (inhibits depolymerization) and with the microtubule-depolymerizing herbicide oryzalin for 1 h before n-butanol treatment. As shown in Figure 5A, taxol (10 μM) stabilized the plant microtubules by arranging them into transverse cables. The effect of n-butanol on taxol-pretreated cells (Figure 5B) was not as dramatic as when taxol was absent (Figure 1D), but still the microtubules detached from the plasma membrane (Figures 5C and 5D). This finding suggests that the more severe microtubular rearrangements normally seen with n-butanol probably are attributable to depolymerization after their release from the membrane. In oryzalin-pretreated cells, the microtubular structures had disappeared almost completely, reflecting their depolymerization (Figures 5E and 5F). Consequently, the subsequent n-butanol treatment had no significant effect (Figures 5E to 5H).
Figure 5.
Effects of Microtubule-Stabilizing and -Depolymerizing Drugs on n-Butanol–Triggered Microtubule Release and PBut Formation in GFP-MAP4–Transformed BY-2 Cells.
(A) to (D) Confocal laser scanning micrographs of interphase cells preincubated with 10 μM taxol ([A] and [C]) and subsequent treatment with 0.5% n-butanol for 15 min ([B] and [D]).
(E) to (H) Confocal laser scanning micrographs of interphase cells preincubated with 10 μM oryzalin to depolymerize the microtubules ([E] and [G]) and subsequent treatment with 0.5% n-butanol for 15 min ([F] and [H]).
(A), (B), (E), and (F) show maximum projections of 40 confocal slices covering a depth of 15 μm and denoting approximately a hemicylinder of each cell. (C), (D), (G), and (H) show xz cross-sections of the cells shown in (A), (B), (E), and (F), respectively. Bars = 5 μm.
(I) TLC analyses of lipid extracts from 32P-prelabeled and taxol- or oryzalin-pretreated or untreated BY-2 cells with 15 min of incubation in 0.5% n-butanol.
As is evident from the TLC analysis shown in Figure 5I, no changes in PBut production were observed in either taxol- or oryzalin-pretreated cells. Together, these results suggest that PLD activity does not require dynamic microtubules but that microtubule detachment does require PLD activity.
To determine if the PLC pathway had any influence on the observed microtubular effects, GFP-MAP4–transformed BY-2 cells were treated with the PLC antagonists neomycin (200 μM) and U73122 (10 μM). However, no microtubular effects were found, nor could the effect of n-butanol on microtubules be inhibited (see supplemental data online). In addition, pertussis toxin (10 μg/mL), an inhibitor of certain G-proteins, affected neither the microtubular organization nor the n-butanol effect (see supplemental data online).
Water-Soluble Synthetic PA Does Not Influence Microtubule Reorganization
Because PLD generates PBut at the cost of its natural product, PA, n-butanol also could be considered an inhibitor of PA formed via the PLD pathway. To determine if a change in PA had any influence on the reorganization of microtubules, GFP-MAP4–transformed BY-2 cells were treated with a mixture (20 μM) of dioctanoyl-PA and BODIPY 558/568-C12–labeled PA (BODIPY-PA), which allowed us to simultaneously monitor the microtubules (in green) and PA (in red) in the same cell. As shown in Figures 6A to 6C, the cells and their microtubules remained unaffected in the presence of increased PA levels, even 30 min after BODIPY-PA application. Nonetheless, 0.5% n-butanol was able to trigger the reorganization of microtubules. To determine whether the BODIPY-PA was metabolized during the experiment, cells were extracted and their lipids analyzed by TLC. As shown in Figure 6G, the intact fluorescent PA still was present and was not affected by treatment with n-butanol. Because the PLC pathway can contribute to the production of PA, these BODIPY-PA experiments also were performed in the presence of the PLC inhibitors neomycin (200 μM) and U73122 (10 μM). However, again, no effects were found (data not shown). These results suggest that PA (a natural product of PLD activation) does not influence microtubule reorganization, although it has to be considered that these synthetic PAs have a different acyl chain composition compared with the naturally occurring species.
Figure 6.
PA Does Not Induce Microtubular Rearrangements.
(A) to (F) Confocal laser scanning micrographs of GFP-MAP4–transformed interphase BY-2 cells pretreated with 20 μM BODIPY-labeled PA (red) in the presence ([D] to [F]) or absence ([A] to [C]) of 0.5% n-butanol. These images are maximum projections of 40 confocal slices covering a depth of ∼20 μm and denoting approximately a hemicylinder of each cell. Bars = 5 μm.
(G) TLC analyses of lipid extracts from BODIPY-labeled, PA-treated BY-2 cells in the presence or absence of 0.5% n-butanol with controls. PC, phosphatidylcholine.
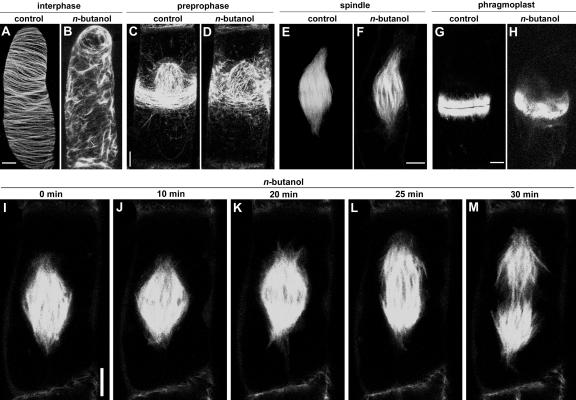
PLD Affects Interphase, Preprophase, and Phragmoplast Microtubules but Not Spindle Microtubules
Immunofluorescence colocalization studies with antibodies against tubulin and p90 have shown that PLD binds microtubules in all four major arrays (Gardiner et al., 2001). To investigate the effect of n-butanol on microtubular arrays formed at successive mitotic stages, GFP-MAP4 distribution was monitored in synchronized BY-2 cells immediately after the addition of 0.5% n-butanol. As shown in Figures 7A to 7H, n-butanol induced microtubule rearrangements at all stages except the spindle phase. To confirm the latter finding, spindle microtubules were followed in time in the presence of n-butanol. As shown in Figures 7I to 7M, their ability to separate chromosomes and advance to the telophase was not affected by n-butanol. This result suggests that only the microtubules in close proximity to membranes are affected, consistent with PLD's role as a membrane anchor for microtubules.
Figure 7.
Effect of n-Butanol on Different Microtubular Arrays in Dividing BY-2 Cells.
(A) to (H) Confocal laser scanning micrographs of GFP-MAP4–transformed BY-2 cells at various stages of the cell cycle recorded after 15 min of treatment with or without 0.5% n-butanol. The effects on interphase cortical microtubules ([A] and [B]), microtubules in the preprophase band ([C] and [D]), spindle microtubules ([E] and [F]), and phragmoplast microtubules ([G] and [H]) are shown. These images are maximum projections of 40 confocal slices covering a depth of almost 20 μm and denoting approximately a hemicylinder of each cell. Bars = 5 μm.
(I) to (M) Confocal laser scanning micrographs of a time-lapse study of spindle microtubules after treatment with 0.5% n-butanol. (I) corresponds to prophase, and (M) corresponds to telophase. The images are single median confocal sections of the cell. Bar = 5 μm.
Analysis of in vivo PLD activity in synchronized 32Pi-labeled BY-2 cells revealed no significant differences in PBut formation during the spindle and other stages of the cell cycle (data not shown). This finding might be attributable to incomplete synchronization, because the percentage of cells simultaneously displaying a spindle structure was <40%, whereas the percentage undergoing division was much higher, and although mitosis took ∼200 min, spindle formation was completed in 30 min. This failed to provide a sufficient time span to effectively separate the spindle stage from the overlapping influence of the PPB and phragmoplast stages in the cell population, which would be required for proper lipid analysis.
Cell Growth and Division of BY-2 Cells Are Impaired by n-Butanol
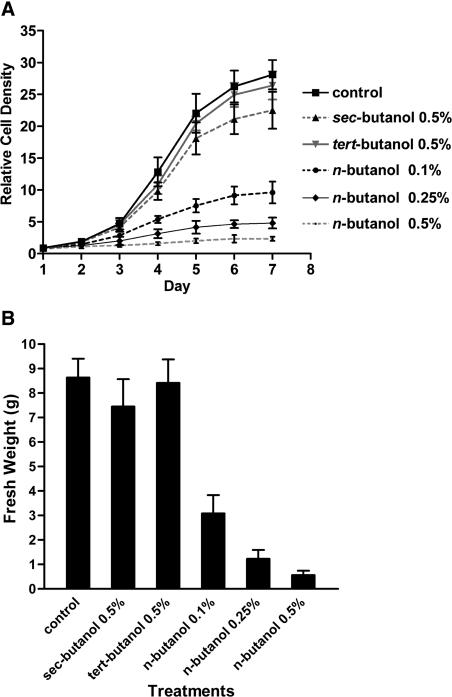
If n-butanol dissociates membrane-microtubule links during mitosis, treated cultures should grow more slowly. To investigate the effects of different butanol isomers, BY-2 cells subcultured every week were inoculated into medium containing 0.1, 0.25, or 0.5% n-butanol or 0.5% sec- or tert-butanol. Relative cell density profiles were generated from daily cell counts with a hemocytometer (see Methods), and fresh weights were measured after 1 week. As shown in Figure 8, n-butanol severely inhibited cell growth, even at the lower concentrations. By contrast, nontransphosphatidylating sec- and tert-alcohols had no effect, and the cultures exhibited sigmoidal growth curves similar to those of the control cells. These results show that prolonged exposure to n-butanol has a concentration-dependent inhibitory effect on cell growth and division, although the cells remain viable.
Figure 8.
Effect of Butanol Isomers on Cell Growth.
GFP-MAP4–transformed BY-2 cells were subcultured in the presence or absence of n-, sec-, or tert-butanol at the concentrations indicated. Relative cell numbers (A) and fresh weights (B) are shown. Means and standard deviations are indicated (n = 3).
DISCUSSION
PLD Activation Triggers Plant Microtubule Reorganization
How microtubules are bound to membranes was a long-standing mystery until Marc and colleagues recently identified a potential linker. It is a 90-kD protein isolated originally from the membrane fraction of BY-2 cells (Marc et al., 1996) that was shown subsequently to decorate the microtubules from all cell cycle stages but that in their absence binds to the plasma membrane (Gardiner et al., 2001). It does not seem to be a hydrophobic, transmembrane protein, because it also decorates the spindle microtubules that have no obvious connection with the membrane. Using a monoclonal antibody and an Arabidopsis cDNA expression library, p90 was identified as a PLD (Gardiner et al., 2001). This finding was confirmed by peptide microsequencing of immunoprecipitated tobacco protein and by showing that it had in vitro PLD activity. Considering PLD's role in signal mechanisms, the authors suggested that this candidate microtubule PLD could respond to membrane receptors and transmit information to the microtubular cytoskeleton. We have now shown that PLD-activating pharmaceuticals and biotic factors dramatically reorganize the microtubule cytoskeleton in tobacco BY-2 cells. These results not only support the proposal by Gardiner et al. (2001) that this 90-kD PLD is the microtubule's link to the membrane but also provide evidence that activation of this PLD triggers microtubule reorganization.
Although we concentrated on microtubules in interphase cells, those in the PPB and those in the phragmoplast also were affected. Significantly, those in the spindle were not. This finding implies that only the microtubules associated with membranes (plasma or Golgi) are susceptible to PLD-activating treatments. The effects were twofold: microtubule release from the membrane and their subsequent disruption, with the extent being dependent on treatment. A combination of the two PLD activators Mas7 and n-butanol was particularly dramatic. As a result, the microtubules appeared to be highly fragmented, although the GFP signal was not dispersed throughout the cytosol, as after oryzalin treatment. By contrast, when the microtubules were stabilized with taxol, after being detached from the membrane they remained intact. These results suggest that microtubules attached to a membrane via their linker proteins remain more stable. On release, they fragment and perhaps even cycle through depolymerization/repolymerization states at different locations in the cytoplasm, but they do not depolymerize completely.
Molecular Nature of the PLD Link between Microtubule and Membrane
We emphasize that the treatments used to activate PLD will activate other signaling pathways as well. For example, most of them have been shown to activate Ca2+ signaling (Munnik et al., 1998a; Knight, 2000). Although they could play a role in the effects illustrated, the evidence for PLD being the pertinent factor is the strongest. First, Gardiner et al. (2001) identified the 90-kD linker as a PLD. Second, all treatments that reorganized the microtubules in BY-2 cells were shown to activate PLD, whereas compounds such as Mas17 and sec- and tert-butanol that do not activate PLD left the microtubular cytoskeleton unaffected. Third, the release of microtubules from membranes was hypothesized based on PLD's unique ability to covalently bind membrane lipids (Munnik and Musgrave, 2001).
To emphasize the importance of the latter, consider a sensible alternative, namely that PLD binds microtubules to membranes via its lipid binding domains. Gardiner et al. (2001) proposed that the C2 domain, which is present in most plant PLDs, binds microtubules to membranes. The C2 domain is activated by binding Ca2+ (Zheng et al., 2000), such as when the Ca2+ concentration increases during treatment with pathogen elicitors, osmotic shock, Mas7, and n-butanol (Munnik et al., 1998a). Under these conditions, the PLD-microtubule complex should bind to the membranes, but in practice, it was released from them, making this option unlikely. There are two plant PLDs (PLDζ1 and PLDζ2) that do not have a C2 domain but instead have a PH/PX lipid binding domain (Qin and Wang, 2002). Although not yet shown for these PLDs, such domains are known to bind polyphosphoinositides (Meijer and Munnik, 2003). As such, polyphosphoinositides could mediate microtubule anchorage to the membrane via PLD. However, this also is unlikely for three reasons: (1) plant cells have extremely low polyphosphoinositide levels (Meijer and Munnik, 2003); (2) of the drugs that increase their levels (neomycin and U73122), neither induced microtubular reorganization or inhibited the n-butanol effect (see supplemental data online); and (3) recent expression data on Arabidopsis PLDζ1 are not in agreement with a microtubule membrane localization (Ohashi et al., 2003).
PLD has been implicated in membrane biogenesis and trafficking (reviewed by Wang, 2001; Meijer and Munnik, 2003). Hence, butanol could inhibit these processes and somehow induce microtubule rearrangements. However, Dixit and Cyr (2002) showed that the disruption of membrane trafficking and the inhibition of secretion by brefeldin A in BY-2 cells had no effect on PPB formation, PPB disappearance, and phragmoplast morphology, whereas cell plate formation was retarded threefold. By contrast, our experiments showed that butanol markedly changed PPB and phragmoplast morphology (Figure 7). Therefore, we believe that disruption of membrane trafficking and secretion cannot account for the microtubule rearrangements, necessitating another model.
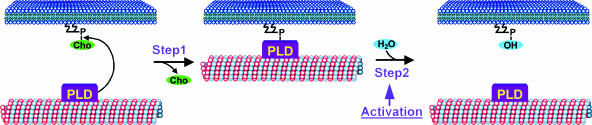
In Figure 9, we propose a new model by which PLD regulates microtubule reorganization that is consistent with our results. The model is based on the two-step transphosphatidylation reaction catalyzed by PLD (Yang et al., 1967) and its ability to form a covalent intermediate with the phosphatidyl moiety connected to a His residue in the catalytic site of the enzyme (Iwasaki et al., 1999; Munnik and Musgrave, 2001). In the first step, the choline group is removed, leaving PLD and its associated microtubule covalently attached to the phosphatidyl moiety. They remain anchored to the membrane until transphosphatidylation occurs in the second step. At that moment, the PLD-microtubule complex releases from the membrane, but the neighboring PLDs still could be attached, maintaining the membrane link. The whole complex is released only when most of the associated PLDs are no longer in step 1 (Figure 9) but are activated to transfer the phosphatidyl group to water (step 2). Therefore, the direct consequences of PLD activation are twofold: (1) microtubules are released to be reorganized; and (2) PA is formed that can trigger downstream targets (Munnik, 2001; Meijer and Munnik, 2003). Note that both effects are simultaneous products of PLD's second activity step. This means that microtubule release from the membrane does not occur downstream from PA formation, unlike most PLD-dependent effects. This finding is in agreement with the observation that PA treatment had no effect on the microtubule organization in interphase cells, although we must remember that the short-chain PA species we used may behave differently in membranes than naturally occurring PAs.
Figure 9.
Model for PLD-Activated Plant Microtubule Reorganization.
For details, see Discussion. Cho, choline.
Additional results obtained from PLC and G-protein inhibitor studies also suggest that the event is related mainly to the activation of PLD. Our “transphosphatidylation” model is further supported by the fact that sec- and tert-butanol, two nontransphosphatidylating alcohols, were unable to induce a microtubular reorganization. Importantly, experimental support for a long-lived PLD-PA intermediate has been provided in an elegant study by Iwasaki et al. (1999) using Streptomyces PLD.
The first crystal structure of that PLD was resolved recently (Leiros et al., 2000). It was seen to contain two flexible loops, both positioned close to the active site, which the authors think function as flexible lids that shield the hydrophobic active site from the aqueous environment. This may prevent water entry and thus maintain PLD covalently bound to the phosphatidyl group. Presumably, more apolar molecules such as n-butanol can penetrate more readily, providing an acceptor for the phosphatidyl group. This could explain why PLD prefers a primary alcohol to water as a nucleophilic acceptor (Munnik et al., 1995). sec- and tert-butanol also will enter the active site, but they are structurally unable to accept the phosphatidyl moiety. This could be the basis for the significant correlation between the ability of each butanol isomer to activate PLD and the ability to release microtubules from membranes. In support of this notion, competition studies using plant PLD have indicated that secondary alcohols cannot access the binding pocket (Ella et al., 1997).
Biological Significance of PLD-Induced Microtubule Rearrangements
PLD was first recognized as a catabolic enzyme that degrades lipids for mobilization. Then it became a signaling enzyme, translating stress into the second messenger PA that further activates downstream responses. We have now presented it in a new role, as a dynamic membrane linker for microtubules.
Nonmotile plants cannot escape stress; therefore, they must mobilize their cell machinery for adaptation. This often involves remodeling cell structure, in which microtubule-membrane associations play a role. For example, cellulose microfibrils around the protoplast must take on a new orientation to compensate for a change in physical stress, and Golgi vesicles must be directed to a cell wall site to repel a pathogen or envelop a symbiont there. The first step in rapid microtubular reorganization is to dismantle the present structure (e.g., break the microtubule-membrane bond). This could explain why stress treatments such as osmotic shock and pathogen elicitors both stimulate PLD and reorganize microtubules, as we have shown for BY-2 cells. However, many other examples have been reported. For example, wounding affects PLD activity (Ryu and Wang, 1998; Zien et al., 2001) and reorganizes cortical microtubules (Hush et al., 1990). Similarly, bacterial infection increases PLD levels (Young et al., 1996; de Torres Zabela et al., 2002) and induces the reorganization of microtubules (Kobayashi et al., 1994). Also, hormones display simultaneous effects on PLD and microtubules. For example, abscisic acid facilitates stomatal closure through PLD activation (Jacob et al., 1999; Sang et al., 2001) and causes microtubule depolymerization (Jiang et al., 1996). We do not claim that all of these dual effects are functionally coupled, but they should be considered in the light of our hypothesis that PLD activation can uncouple microtubules from membranes.
The disruption of PPB from the cellular cortex will seriously interfere with cell division, which also is indicated from our data, because the cells incubated with n-butanol did not progress or progressed very slowly to prophase. We think that this response is biologically significant, because it may enable cells to shift their efforts from cell division to defense. Vesicle transport provides the best example of transient membrane-microtubule associations in the cell (Verma, 2001). One of the most dramatic examples in biology is the coordinated mass transport of Golgi vesicles to form the new cell wall and plasma membranes between dividing plant cells (Samuels et al., 1995; Smith, 1999, 2002). PLD also could be involved here, because the phragmoplast structure was disrupted in BY-2 cells after activating PLD, affecting the formation of a new cell wall, presumably by disrupting the interaction between the vesicles and microtubules. In support of this idea, cells cultivated in low concentrations of n-butanol grew slowly, whereas those cultivated in the same concentrations of sec- or tert-butanol were unaffected. Interestingly, the yeast PLD called Spo14p colocalizes with Golgi vesicles and associates with microtubules at the start of meiosis. Significantly, mutants lacking Spo14 activity fail to complete the second meiotic division, which results in the failure of wall formation between the daughter spores (Rudge et al., 1998). The first meiotic division of the chromosomes, which does not involve wall formation, is completed normally, just as the spindle progression in BY-2 cells seemed unaffected by PLD activation. We do not know whether the PLD along the spindle is just silent, waiting to be transported to the sites in the phragmoplast that might be active upon mitosis, or whether it functions at targets other than microtubules.
Under normal circumstances, vesicles have to be coupled to microtubules for transport and uncoupled at their sites of use. If PLD is the linker, then uncoupling must be regulated as a normal aspect of cell metabolism. Significantly, two small G-proteins, Rho and Arf, that are associated with the cytoskeleton and vesicles in mammalian cells, are able to activate PLD (Powner and Wakelam, 2002). Equivalent plant proteins could be the factors that regulate uncoupling in unstressed cells.
Will the Real Microtubule-Membrane Linker PLD Stand Up?
Although Gardiner et al. (2001) identified the 90-kD tobacco protein as a PLD, it is not clear which one it is, and it is even possible that more than one is activated. Plants have many PLDs; for example, the Arabidopsis genome is predicted to contain 12 isoforms (Elias et al., 2002; Qin and Wang, 2002). The problem appeared to be solved, because Gardiner et al. (2001) sequenced part of their tobacco protein and suggested that it was a PLDδ isoform. However, PLDδ-silenced Arabidopsis plants did not display a clear phenotype (Katagiri et al., 2001). This is unusual, because defects in MAPs usually exhibit a dramatic phenotype. Therefore, the lack of an identifiable phenotype in PLDδ-silenced plants indicates that either the microtubule PLD is a different isoform or that other PLDs can assume the silenced gene's function.
We have tested whether N-acylethanolamine, a compound that inhibits the PLDα subclass (Austin-Brown and Chapman, 2002), affected microtubular arrays. No effect was found, nor did we detect an effect on n-butanol–induced microtubule rearrangements (see supplemental data online). Although this finding suggests that PLDα isoforms are not involved, it leaves the true identity a fairly open and pressing question. It is hoped that further analysis of the 90-kD tobacco protein, Arabidopsis T-DNA inserts in PLD genes, and PLD overexpression lines will reveal the real hero.
METHODS
BY-2 Cells Expressing Fluorescent Protein–Based Microtubule Reporters
The tobacco (Nicotiana tabacum) plant microtubule-reporter fusions GFP-MAP4 and YFP-CLIP1701-1240 in vector pBINPLUS, and GFP-TUA6 in vector pB121, were transformed into tobacco BY-2 cells using Agrobacterium tumefaciens as described (Dhonukshe and Gadella, 2003). Stable transformants were maintained as weekly subcultured cell suspensions grown in the dark at 25°C on a rotary shaker at 125 rpm in 250-mL Erlenmeyer flasks containing 50 mL of BY-2 medium (Dhonukshe and Gadella, 2003).
Fluorescence Microscopy
Samples were prepared in NUNC chambers (Nunc, Inc., Naperville, IL) containing eight wells designed especially for microscopy. For all treatments (except cell cycle studies), 5-day-old weekly subcultured BY-2 cells were used. Cells (200 μL) were treated by adding an equal volume of agonist-dissolved BY-2 medium for the times and concentrations indicated in the figure legends and observed immediately using fluorescence microscopy. Images were acquired using confocal laser scanning microscopy based on the Zeiss LSM 510 system (Jena, Germany) composed of an Axiovert inverted microscope equipped with argon ion and HeNe lasers as excitation sources. BY-2 cells expressing GFP were excited with the 488-nm laser line, and GFP emission was detected using a 505- to 530-nm band-pass filter. Additional filtering was obtained using the 488-nm primary dichroic mirror, reflecting the laser light and transmitting fluorescence.
In the case of dual-color GFP and BODIPY imaging, GFP was excited with the 488-nm argon line and BODIPY was excited with the 543-nm HeNe line. GFP was detected with the 505- to 530-nm band-pass filter, and BODIPY was detected with a 560-nm long-pass filter in another detection channel. Excitation was separated from emission using a 488/543-nm primary dichroic mirror, and BODIPY fluorescence was separated from GFP fluorescence using a 545-nm secondary dichroic mirror. Crosstalk-free fluorescence images were acquired by operating in the multiple-track mode. A Zeiss ×40 water-immersion objective (numerical aperture 1.3) with correction for the NUNC chamber bottom thickness was used to scan samples. Images were captured, and maximum projections and cross-sections were obtained using LSM 510 image-acquisition software version 3.03 (Zeiss). For each sample, ∼40 optical sections spaced 0.4 μm apart were taken to cover at least a hemisphere of a cell. Text was added, and images were sized using Adobe Photoshop 5.0 (Mountain View, CA). No filtering or pixel manipulations were performed, so images correspond to the raw image data.
Cell Synchronization
BY-2 cells were synchronized with a two-step procedure performed by applying 5 μg/mL aphidicolin (Sigma-Aldrich) followed by 3 μM propyzamide (Sigma-Aldrich) as described previously (Nagata and Kumagai, 1999).
Pharmacological Microtubule Treatments
For microtubule stabilization, 10 μM taxol from Nigrospora sphaercia (Sigma-Aldrich) was used, and for microtubule depolymerization, 10 μM oryzalin (Greyhound Chromatography and Allied Chemicals, Merseyside, UK) was used. For inhibition of the PLC pathway, either 200 μM neomycin (Sigma-Aldrich) or 10 μM U73122 (Sigma-Aldrich) was used, and for G-protein inhibition, 10 μg/mL pertussis toxin (Sigma-Aldrich) was used. All pharmacological agents were applied to BY-2 cells at least 1 h before their microscopic analysis.
Measurement of Cell Density and Fresh Weight
Relative cell density measurements were performed as described previously (Dhonukshe and Gadella, 2003). Fresh weights were obtained 7 days after treatment by centrifuging 15 mL of cell suspension at 796g for 5 min. Mean values, standard deviations, and graphs were obtained using Microsoft Excel (Redmond, WA).
In Vivo Phospholipase D Measurements
To assay phospholipase D (PLD) activity in living cells, the production of phosphatidylbutanol was measured (Munnik et al., 1995). In brief, cells were metabolically labeled by incubating them for 3 h with 100 μCi of carrier-free PO43− (32Pi) (Amersham International) per milliliter of cells. They were then divided into aliquots of 85 μL, treated by adding an equal volume of agonist, and dissolved in cell-free medium containing 0.5% (v/v, final concentration) n-butanol for the times indicated in the figure legends. Incubations were stopped by adding 20 μL of 50% (v/v) perchloric acid and snap-freezing them in liquid nitrogen. After 5 to 30 min, samples were centrifuged and the lipids were extracted by adding 750 μL of CHCl3:methanol:HCl (50:100:1, v/v) while vortexing for 15 s. A two-phase system was induced by adding 200 μL of 0.9% (w/v) NaCl and 750 μL of CHCl3. Lipids were extracted further as described previously (van der Luit et al., 2000) and chromatographed on heat-activated silica 60 thin layer chromatography (TLC) plates (20 × 20 cm; Merck, Darmstadt, Germany) using the organic phase of a mixture of ethyl acetate:iso-octane:formic acid:water (13:2:3:10, v/v). Radiolabeled phospholipids were visualized by autoradiography and quantified by phosphorimaging (Storm; Molecular Dynamics, Sunnyvale, CA).
Synthesis of BODIPY-PA and BY-2 Cell Labeling
Acylation of 5 μM lysophosphatidylcholine (Sigma-Aldrich) was performed according to Gupta et al. (1977) by reacting it with the anhydride of 10 μM BODIPY 558/568-C12 (Molecular Probes, Leiden, The Netherlands) prepared in dry CHCl3 (Selinger and Lapidot, 1966). Purification of the fluorescent phosphatidylcholine was performed on CM-cellulose (Comfurius and Zwaal, 1977), from which the product was eluted with 4% methanol in CHCl3, as verified by TLC. To synthesize BODIPY 558/568-C12-PA, BODIPY 558/568-C12-phosphatidylcholine was suspended in 50 mM Tris, pH 8.0, and 10 mM CaCl2 incubated with 1000 units of PLD from Streptomyces chromofuscus (Sigma-Aldrich) at 37°C for 60 min. The resulting fluorescent BODIPY 558/568-C12-PA was purified on silica gel with chloroform and increasing amounts of methanol. Aqueous PA suspensions were made by evaporating chloroform stocks of dioctanoyl-PA and BODIPY-PA and dispersing them into calcium-free medium by sonication with a Branson B-12 tip sonicator (Danbury, CT) at 40 W for 10 s three times while cooling on ice water. To load BY-2 cells with BODIPY-PA, they were washed three times with calcium-free BY-2 medium and then incubated with PA in a final concentration of 20 μM. For simultaneous visualization of microtubules and BODIPY 558/568-C12-PA, cells were observed by fluorescence microscopy in multitracking imaging mode.
To check for BODIPY-PA uptake and metabolism, BY-2 cells were washed three times in calcium-free medium. After the last wash, the medium was removed and 1 mL of cold (−20°C) Folch extraction mix (CHCl3:methanol:0.6 M HCl [1:2:0.8]) and 250 μL of chloroform were added to the cell pellet. After vigorous shaking and phase separation, the lower phase was isolated and the volume was reduced by evaporation. Lipids were analyzed on silica gel 60 TLC plates using CHCl3:methanol:5% NH4OH (45:35:10, v/v) as a solvent.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Teun Munnik, munnik@science.uva.nl.
NOTE ADDED IN PROOF
An effect of 1-butanol on Arabidopsis seedling development and microtubule organization has recently been published (Gardiner, J., Collings, D.A., Harper, J.D., and Marc J. [2003]. The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol. 44, 687–696).
Supplementary Material
Acknowledgments
We thank Kent Chapman (University of North Texas, Denton) for his generous gift of N-acylethanolamine and our colleagues for many stimulating discussions, especially Alan Musgrave. The latter also is gratefully acknowledged for his critical comments on the manuscript. P.D and T.W.J.G were supported by the Netherlands Organization for Scientific Research (NWO-FOM-ALW 805.47.012), J.G. and T.W.J.G. were supported by NWO-CW (700.50.513), T.W.J.G. was supported by NWO-ALW-Van der Leeuw (835.25.004), A.M.L. was supported by the Fundación Antorchas and the Consejo Nacional de Investigaciones Científicas y Técnicas, and T.M. was supported by NWO (810.66.011, 810-36.005, and 99002), the European Community (HPRN-CT-2000-00093), and the Royal Netherlands Academy of Arts and Sciences.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014977.
Footnotes
Online version contains Web-only data.
References
- Akashi, T., Kawasaki, S., and Shibaoka, H. (1990). Stabilization of cortical microtubules by the cell wall in cultured tobacco cells. Planta 182, 363–369. [DOI] [PubMed] [Google Scholar]
- Akashi, T., and Shibaoka, H. (1991). Involvement of transmembrane proteins in the association of cortical microtubules with the plasma membrane in tobacco BY-2 cells. J. Cell Sci. 98, 169–174. [Google Scholar]
- Austin-Brown, S.L., and Chapman, K.D. (2002). Inhibition of phospholipase Dα by N-acylethanolamines. Plant Physiol. 129, 1892–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., and Cande, W.Z. (1990). The structure and function of the mitotic spindle in flowering plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 277–315. [Google Scholar]
- Brown, R.M.J., and Montezinos, D. (1976). Cellulose microfibrils: Visualization of biosynthetic and oriented complexes in association with the plasma membrane. Proc. Natl. Acad. Sci. USA 73, 143–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfurius, P., and Zwaal, R.F. (1977). The enzymatic synthesis of phosphatidylserine and purification by CM-cellulose column chromatography. Biochim. Biophys. Acta 488, 36–42. [DOI] [PubMed] [Google Scholar]
- Cyr, R.J. (1994). Microtubules in plant morphogenesis: Role of the cortical array. Annu. Rev. Cell Biol. 10, 153–180. [DOI] [PubMed] [Google Scholar]
- den Hartog, M., Musgrave, A., and Munnik, T. (2001). Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: A role for phospholipase C and D in root hair deformation. Plant J. 25, 55–65. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117. [DOI] [PubMed] [Google Scholar]
- de Torres Zabela, M., Fernandez-Delmond, I., Niittyla, T., Sanchez, P., and Grant, M. (2002). Differential expression of genes encoding Arabidopsis phospholipases after challenge with virulent or avirulent Pseudomonas isolates. Mol. Plant-Microbe Interact. 15, 808–816. [DOI] [PubMed] [Google Scholar]
- de Vrije, T., and Munnik, T. (1997). Activation of phospholipase D by calmodulin agonists and mastoparan in carnation petal tissue. J. Exp. Bot. 48, 1631–1637. [Google Scholar]
- Dhonukshe, P., and Gadella, T.W., Jr. (2003). Alteration of microtubule dynamic instability during preprophase band formation revealed by yellow fluorescent protein–CLIP170 microtubule plus-end labeling. Plant Cell 15, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit, R., and Cyr, R. (2002). Golgi secretion is not required for marking the preprophase band site in cultured tobacco cells. Plant J. 29, 99–108. [DOI] [PubMed] [Google Scholar]
- Elias, M., Potocky, M., Cvrckova, F., and Zarsky, V.V. (2002). Molecular diversity of phospholipase D in angiosperms. BMC Genomics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ella, K.M., Meier, K.E., Kumar, A., Zhang, Y., and Meier, G.P. (1997). Utilization of alcohols by plant and mammalian phospholipase D. Biochem. Mol. Biol. Int. 41, 715–724. [DOI] [PubMed] [Google Scholar]
- Fisher, D.D., and Cyr, R.J. (2000). Mechanical forces in plant growth and development. Gravit. Space Biol. Bull. 13, 67–73. [PubMed] [Google Scholar]
- Frank, W., Munnik, T., Kerkmann, K., Salamini, F., and Bartels, D. (2000). Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell 12, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J.C., Harper, J.D., Weerakoon, N.D., Collings, D.A., Ritchie, S., Gilroy, S., Cyr, R.J., and Marc, J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13, 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., Jr., Brower, D.L., and Staehelin, L.A. (1980). Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J. Cell Biol. 84, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., Jr., and Staehelin, L.A. (1988). Spatial relationship between microtubules and plasma-membrane rosettes during the deposition of primary wall microfibrils in Closterium sp. Planta 173, 22–30. [DOI] [PubMed] [Google Scholar]
- Goddard, R.H., Wick, S.M., Silflow, C.D., and Snustad, D.P. (1994). Microtubule components of the plant cell cytoskeleton. Plant Physiol. 104, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, C.L., and Cyr, R.J. (2000). Microtubule reorganization in tobacco BY-2 cells stably expressing GFP-MBD. Planta 210, 502–509. [DOI] [PubMed] [Google Scholar]
- Green, P.B. (1962). Mechanism for plant cellular morphogenesis. Science 138, 1404–1405. [DOI] [PubMed] [Google Scholar]
- Gupta, C.M., Radhakrishnan, R., and Khorana, H.G. (1977). Glycerophospholipid synthesis: Improved general method and new analogs containing photoactivable groups. Proc. Natl. Acad. Sci. USA 74, 4315–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallouin, M., Ghelis, T., Brault, M., Bardat, F., Cornel, D., Miginiac, E., Rona, J.P., Sotta, B., and Jeannette, E. (2002). Plasmalemma abscisic acid perception leads to RAB18 expression via phospholipase D activation in Arabidopsis suspension cells. Plant Physiol. 130, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham, A.R., and Gunning, B.E. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77, 14–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasezawa, S., and Nozaki, H. (1999). Role of cortical microtubules in the orientation of cellulose microfibril deposition in higher-plant cells. Protoplasma 209, 98–104. [DOI] [PubMed] [Google Scholar]
- Heath, I.B. (1974). A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J. Theor. Biol. 48, 445–449. [DOI] [PubMed] [Google Scholar]
- Herth, W. (1985). Plasma-membrane rosettes involved in localized wall thickening during xylem vessel formation of Lepidium sativum L. Planta 164, 12–21. [DOI] [PubMed] [Google Scholar]
- Himmelspach, R., Wymer, C.L., Lloyd, C.W., and Nick, P. (1999). Gravity-induced reorientation of cortical microtubules observed in vivo. Plant J. 18, 449–453. [DOI] [PubMed] [Google Scholar]
- Hush, J.M., Hawes, C.R., and Overall, R.L. (1990). Interphase microtubule re-orientation predicts a new cell polarity in wounded pea roots. J. Cell Sci. 96, 47–61. [Google Scholar]
- Hush, J.M., and Overall, R.L. (1991). Electrical and mechanical fields orient cortical microtubules in higher plant tissues. Cell Biol. Int. Rep. 15, 551–560. [Google Scholar]
- Iwasaki, Y., Horiike, S., Matsushima, K., and Yamane, T. (1999). Location of the catalytic nucleophile of phospholipase D of Streptomyces antibioticus in the C-terminal half domain. Eur. J. Biochem. 264, 577–581. [DOI] [PubMed] [Google Scholar]
- Jacob, T., Ritchie, S., Assmann, S.M., and Gilroy, S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 96, 12192–12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.-J., Nakajima, N., and Kondo, N. (1996). Disruption of microtubules by abscisic acid in guard cells of Vicia faba L. Plant Cell Physiol. 37, 697–701. [Google Scholar]
- Katagiri, T., Takahashi, S., and Shinozaki, K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26, 595–605. [DOI] [PubMed] [Google Scholar]
- Knight, H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195, 269–324. [DOI] [PubMed] [Google Scholar]
- Kobayashi, I., Kobayashi, Y., and Hardham, A.R. (1994). Dynamic reorganization of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta 195, 237–247. [Google Scholar]
- Kost, B., and Chua, N.H. (2002). The plant cytoskeleton: Vacuoles and cell walls make the difference. Cell 108, 9–12. [DOI] [PubMed] [Google Scholar]
- Law, G.J., and Northrop, A.J. (1994). Synthetic peptides to mimic the role of GTP binding proteins in membrane traffic and fusion. Ann. N.Y. Acad. Sci. 710, 196–208. [DOI] [PubMed] [Google Scholar]
- Laxalt, A.M., and Munnik, T. (2002). Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5, 332–338. [DOI] [PubMed] [Google Scholar]
- Laxalt, A.M., ter Riet, B., Verdonk, J.C., Parigi, L., Tameling, W.I., Vossen, J., Haring, M., Musgrave, A., and Munnik, T. (2001). Characterization of five tomato phospholipase D cDNAs: Rapid and specific expression of LePLDβ1 on elicitation with xylanase. Plant J. 26, 237–247. [DOI] [PubMed] [Google Scholar]
- Ledbetter, M.C., and Porter, K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros, I., Secundo, F., Zambonelli, C., Servi, S., and Hough, E. (2000). The first crystal structure of a phospholipase D. Struct. Fold. Des. 8, 655–667. [DOI] [PubMed] [Google Scholar]
- Lloyd, C., and Chan, J. (2002). Helical microtubule arrays and spiral growth. Plant Cell 14, 2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, C., and Hussey, P. (2001). Microtubule-associated proteins in plants: Why we need a MAP. Nat. Rev. Mol. Cell Biol. 2, 40–47. [DOI] [PubMed] [Google Scholar]
- Marc, J., Granger, C.L., Brincat, J., Fisher, D.D., Kao, T., McCubbin, A.G., and Cyr, R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc, J., Sharkey, D.E., Durso, N.A., Zhang, M., and Cyr, R.J. (1996). Isolation of a 90-kD microtubule-associated protein from tobacco membranes. Plant Cell 8, 2127–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201, 252–260. [DOI] [PubMed] [Google Scholar]
- Meijer, H.J., and Munnik, T. (2003). Phospholipase-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y. (1999). The preprophase band of microtubules: Its function as a cytokinetic apparatus in higher plants. Int. Rev. Cytol. 187, 1–49. [Google Scholar]
- Mitchison, T., and Kirschner, M. (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- Mueller, S.C., and Brown, R.M.J. (1982. a). The control of cellulose microfibril deposition in the cell wall of higher plants. I. Can directed membrane flow orient cellulose microfibrils? Indirect evidence from freeze-fractured plasma membranes of maize and pine seedlings. Planta 154, 489–500. [DOI] [PubMed] [Google Scholar]
- Mueller, S.C., and Brown, R.M.J. (1982. b). The control of cellulose microfibril deposition in the cell wall of higher plants. II. Freeze-fracture microfibril patterns in maize seedling tissues following experimental alteration with colchicine and ethylene. Planta 154, 501–515. [DOI] [PubMed] [Google Scholar]
- Munnik, T. (2001). Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Arisz, S.A., De Vrije, T., and Musgrave, A. (1995). G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7, 2197–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik, T., Irvine, R.F., and Musgrave, A. (1998. a). Phospholipid signalling in plants. Biochim. Biophys. Acta 1389, 222–272. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Meijer, H.J., Ter Riet, B., Hirt, H., Frank, W., Bartels, D., and Musgrave, A. (2000). Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22, 147–154. [DOI] [PubMed] [Google Scholar]
- Munnik, T., and Musgrave, A. (2001). Phospholipid signaling in plants: Holding on to phospholipase D. Sci. STKE 111, PE42. [DOI] [PubMed] [Google Scholar]
- Munnik, T., van Himbergen, J.A., ter Riet, B., Braun, F., Irvine, R.F., van den Ende, H., and Musgrave, A. (1998. b). Detailed analysis of the turnover of polyphosphoinositides and phosphatidic acid upon activation of phospholipase C and D in Chlamydomonas cells treated with non-permeabilizing concentrations of mastoparan. Planta 207, 133–145. [Google Scholar]
- Nagata, T., and Kumagai, F. (1999). Plant cell biology through the window of the highly synchronized tobacco BY-2 cell line. Methods Cell Sci. 21, 123–127. [DOI] [PubMed] [Google Scholar]
- Newcomb, E.H. (1969). Plant microtubules. Annu. Rev. Plant Physiol. 20, 253–288. [Google Scholar]
- Nick, P. (1998). Signaling to the microtubular cytoskeleton in plants. Int. Rev. Cytol. 184, 33–81. [Google Scholar]
- Nogales, E. (2000). Structural insights into microtubule function. Annu. Rev. Biochem. 69, 277–302. [DOI] [PubMed] [Google Scholar]
- Nogales, E., Whittaker, M., Milligan, R.A., and Downing, K.H. (1999). High-resolution model of the microtubule. Cell 96, 79–88. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Rodrigues-Pousada, R., Possenti, M., Ruberti, I., Morelli, G., and Aoyama, T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D. (1967). The effects of colchicine on the ultra-structure of dividing plant cells, xylem wall differentiation and distribution of cytoplasmic microtubules. Dev. Biol. 15, 206–236. [Google Scholar]
- Powner, D.J., and Wakelam, M.J. (2002). The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS Lett. 531, 62–64. [DOI] [PubMed] [Google Scholar]
- Qin, C., Wang, C., and Wang, X. (2002). Kinetic analysis of Arabidopsis phospholipase Dδ: Substrate preference and mechanism of activation by Ca2+ and phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 20, 49685–49690. [DOI] [PubMed] [Google Scholar]
- Qin, C., and Wang, X. (2002). The Arabidopsis phospholipase D family: Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol. 128, 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quader, H. (1986). Cellulose microfibril orientation in Oocystis solitaria: Proof that microtubules control the alignment of the terminal complexes. J. Cell Sci. 83, 223–234. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S. (2001). Molecular motors and their functions in plants. Int. Rev. Cytol. 204, 97–178. [DOI] [PubMed] [Google Scholar]
- Ritchie, S., and Gilroy, S. (1998). Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 95, 2697–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.G., and Quader, H. (1982). The microtubule-microfibril syndrome. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (London: Academic Press), pp. 109–126.
- Rudge, S.A., Morris, A.J., and Engebrecht, J. (1998). Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelland, E., Cantrel, C., Gawer, M., Kader, J.C., and Zachowski, A. (2002). Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, S.B., and Wang, X. (1998). Increase in free linolenic and linoleic acids associated with phospholipase D-mediated hydrolysis of phospholipids in wounded castor bean leaves. Biochim. Biophys. Acta 1393, 193–202. [DOI] [PubMed] [Google Scholar]
- Samuels, A.L., Giddings, T.H., Jr., and Staehelin, L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Y., Zheng, S., Li, W., Huang, B., and Wang, X. (2001). Regulation of plant water loss by manipulating the expression of phospholipase Dα. Plant J. 28, 135–144. [DOI] [PubMed] [Google Scholar]
- Seagull, R.W., and Heath, I.B. (1980). The organization of cortical microtubule arrays in the radish root hair. Protoplasma 103, 205–229. [Google Scholar]
- Selinger, Z., and Lapidot, Y. (1966). Synthesis of fatty acid anhydrides by reaction with dicyclohexylcarbodiimide. J. Lipid Res. 7, 174–175. [PubMed] [Google Scholar]
- Shibaoka, H. (1994). Plant hormone-induced changes in the orientation of cortical microtubules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 527–544. [Google Scholar]
- Smith, L.G. (1999). Divide and conquer: Cytokinesis in plant cells. Curr. Opin. Plant Biol. 2, 447–453. [DOI] [PubMed] [Google Scholar]
- Smith, L.G. (2001). Plant cell division: Building walls in the right places. Nat. Rev. Mol. Cell Biol. 2, 33–39. [DOI] [PubMed] [Google Scholar]
- Smith, L.G. (2002). Plant cytokinesis: Motoring to the finish. Curr. Biol. 12, R206–R208. [DOI] [PubMed] [Google Scholar]
- Sonesson, A., Berglund, M., Staxen, I., and Widell, S. (1997). The characterization of plasma membrane-bound tubulin of cauliflower using Triton X-114 fractionation. Plant Physiol. 115, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonobe, S., and Takahashi, S. (1994). Association of microtubules with the plasma membrane of tobacco BY-2 cells in vitro. Plant Cell Physiol. 35, 451–460. [Google Scholar]
- Sonobe, S., Yamamoto, S., Motomura, M., and Shimmen, T. (2001). Isolation of cortical MTs from tobacco BY-2 cells. Plant Cell Physiol. 42, 162–169. [DOI] [PubMed] [Google Scholar]
- van der Luit, A.H., Piatti, T., van Doorn, A., Musgrave, A., Felix, G., Boller, T., and Munnik, T. (2000). Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 123, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Himbergen, J.A.J., ter Riet, B., Meijer, H.J.G., van den Ende, H., Musgrave, A., and Munnik, T. (1999). Mastoparan analogues activate phospholipase C and phospholipase D activity in Chlamydomonas: A comparative study. J. Exp. Bot. 50, 1735–1742. [Google Scholar]
- Verma, D.P. (2001). Cytokinesis and building of the cell plates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 751–784. [DOI] [PubMed] [Google Scholar]
- Vesk, P.A., Vesk, M., and Gunning, B.E. (1996). Field emission scanning electron microscopy of microtubule arrays in higher plant cells. Protoplasma 195, 168–182. [Google Scholar]
- Wang, C., Zien, C.A., Afitlhile, M., Welti, R., Hildebrand, D.F., and Wang, X. (2000). Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12, 2237–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. (2001). Plant phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 211–231. [DOI] [PubMed] [Google Scholar]
- Wang, X. (2002). Phospholipase D in hormonal and stress signaling. Curr. Opin. Plant Biol. 5, 408–414. [DOI] [PubMed] [Google Scholar]
- Wang, X., Wang, C., Sang, Y., Qin, C., and Welti, R. (2002). Networking of phospholipases in plant signal transduction. Physiol. Plant. 115, 331–335. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115, 1345–1354. [DOI] [PubMed] [Google Scholar]
- Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H.E., Rajashekar, C.B., Williams, T.D., and Wang, X. (2002). Profiling membrane lipids in plant stress responses: Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002. [DOI] [PubMed] [Google Scholar]
- Williamson, R.E. (1991). Orientation of cortical microtubules in interphase plant cells. Int. Rev. Cytol. 129, 135–206. [Google Scholar]
- Yang, S.F., Freer, S., and Benson, A.A. (1967). Transphosphatidylation by phospholipase D. J. Biol. Chem. 242, 477–484. [PubMed] [Google Scholar]
- Young, S.A., Wang, X., and Leach, J.E. (1996). Changes in the plasma membrane distribution of rice phospholipase D during resistant interactions with Xanthomonas oryzae pv oryzae. Plant Cell 8, 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Krishnamoorthi, R., Zolkiewski, M., and Wang, X. (2000). Distinct Ca2+ binding properties of novel C2 domains of plant phospholipase Dα and β. J. Biol. Chem. 275, 19700–19706. [DOI] [PubMed] [Google Scholar]
- Zien, C.A., Wang, C., Wang, X., and Welti, R. (2001). In vivo substrates and the contribution of the common phospholipase D, PLDα, to wound-induced metabolism of lipids in Arabidopsis. Biochim. Biophys. Acta 1530, 236–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.