Abstract
Background
The formation of fusion genes plays roles in both oncogenesis and evolution by facilitating the acquisition of novel functions. Here we describe the first example of a human polymorphic in-frame fusion of two unrelated genes associated with a copy number variant.
Design and Methods
Array comparative genomic hybridization was used to identify cryptic oncogenic fusion genes. Fusion gene structure and origin was examined using molecular biological and computational methods. Phenotype associations were examined using PopGen cohorts.
Results
Targeted array comparative genomic hybridization to identify cryptic oncogenic fusion genes in patients with atypical myeloproliferative neoplasms identified a 111 kb amplification with breakpoints within the TRK-fused gene (TFG, a target of translocations in lymphoma and thyroid tumors) and G-protein-coupled receptor 128 (GPR128) resulting in an expressed in-frame TFG-GPR128 fusion transcript. The fusion gene was also identified in healthy individuals at a frequency of 0.02 (3/120). Normally both genes are in identical orientations with TFG immediately downstream of GPR128. In individuals with a copy number variant amplification, one or two copies of the TFG-GPR128 fusion are found between the two parental genes. The breakpoints share a region of microhomology, and haplotype and microsatellite analysis indicate a single ancestral origin. Analysis of PopGen cohorts showed no obvious phenotype association. An in silico search of EST databases found no other copy number variant amplification-associated fusion transcripts, suggesting that this is an uncommon event.
Conclusions
The finding of a polymorphic gene fusion in healthy individuals adds another layer to the complexity of human genome variation and emphasizes the importance of careful discrimination of oncogenic changes found in tumor samples from non-pathogenic normal variation.
Keywords: fusion gene, oncogenesis, targeted array, array genomic comparative hybridization
Introduction
The role of genomic rearrangements, primarily segmental and tandem duplications, in recent human evolution is now well recognized. Several examples of human genes that have recently arisen by fusion of separate parental genes have been documented.1
Somatic acquisition of fusion genes is also a well-described pathogenic mechanism in hematologic and soft tissue malignancies. Typically, these fusions encode oncogenic, chimeric proteins that play a critical role in driving malignant growth. Consequently, they are excellent targets for therapy, as exemplified by the dramatic clinical effects of the tyrosine kinase inhibitor imatinib against the BCR-ABL fusion in chronic myeloid leukemia.
In hematologic malignancies, the presence of a particular gene fusion is usually signaled by the finding of a recurrent, acquired chromosomal rearrangement in bone marrow-derived metaphases. However some fusions are cytogenetically cryptic and thus may evade detection. Although the incidence of gene fusions associated with visible karyotypic abnormalities is well established, the incidence of cryptic fusions is unknown, The initial aim of our study was a systematic search for cryptic fusion genes in patients with atypical myeloproliferative neoplasms using array comparative genomic hybridization analysis, exploiting the fact that most of these abnormalities are associated with small genomic copy number changes that interrupt the target genes. For example, FIP1L1-PDGFRA in chronic eosinophilic leukemia is formed by an 800 kb intrachromosomal deletion2 and NUP214-ABL in T-cell acute lymphoblastic leukemia is formed by a 500 kb episomal amplification.3
The use of high-resolution array comparative hybridizaton has revealed substantial constitutional copy number variation in large stretches of the human genome4–6 and, therefore, in any analysis of malignancy it is essential to distinguish acquired copy number variants (CNV) from those that are inherited. In this study we describe the unexpected finding of a constitutional polymorphic gene fusion occurring at a relatively high frequency in apparently healthy Europeans.
Design and Methods
Samples
Samples of bone marrow or peripheral blood from patients with atypical myeloproliferative neoplasms were analyzed by CGH array (37 samples) and by reverse transcriptase polymerase chain reaction (RT-PCR) (575 samples). All were accrued between 1999 and 2007 at the Wessex Regional Genetics Laboratory (WRGL), UK. A cohort of 120 anonymized control peripheral blood samples were from healthy first-degree relatives of individuals investigated for a variety of genetic conditions at the WRGL. DNA samples from ten individuals, previously identified as having a CNV in the TFG-GPR128 region7 from data deposited at the Database of Genomic Variants (http://projects.tcag.ca/variation/), were obtained from the PopGen Project based in Kiel, Germany.8 Two PopGen population cohorts were used as controls, cohort 1 comprised 737 anonymous blood donors of German descent collected by the PopGen biobank,8 cohort 2 comprised 539 individuals with phenotypic and Affymetrix SNP 6.0 array data recruited through the Blood Service of the University Hospital Schleswig-Holstein. All individuals completed questionnaires and underwent physical examination at PopGen facilities.
Comparative genomic hybridization array analysis
Targeted Agilent arrays (Agilent Technologies, Palo Alto, CA, USA) were designed using the Agilent online array design program eArray (https://earray.chem.agilent.com/earray). An initial array targeted approximately 500 candidate genes for involvement in atypical myeloproliferative neoplasms, including a 60 kb region comprising 50 oligonucleotides centered upon TFG. A second array included 1100 oligonucleotides targeting 500 kb centered upon the TFG-GPR128 CNV. All procedures were performed according to the manufacturer’s protocols. Array data were analyzed using CGH Analytics software (Agilent Technologies).
Microsatellite and genotype analysis
A 3100 Genetic analyzer (Applied Biosystems, Foster City, CA, USA) was used for Genescan analysis and genomic sequencing. Sequences were analyzed using Mutation Surveyor (Softgenetics, State College, PA, USA) and Genescan results by Genescan and Genotype software (Applied Biosystems). Primer sequences are provided in Online Supplementary Table S1.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) was performed using standard techniques, as previously described.9
Bioinformatic analysis
We used Galaxy (http://main.g2.bx.psu.edu),10 an interactive web-based portal which allows users to carry out computational operations on large data sets from remote databases, to perform an in silico screen for other examples of fusion genes resulting from CNV amplifications. We chose to screen only for CNV-amplification derived fusion expressed sequence tags (EST) for two reasons: (i) fusion EST caused by CNV deletion would be mimicked by fusion EST arising from the splicing together of exons from neighboring genes by exon skipping, thereby increasing the false discovery rate, (ii) theoretically, the chance of CNV breakpoints lying within genes and therefore potentially giving rise to fusions is the same for both amplifications and deletions, therefore only identifying CNV-amplification derived fusions would still give an indication of the incidence of these events. We were aided in the screen by the fact that EST such as those from TFG-GPR128 which map to two different genes, have separate entries in the University of California, Santa Cruz (UCSC) Genome Bioinformatics table all_est (http://genome.ucsc.edu) for each locus. EST from the UCSC all_est table were, therefore, filtered to select those with two entries, for which the two entries satisfied the following conditions: (i) on the same chromosome; (ii) running in the same orientation; (iii) no overlap of more than 10 bp within the EST sequence; (iv) the 5′ table entry mapping to a genomic position ‘downstream’ of the 3′ entry; and (v) mapping to genomic positions not separated by more than 5 Mb (this would include all CNV, larger rearrangements would be detectable cytogenetically) and falling within two different genes. Resulting EST were then visualized on the UCSC browser to select those mapping to exons with canonical splice sites at the fusion gene junction. An explanatory diagram is shown in Figure 1.
Figure 1.
Relationship between gene, EST and entries in the UCSC all_est table and criteria for EST selection.
Results
Identification of TFG-GPR128
To search for copy number changes associated with cryptic fusion genes, we designed an Agilent 44K custom array that targeted 500 candidate genes, including all tyrosine kinases, known gene targets of oncogenic chromosomal rearrangements plus a selection of other oncogenes and signaling molecules. Each gene was targeted with 50–75 probes to ensure sensitive detection of mosaic acquired abnormalities in a variable background of normal cells. This report focuses on the results of one of the array gene targets, TFG (TRK-fused gene).
Of 37 samples from patients with atypical myeloproliferative neoplasms that we initially analyzed, one case with hypereosinophilic syndrome showed an amplified region with a breakpoint within TFG. This gene is a known target of acquired chromosomal translocations that generate fusions with ALK, NTRK1 and NR4A3 (NOR1) in anaplastic large cell lymphoma,11 thyroid carcinoma12 and skeletal myxoid chondrosarcoma,13 respectively.
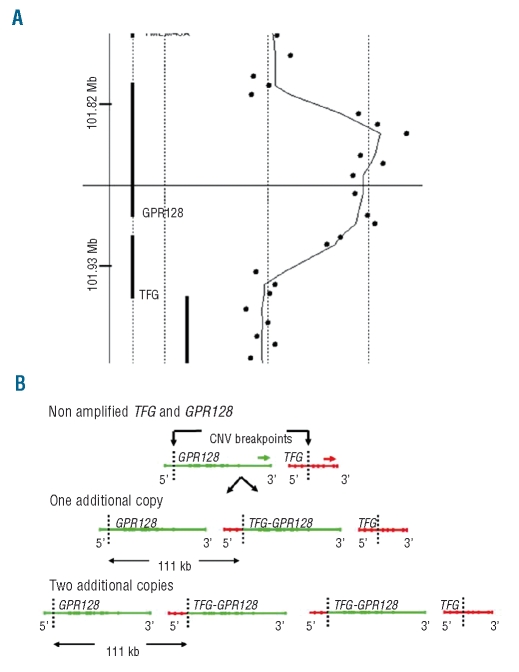
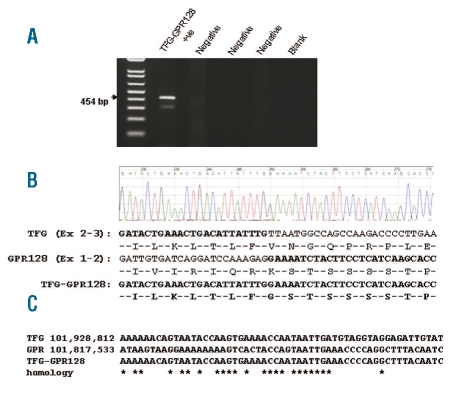
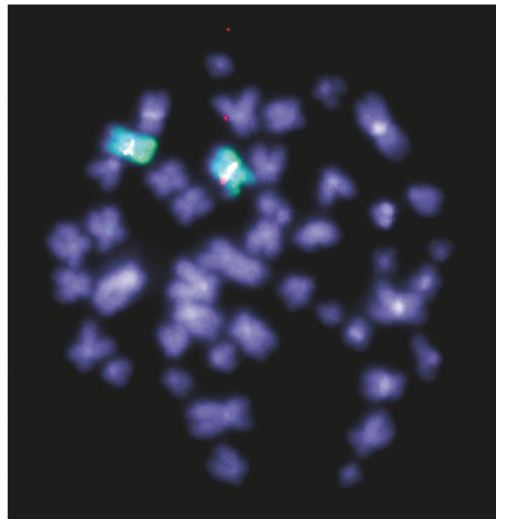
We clarified the breakpoints by hybridization of the same sample to an Agilent 244K whole genome array, which demonstrated an amplification of 111 kb with breakpoints within intron 3 of TFG and intron 1 of the proximal gene, GPR128 (Figure 2A). A tandem duplication of this genomic region would, therefore, place 5′ TFG sequences upstream of the 3′ part of GPR128 with the possibility of forming a fusion gene (Figure 2B). RT-PCR on cDNA from the same patient demonstrated expression of a fusion transcript (Figure 3A) and sequencing showed this to be in frame (Figure 3B). To identify the genomic breakpoint, we employed PCR with primers placed at several kilobase intervals within the breakpoint introns. The breakpoints were found to lie at regions of homology in TFG at chr3:101,928,847 and GPR128 at chr3:101,817,568 (UCSC hg18) (Figure 3C). FISH with BAC RP11-398O21 on metaphases from a TFG-GPR128-positive individual, which entirely overlapped the amplification, showed single signals on each chromosome 3 (Figure 4) indicating that the region of amplification had not been excised and relocated to another part of the genome.
Figure 2.
Comparative genomic hybridization (CGH) array results and proposed structure of a 111 kb amplification. (A) Hybridization to an Agilent 244K CGH array allowed breakpoints to be positioned within TFG intron 3 and GPR128 intron 1. The log2 ratio of signals within the amplification suggested two additional copies of the CNV region. (B) Non-rearranged GPR128 lies upstream of TFG (top). The CNV amplification, with breakpoints within TFG and GPR128, results in one (middle) or two (bottom) copies of the TFG-GPR128 fusion gene. Arrows show direction of transcription.
Figure 3.
Analysis of TFG-GPR128 DNA and RNA. (A) RT-PCR with primers within TFG and GPR128 identified a fusion transcript. (B) Sequencing showed TFG exon 3 to be fused in frame to GPR128 exon 2. (C) Genomic sequencing demonstrated microhomology at the breakpoint regions. Genomic coordinates according to UCSC hg18.
Figure 4.
FISH on TFG-GPR128 positive cells with BAC RP11-398O21 (which entirely contains the CNV) and a chromosome 3 painting probe showing a single region of hybridization indicating that the amplified region has not been excised and relocated to another chromosome.
TFG-GPR128 is constitutional and associated with a copy number variant
Initially, two factors pointed to TFG-GPR128 being a novel leukemia fusion gene: (i) no CNV at that time was present in the relevant databases, and (ii) the predicted fusion protein was structurally similar to known oncogenic fusions, i.e. a partner (TFG) that was a known translocation target and which contributed a coiled-coil self-association motif, fused to a gene for a protein with signaling potential (GPR128).
A cohort of 575 cDNA samples from patients with atypical myeloproliferative neoplasms was screened by RT-PCR, thereby identifying a further seven (1.2%) TFG-GPR128-positive cases. However, a screen of 120 DNA samples from healthy control individuals, using an amplification refractory mutation system (ARMS) PCR with primers flanking the breakpoints, identified three positive individuals (2.5%), indicating that the amplification is a constitutional CNV and not an acquired mutation. Supporting evidence was obtained in one patient who presented with an ETV6-PDGFRB fusion and became ETV6-PDGFRB-negative during imatinib therpay, as determined by RT-PCR, but in whom both presentation and remission samples were found to be positive for TFG-GPR128. In addition, all TFG-GPR128-positive samples were found to share precisely the same genomic breakpoints. Constitutional DNA was not available from the other cases with atypical myeloproliferative neoplasms who were positive.
Prevalence of TFG-GPR128
Subsequent to this analysis, a CNV at this location was identified in three large genome-wide studies7,14,15 and data deposited in the Database of Genomic Variants (http://projects.tcag.ca/variation/). In one of these studies,7 506 unrelated individuals from the population-based PopGen biobank of samples from Schleswig-Holstein, Germany were genotyped on the Affymetrix 500K single nucleotine polymorphism (SNP) array, which led to the identification of ten individuals (2.0%) with CNV overlapping the TFG-GPR128 CNV region. We analyzed DNA from these ten individuals by ARMS PCR and found all to be positive for the fusion. Taken together, the data (3/120 local controls and 10/506 PopGen samples) suggest an incidence of around 2% in the UK and German populations.
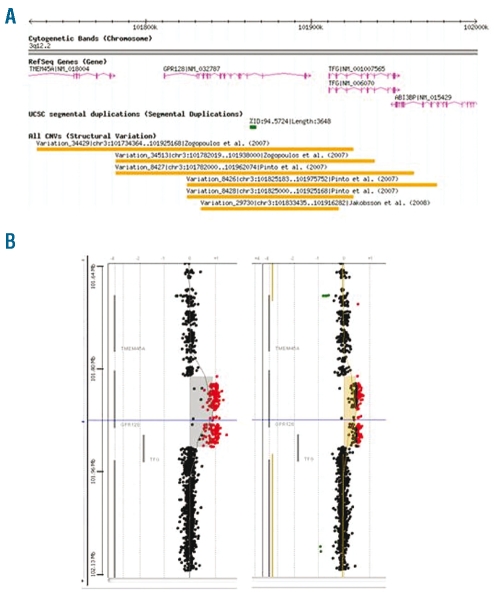
Variability of the TFG-GPR128 copy number variant
Although we were able to confirm the presence of the TFG-GPR128 fusion in all ten PopGen samples, the 500K SNP array data from Pinto7 placed the breakpoints in nine of the ten samples several kilobases from the breakpoints we had identified (Figure 5A). It was, therefore, possible that the TFG-GPR128 CNV lay closely adjacent to other CNV. To clarify the precise distribution of CNV in this region, we examined all ten PopGen samples using a custom 4x44K Agilent array with 1100 probes targeted to lie within a 500 kb region centered on the TFG-GPR128 CNV. Eight samples showed two additional copies and two showed a single additional copy with no evidence of any adjacent CNV (Figure 5B). Individuals with two copies of TFG-GPR128 may have either two copies of the chromosome carrying the fusion or one chromosome without the fusion plus a chromosome with two copies of the fusion. Since the frequency of TFG-GPR128 homozygotes is predicted to be 0.0001, the latter explanation seems more likely; however, in the absence of parental DNA this remains speculative.
Figure 5.
Variability of the TFG-GPR128 CNV region. (A) Previously published SNP array data demonstrated apparently variable CNV boundaries (from the Database of Genomic Variants). (B) Hybridization of ten TFG-GPR128 positive samples identified by Pinto7 to a custom Agilent 44K array (1100 probes targeted to lie within a 500 kb region centered on the TFG-GPR128 CNV) showed identical breakpoints with either two copies (left-hand array, eight cases) or one copy (right-hand array, two cases) of TFG-GPR128 with no evidence of adjacent CNV. Signal intensities are plotted on the x-axis as log2 ratio.
No evidence of association with an abnormal phenotype
The identification of ten TFG-GPR128-positive individuals from the PopGen biobank allowed us to examine whether the fusion is associated with any common disease phenotype (Online Supplementary Table S2). We compared the incidence of common diseases in the ten TFG-GPR128-positive individuals with that in 737 controls and an apparent excess of neuropathies (stroke, migraine, Parkinson’s disease, essential tremor, epilepsy or other unclassified neuropathy) was observed (5/10 TFG-GPR128-positive individuals versus 110/737 controls; P=0.01, Fisher’s exact test, two-tailed, uncorrected for multiple testing). However, this association could not be confirmed in a second cohort of 539 individuals for whom Affymetrix SNP 6.0 data were available. Using conservative criteria to avoid false positives, i.e. with amplification breakpoints closely overlapping the known CNV breakpoints, five TFG-GPR128-positive cases were identified, none of whom was classed as having neuropathies. Notably, there was no association of TFG-GPR128 positivity and malignancy; however, this does not exclude the possibility of a low penetrance predisposition of malignancy or other clinical phenotype.
TFG-GPR128-positive cases share a common haplotype
To investigate the origin of TFG-GPR128, we examined markers within the CNV. A microsatellite at chr3:101,840,545-101,840,591 (UCSC hg18) was particularly informative, showing an allele with 12 TG nucleotide repeats in all 19 TFG-GPR128-positive individuals examined whereas only 2/52 controls without TFG-GPR128 had a 12-repeat allele. To investigate the haplotype structure of the CNV region a linkage disequilibrium map was constructed of the TFG-GPR128 region using the LDMAP program16 and data from the 539 samples (534 controls and five TFG-GPR128-positive cases) that had Affymetrix SNP 6.0 genotypes. A core region of high linkage disequilibrium and relatively low haplotype diversity was determined as spanning 0.5 linkage disequilibrium units either side of the TG repeat. This region covered about 250 kb and contained 71 SNP between rs6777810 (chr3:101,729,832, UCSC hg18) and rs9850273 (chr3:101,982,055, UCSC hg18). Haplotype analysis was undertaken with PHASE,17 which identified 121 distinct haplotypes, with 15 having ten or more copies in the sample, collectively accounting for 81% of the assigned haplotypes. The five TFG-GPR128-positive cases were found to all share the fourth most common haplotype which was also found in 95/534 (17.8%) of controls (Online Supplementary Table S3). The sharing of an extended haplotype spanning more than 250 kb in the cases is consistent with a single ancestral origin as indicated by microsatellite analysis.
Distribution of TFG-GPR128
Considerable diversity in CNV distribution has been described both within and between populations.15 A study of the original 270 HapMap samples (30 Yoruba trios from Nigeria; 30 trios of European descent from Utah, USA (CEPH); 45 unrelated Japanese from Tokyo, Japan and 45 unrelated Han Chinese from Beijing, China) using Affymetrix 500K SNP arrays5 did not find any examples of the TFG-GPR128 CNV. Because of the potential variability in CNV calls highlighted by the analysis of PopGen samples, we reanalyzed these data [using data deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession GSE5173], focusing specifically on the TFG-GPR128 region and confirmed the absence of the CNV. In one worldwide study of 29 human populations comprising a total of 485 individuals,15 two TFG-GPR128-associated CNV positive individuals were identified, one from a Middle Eastern Bedouin population (n=47) and a second from a European Basque population (n=13). In the study by Zogopoulos,14 of a total of 1190 control individuals, six TFG-GPR128 CNV-positive individuals, all of Caucasian origin, were identified. All TFG-GPR128 cases identified so far are, therefore, of European or Middle Eastern ancestry. In our haplotype analysis, the SNP rs9873555 was identified as tagging the extended haplotype common to the TFG-GPR128-positive cases and several minor haplotypes. From International HapMap Data (http://www.hapmap.org), the minor allele frequencies for rs9873555 in Japanese, Han Chinese, Yoruba and CEPH populations are 0, 0.006, 0.137 and 0.161, respectively, therefore providing further evidence that the background upon which TFG-GPR128 has arisen is restricted. However, the possibility that TFG-GPR128 might also have arisen independently in other populations cannot be excluded.
Fusion gene formation by copy number variant amplification is uncommon
We sought to determine whether other CNV give rise to fusion genes. We used an in silico strategy to search for chimeric mRNA associated with known constitutional genomic copy number gains similar to the TFG-GPR128 CNV. A search of the UCSC Genome Bioinformatics database table all_est (http://genome.ucsc.edu) using Galaxy (http://main.g2.bx.psu.edu) and the strategy outlined in Figure 1, resulted in the identification of 122 fusion EST. These were examined manually using the UCSC browser to select those chimeric EST entirely overlapping with exons and with canonical splice sites at the junction of the fusion partner genes i.e. formed by precise fusion of intact exons from the two genes. This resulted in 26 fusion EST from 19 different fusions, including five EST from TFG-GPR128 (Table 1), which confirmed the validity of our method. An additional search using the NCBI Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov) with each fusion EST identified two further TFG-GPR128 EST and a single additional MSH5-AIF1 fusion EST. None of the EST fully overlapped with known CNV but three showed partial overlaps: CD44-PDHX, NRG4-SCAPER, SLC38A7-GOT2. Currently there is no clear evidence that these are CNV-derived rather than examples of trans-splicing or technical artifacts.
Table 1.
Fusion EST identified by an in silico search.
Discussion
We describe here the finding of a polymorphic in-frame gene fusion, TFG-GPR128, associated with a CNV. The fusion appears to have been derived from a single ancestral event and, in a limited analysis, is not associated with any obvious clinical phenotype. The precise normal cellular roles of TFG and GPR128 are unclear. GPR128 is an orphan member of the adhesion subfamily of G-protein coupled receptors which are characterized by a long serine/threonine-rich N-terminus,18 most of which is predicted to be retained in the TFG-GPR128 fusion protein. TFG has SH2 and SH3 binding motifs plus an N-terminal coiled-coil domain that may mediate self-association. The C. elegans homolog of TFG suppresses apoptosis and is required for normal cell-size control.19 Furthermore, a role for TFG in signaling is supported by the observations that TFG interacts with PTEN,20 the NF-κB pathway proteins (IKBKG) NEMO and TANK21 and regulates (PTPN6) SHP-1 activity.21 From NCBI Unigene EST profile data (http://www.ncbi.nlm.nih.gov/UniGene), expression of GPR128 appears to be restricted to the adrenal gland, intestine, kidney, liver, lung, placenta, skin, stomach and testis whereas TFG is strongly and ubiquitously expressed. Consequently the TFG-GPR128 fusion protein, if indeed it is translated from the chimeric mRNA, would be expected to have similar expression to TFG and potentially may affect signaling in diverse tissues. Although we found no obvious clinical phenotype associated with the presence of TFG-GPR128, it is possible that relatively subtle changes may emerge on more detailed analysis, for example once the normal functions of TFG and GPR128 are better understood.
It is remarkable that TFG is also the target of acquired, oncogenic translocations in malignancy and suggests there may be a common mechanism of fusion gene formation. Currently, however, there is no evidence for this: the TFG breakpoint in TFG-GPR128 occurs in intron 3 while TFG-ALK breakpoints occur in introns 3, 4 and 6,22 and TFG-NOR1 in intron 7.13 Moreover, although we found microhomology at the TFG and GPR128 breakpoints, which suggests formation by non-allelic homologous recombination, no evidence of homology was reported at the breakpoint sites in ALK-TFG fusion genes.22 It seems likely that involvement of TFG in both oncogenic and non-oncogenic fusion genes is coincidental, although it is possible that some unknown local structural feature may increase the probability of translocations at the TFG locus.
Gene fusions are well known in evolution and may provide the opportunity for the acquisition of novel functions. Examples include POMZP3, formed by fusion of the POM121 membrane glycoprotein and ZP3 (zona pellucida glycoprotein 3) 3–5 million years ago,23 USP6, a hominoid-specific fusion formed by the fusion of USP32 and TBC1D3 approximately 21–33 million years ago,24 and LRTOMT, a candidate catechol-O-methyltransferase that is mutated in non-syndromic deafness.25 The finding of the TFG-GPR128 polymorphism may thus be viewed as an intermediate step in evolution that could theoretically culminate in fixation or elimination.
Given the high frequency of CNV in the human genome, we searched for other polymorphic gene fusions in EST databases. Although we failed to find any other examples, EST are derived predominantly from the 5′ and 3′ ends of genes and thus fusions involving the central regions of genes may be under-represented. High throughput transcriptome analysis using sequencing26 or paired end ditag analysis27 on a population basis may yield further examples of polymorphic gene fusions. Indeed, paired end ditag analysis of two cancer cell lines yielded a surprisingly large number of candidate chimeric transcripts that may potentially have been acquired or inherited.27
In summary, the work presented here describes a further example of genomic complexity at the population level which may have implications for understanding human evolution. Our work also emphasizes the importance of careful discrimination of oncogenic changes found in tumor samples from non-pathogenic normal variation.
Supplementary Material
Footnotes
Funding: this work was supported by Leukemia Research (UK) (A Chase, FG and NCPC), the Deutsche José Carreras Leukämie-Stiftung, Germany (AR), the Dr. Mildred Scheel Stiftung für Krebsforschung, Germany (TE), the Capio Deutsche Klinik GmbH and by the DFG/BMBF Excellence Cluster Inflammation at Interfaces (AF). PopGen patient recruitment and the Affymetrix chip experiments were funded by the German National Genome Research Network (NGFN-2; Federal Ministry of Education and Research).
The online version of this article has a supplementary appendix.
Authorship and Disclosures
ACh, TE, FG, PE and AR performed experiments; ACo and ACh analyzed data; AF and SS provided PopGen samples and data; ACh and NC designed the research and wrote the paper; all authors contributed to and approved the final manuscript.
The authors reported no potential conflicts of interests.
References
- 1.Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17(11):661–9. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 2.Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348(13):1201–14. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 3.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36(10):1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 4.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(1):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 7.Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;16(Spec No. 2):R168–R73. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- 8.Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9(1):55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 9.Sohal J, Chase A, Goldman JM, Cross NC. Assignment of ZNF262 to human chromosome band 1p34-->p32 by in situ hybridization. Cytogenet Cell Genet. 1999;85(3–4):306–7. doi: 10.1159/000015282. [DOI] [PubMed] [Google Scholar]
- 10.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94(9):3265–8. [PubMed] [Google Scholar]
- 12.Greco A, Mariani C, Miranda C, Lupas A, Pagliardini S, Pomati M, et al. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol. 1995;15(11):6118–27. doi: 10.1128/mcb.15.11.6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hisaoka M, Ishida T, Imamura T, Hashimoto H. TFG is a novel fusion partner of NOR1 in extraskeletal myxoid chondrosarcoma. Genes Chromosomes Cancer. 2004;40(4):325–8. doi: 10.1002/gcc.20044. [DOI] [PubMed] [Google Scholar]
- 14.Zogopoulos G, Ha KC, Naqib F, Moore S, Kim H, Montpetit A, et al. Germ-line DNA copy number variation frequencies in a large North American population. Hum Genet. 2007;122(3–4):345–53. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- 15.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 16.Lau W, Kuo TY, Tapper W, Cox S, Collins A. Exploiting large scale computing to construct high resolution linkage disequilibrium maps of the human genome. Bioinformatics. 2007;23(4):517–9. doi: 10.1093/bioinformatics/btl615. [DOI] [PubMed] [Google Scholar]
- 17.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yona S, Lin HH, Siu WO, Gordon S, Stacey M. Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem Sci. 2008;33(10):491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, McCloskey T, Joshi PM, Rothman JH. ced-4 and proto-oncogene TFG-1 antagonistically regulate cell size and apoptosis in C. elegans. Curr Biol. 2008;18(14):1025–33. doi: 10.1016/j.cub.2008.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herlevsen M, Oxford G, Ptak C, Shabanowitz J, Hunt DF, Conaway M, et al. A novel model to identify interaction partners of the PTEN tumor suppressor gene in human bladder cancer. Biochem Biophys Res Commun. 2007;352(2):549–55. doi: 10.1016/j.bbrc.2006.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miranda C, Roccato E, Raho G, Pagliardini S, Pierotti MA, Greco A. The TFG protein, involved in oncogenic rearrangements, interacts with TANK and NEMO, two proteins involved in the NF-kappaB pathway. J Cell Physiol. 2006;208(1):154–60. doi: 10.1002/jcp.20644. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez L, Bea S, Bellosillo B, Pinyol M, Falini B, Carbone A, et al. Diversity of genomic breakpoints in TFG-ALK translocations in anaplastic large cell lymphomas: identification of a new TFG-ALK(XL) chimeric gene with transforming activity. Am J Pathol. 2002;160(4):1487–94. doi: 10.1016/S0002-9440(10)62574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kipersztok S, Osawa GA, Liang LF, Modi WS, Dean J. POM-ZP3, a bipartite transcript derived from human ZP3 and a POM121 homologue. Genomics. 1995;25(2):354–9. doi: 10.1016/0888-7543(95)80033-i. [DOI] [PubMed] [Google Scholar]
- 24.Paulding CA, Ruvolo M, Haber DA. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc Natl Acad Sci USA. 2003;100(5):2507–11. doi: 10.1073/pnas.0437015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed ZM, Masmoudi S, Kalay E, Belyantseva IA, Mosrati MA, Collin RW, et al. Mutations of LRTOMT, a fusion gene with alternative reading frames, cause non-syndromic deafness in humans. Nat Genet. 2008;40(11):1335–40. doi: 10.1038/ng.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, et al. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using paired-end ditags (PETs) Genome Res. 2007;17(6):828–38. doi: 10.1101/gr.6018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.