Abstract
Commitment towards megakaryocyte versus erythroid blood cell lineages occurs in the megakaryocyte-erythroid progenitor, where mutually exclusive expression of either EKLF (Klf1) or Fli1 defines alternative outcomes. Here we show there is a marked increase in the number of circulating platelets in mice lacking the erythroid transcription factor EKLF. In addition, committed erythroid cells retain key signatures of megakaryocytes both on the cell surface and at the mRNA level. We also show that the effect of EKLF on megakaryocyte-erythroid progenitor lineage decision and commitment is cell autonomous in bone marrow reconstitution assays where stem cells lacking EKLF favor the megakaryocyte differentiation pathway. We conclude the megakaryocyte program is aberrantly activated in EKLF null erythroid cells.
Keywords: megakaryocyte, EKLF, megakaryocyte differentiation
Introduction
Erythroid Krüppel-like factor (EKLF) is the founding member of the Krüppel-like factor family of transcription factors and is critical for many aspects of erythropoiesis.1–3 Previous cell line and biochemical studies have shown an important role for EKLF in determination of erythroid versus megakaryocyte lineages.4–7 For example, studies performed in mouse ES cells have demonstrated that the forced expression of EKLF reduces megakaryocytic, but increases erythroid differentiation potential.5 Endogenous EKLF is expressed prior to erythroid commitment to push megakaryocyte-erythroid progenitor (MEP) cells towards this pathway,7 a conclusion supported by an observed increase in megakaryocyte numbers during thrombopoietin (TPO) induced differentiation of EKLF−/− primary fetal liver cultures when compared to WT cultures.5 Additional studies using specific siRNAs targeting EKLF in murine erythroleukemia (MEL) cells demonstrated that the loss of EKLF enhances the megakaryocyte program.4
The biological mechanism responsible for lineage determination in the MEP may involve direct functional interactions between Fli1 and EKLF at megakaryocytic and erythroid gene promoters in an elaborate cross-antagonsim.8 It appears that the antagonism of megakaryocytic gene promoters is dependent on a specific EKLF sumoylation event that is critical for its function as a transcriptional repressor.6 To date no studies have examined the consequences of loss of EKLF in vivo with respect to megakaryopoiesis partly because until recently it was not appreciated that platelets are produced very early during development.9
Design and Methods
Mouse lines and procedures
EKLF−/− erythroid tissues were collected from EKLF+/− timed matings and genotypes confirmed as previously described.3 EKLF+/− EGFP-actin transgenic mice were generated by crossing EKLF+/− mice10 with EGFP-actin transgenic mice11 on a congenic Balb/c background. Balb/c bone marrow transplant recipient mice were lethally irradiated (850Gy) and transplanted at eight weeks of age by injection of a 50:50 mix of WT fetal liver cells with EKLF−/− fetal liver cells marked with EGFP (EKLF−/− GFP), or a 50:50 mix of WT and WT-EGFP livers (CON GFP) that served as a control.
Flow cytometry
Analysis of peripheral blood samples was performed using an LSRII flow cytometer (BD Biosciences, NJ, USA) with antibodies for CD71 (CD71-PE, 553267) (BD Pharmingen, CA, USA) and CD41 (CD41-FITC, 553848)(BD Pharmingen). CD71-PE positive cells and EGFP positive cells were sorted from fetal liver and bone marrow preparations, respectively, using an Influx cell sorter (Cytopeia, WA, USA).
Gene expression profiling
cDNA was prepared from CD71 positive sorted cells as previously described.3 Primers for real-time RT-PCR were designed using Primer Express software (Applied Biosystems, CA, USA). Quantitative real time RT-PCR was performed using SYBR Green chemistry on an ABI-Prism 7500 sequence detection system (Applied Biosystems) (Primer sequences are shown in Online Supplementary Table S1).
Histology
Splenic sections were prepared from bone marrow transplanted mice, stained with hematoxylin and eosin and imaged using a BX51 microscope fitted with a DP70 digital camera imaging system (Olympus, Tokyo, Japan).
Methylcellulose colony assays
EGFP positive cells sorted from bone marrow preparations were plated at 3×104 cells/mL in MethoCult GFM3434 methylcellulose-based medium (Stem Cell Technologies, BC, Canada). CFUe were scored after three days. BFUe, BFUe/mk, and CFUmk were scored after 12 days. Colonies were scored based on morphology previously described.12
Results and Discussion
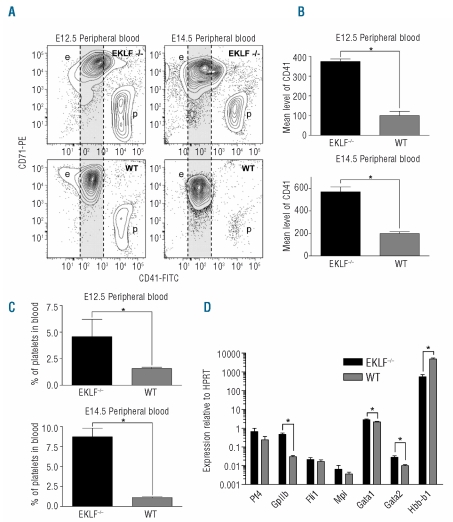
The objective of this study was to characterize the fate of MEPs in EKLF−/− mice which die by E15.5 from anemia.10,13 We initially observed a dramatic increase in the number of circulating CD41+ platelets at E12.5 and E14.5 in EKLF−/− embryos when compared to WT embryos (Figure 1A and C). Surprisingly, further analysis of CD71+ erythroid cells at E12.5 and E14.5 revealed that EKLF−/− cells also expressed CD41 at significant levels on the cell surface (Figure 1A). We found a marked increase in the mean level of CD41 at the cell surface for primitive (E12.5) and definitive (E14.5) EKLF−/− erythroid cells compared to WT cells (Figure 1B). This observation suggested that the loss of EKLF was leading to failure of appropriate erythroid versus megakaryocyte lineage choices in the progeny of the MEP.
Figure 1.
Increased platelets and lineage infidelity in EKLF−/− mice. (A) Representative contour plots generated from E12.5 and E14.5 EKLF−/− or wild-type (WT) peripheral blood by combined staining for CD71 (erythroid) and CD41 (megakaryocyte). The WT level of CD41 staining is indicated by a gray area for comparison (e: erythroid cells; p: platelets). (B) Quantification of the mean level of CD41 staining observed in the peripheral blood of E12.5 and E14.5 embryos for each genotype (Mean±SEM, ≥3 for each genotype, *P<0.05 by Student’s t test). (C) Quantification of the percentage of platelets contained in the peripheral blood of E12.5 and E14.5 embryos for each genotype (Mean±SEM, n≥3 for each genotype, *P<0.05 by Student’s t test). (D) Gene expression determined by RT-PCR for CD71 sorted E14.5 fetal liver cells (Mean±SEM normalized to the housekeeping gene HPRT and presented on a log scale, n=9 for each, *P<0.05 by Student’s t test).
We hypothesized that these biphenotypic cells (CD71+, CD41+) had arisen due to either an expansion of a normal but rare cell type, or an aberrant gene expression program caused by the loss of EKLF. To confirm that the latter was the most likely explanation we sorted CD71 positive erythroid cells from both EKLF−/− and WT fetal livers at E14.5 and analyzed the expression of megakaryocyte genes14,15 at the mRNA level by real-time RT-PCR. We found a significant increase in the expression of GpIIb in the EKLF−/− sorted cells compared to WT sorted cells as expected since this gene encodes the CD41 antigen (Figure 1D). Levels of Gata1 and Gata2 were also significantly increased, although the change was less dramatic (Figure 1D). The genes for platelet factor 4 (Pf4) and the TPO receptor (Mpl) were also mildly increased in EKLF−/− erythroid cells; however, this increase was not found to be significant. Interestingly, we found only a minor change (not significant) in the expression level of the Fli1 gene, a critical determinant of the megakaryocyte lineage. Expression of βMaj-Globin (Hbb-b1) was almost absent in the EKLF−/− sorted cells, and served as a useful control (Figure 1D). We conclude that the loss of EKLF leads to failure to silence megakaryocyte specific genes in erythroid progeny of MEPs; that is a lineage infidelity that results in the majority of cells becoming abnormally biphenotypic. It is possible that the molecular mechanism responsible for the lineage promiscuity found in EKLF−/− erythroid cells involves both the loss of sumoylation dependent EKLF repression and a loss of BKLF repression of megakaryocyte genes, and emerges independently of a change in Fli1 gene expression.6,16
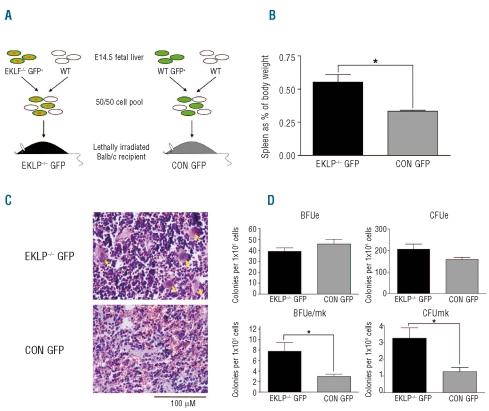
In order to verify these observations, and to demonstrate that the effect of loss of EKLF on lineage choice was cell autonomous, we performed bone marrow reconstitution assays. The other aim of this study was to test whether EKLF acts as a tumor suppressor gene, in a similar manner to that which has been proposed for other Kruppel-like factors.17 EKLF−/− erythroid progenitors are more readily transformed by co-operating oncogenes,18 and we hypothesized EKLF−/− MEPs might similarly obtain additional mutations and generate leukemia. We generated a 50:50 mix of WT fetal liver cells with EKLF−/− fetal liver cells marked with EGFP to reconstitute the bone marrow compartment of lethally irradiated recipient mice (Figure 2A). A mixture of EKLF−/− and WT fetal livers was necessary as reconstitution with EKLF−/− bone marrow alone would lead to rapid death from anemia. Reconstitution of lethally irradiated mice with a 50:50 mix of WT and WT-EGFP livers served as a control for this experiment. Successful reconstitution of the bone marrow was confirmed at eight weeks post transplant by FACS using markers for non-erythroid lineages (B220, Gr1, and Mac1) and fluorescence microscopy of peripheral blood smears to look for EGFP positive cells which were derived from the EKLF−/− compartment. As expected, we found equal reconstitution (~50:50 GFP+ to GFP-) of EKLF−/− cells to each of the non-erythroid lineages investigated (data not shown).
Figure 2.
Lineage promiscuity and increased platelet production is transplantable. (A) A schematic presentation of the experimental design for bone marrow reconstitution assays. Mice receive a mixture of fetal liver cells containing either EKLF−/− cells (EKLF−/− GFP) or WT cells (CON GFP). (B) Spleen weights as a percentage of total body weight for EKLF−/− transplants (EKLF−/− GFP) or control transplants (CON GFP). Mean + SEM, n≥4 for each group, *P<0.05 by Student’s t test. (C) Histological examination of transplant mouse spleens by hematoxylin and eosin staining. Images acquired at 40x magnification with scale bar indicated. (D) Methylcellulose colony assays using GFP positive sorted bone marrow from EKLF−/− GFP or CON GFP mice. CFUe were scored at day 3, other colonies were scored at day 12. Mean + SEM, n=4, *P<0.05 by Student’s t test.
At the conclusion of the experiment (12 months post transplant) mice were sacrificed and the erythroid compartment examined by FACS, histologically, and by methylcellulose colony assays. We did not find a single leukemia suggesting either EKLF does not function as a tumor suppressor or additional mutations are required (n. more than 20 individual mice for experimental and control groups). However, we found a significant increase in spleen weights in the EKLF−/− GFP cohort compared to the control cohort (CON GFP, Figure 2B). We attributed this to an increase in red pulp caused by an expansion of the erythroid compartment (Figure 2C). We believe this was the result of an increase in red cell production by WT HSCs to compensate for the EKLF−/− HSCs which are unable to produce healthy red cells. We also observed an increase in megakaryocyte numbers in the spleens of these mice by hematoxylin and eosin staining (yellow arrows, Figure 2C). In colony assays using FACS sorted GFP positive bone marrow, the EKLF−/− GFP mice produced significantly more megakaryocyte colonies (CFUmk) and mixed erythroid/megakaryocyte colonies (BFUe/mk) than the control mice (Figure 2D), demonstrating an increased megakaryocyte potential for EKLF−/− HSCs and/or MEPs. We found no significant change in the number of BFUe and CFUe as we expected from previous work (Figure 2D).10 We conclude from these observations that the loss of EKLF in vivo leads to megakaryocyte-erythroid lineage promiscuity. Although this may partly result from failure to silence Fli1 in erythroid progenitor cells as suggested previously,4,8 this work suggests other molecular mechanisms may be at play.
Footnotes
Funding: this work was supported by a grant from the Cancer Council Queensland (519718/ACP), and an Australian Research Council Discovery Grant (DP0770471/ACP). MRT is the recipient of an Australian Postgraduate Award.
The online version of this article has a supplementary appendix.
Authorship and Disclosures
MRT designed and performed research, and wrote the paper. ACP advised on research and contributed to manuscript preparation.
The authors reported no potential conflicts of interest.
References
- 1.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13(5):2776–86. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drissen R, von Lindern M, Kolbus A, Driegen S, Steinlein P, Beug H, et al. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol Cell Biol. 2005;25(12):5205–14. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107(8):3359–70. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouilloux F, Juban G, Cohet N, Buet D, Guyot B, Vainchenker W, et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112(3):576–84. doi: 10.1182/blood-2007-07-098996. [DOI] [PubMed] [Google Scholar]
- 5.Frontelo P, Manwani D, Galdass M, Karsunky H, Lohmann F, Gallagher PG, et al. Novel role for EKLF in megakaryocyte lineage commitment. Blood. 2007;110 (12):3871–80. doi: 10.1182/blood-2007-03-082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siatecka M, Xue L, Bieker JJ. Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol Cell Biol. 2007;27 (24):8547–60. doi: 10.1128/MCB.00589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmann F, Bieker JJ. Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development. 2008;135(12):2071–82. doi: 10.1242/dev.018200. [DOI] [PubMed] [Google Scholar]
- 8.Starck J, Cohet N, Gonnet C, Sarrazin S, Doubeikovskaia Z, Doubeikovski A, et al. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol. 2003;23(4):1390–402. doi: 10.1128/MCB.23.4.1390-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KK, et al. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 2007;109(4):1433–41. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perkins AC, Sharpe AH, Orkin SH. Lethal β-thalassemia in mice lacking the erythroid Caccc-transcription factor Eklf. Nature. 1995;375(6529):318–22. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 11.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407(3):313–9. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 12.Metcalf D, Metcalf D. Clonal culture of hemopoietic cells: techniques and applications. Amsterdam, New York, NY, USA: Elsevier Science Publishers; Sole distributors for the USA and Canada, Elsevier Science Pub. Co; 1984. [Google Scholar]
- 13.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375(6529):316–8. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 14.Pang L, Xue HH, Szalai G, Wang X, Wang Y, Watson DK, et al. Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood. 2006;108(7):2198–206. doi: 10.1182/blood-2006-04-019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, et al. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity. 2000;13(2):167–77. doi: 10.1016/s1074-7613(00)00017-0. [DOI] [PubMed] [Google Scholar]
- 16.Funnell AP, Maloney CA, Thompson LJ, Keys J, Tallack M, Perkins AC, et al. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol Cell Biol. 2007;27(7):2777–90. doi: 10.1128/MCB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188(2):143–60. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 18.Coghill E, Eccleston S, Fox V, Cerruti L, Brown C, Cunningham J, et al. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood. 2001;97(6):1861–8. doi: 10.1182/blood.v97.6.1861. [DOI] [PubMed] [Google Scholar]