Abstract
Peripheral arterial disease (PAD) is associated with significant morbidity and mortality, and has a higher prevalence in African Americans than Caucasians. Ankle arm index (AAI) is the ratio of systolic blood pressure in the leg to that in the arm, and, when low, is a marker of PAD. We used an admixture mapping approach to search for genetic loci associated with low AAI. Using data from 1040 African-American participants in the observational, population-based Health, Aging, and Body Composition Study who were genotyped at 1322 single nucleotide polymorphisms(SNPs) that are informative for African versus European ancestry and span the entire genome, we estimated genetic ancestry in each chromosomal region and then tested the association between AAI and genetic ancestry at each locus. We found a region of chromosome 11 that reaches its peak between 80 and 82 Mb associated with low AAI (p<0.001 for rs12289502 and rs9665943, both within this region). 753 African-American participants in the observational, population-based Cardiovascular Health Study were genotyped at rs9665943 to test the reproducibility of this association, and this association was also statistically significant (odds ratio(OR) for homozygous African genotype 1.59 (95% confidence interval (CI) 1.12–2.27)). Another candidate SNP (rs1042602) in the same genomic region was tested in both populations, and was also found to be significantly associated with low AAI in both populations (OR for homozygous African genotype 1.89 (95% CI 1.29–2.76)). This study identifies a novel region of chromosome 11 representing an area with a potential candidate gene associated with PAD in African Americans.
Keywords: peripheral vascular disease, genetics, African-American
INTRODUCTION
Peripheral arterial disease (PAD) is a common and serious medical problem, associated with significant functional limitation due to claudication and leg weakness as well as considerable morbidity and mortality from cardiovascular causes [1–3]. African Americans have increased rates of PAD[1,4]; a recent pooled analysis of several community-based studies found that over 20% of African-American men between the ages of 70 and 79 had PAD, compared with under 9% of non-Hispanic whites of the same age.[5] Prior studies have estimated the heritability of PAD to be between 35 and 48%.[6,7] Numerous candidate genes have been tested for association with PAD [8–11], however the results have been equivocal. Two prior linkage scans have identified separate chromosomal regions on chromosomes 1 and 3 that may contain genes related to PAD susceptibility. [7,12]
Ankle arm index (AAI) is the ratio of systolic blood pressure in the lower extremity to that in the upper extremity. AAI values of less than 0.9 are consistent with the presence of PAD and associated with an increased risk of mortality, particularly from cardiovascular causes.[2, 13–16] The higher prevalence of PAD in African Americans makes it an interesting phenotype for admixture mapping, a technique for detecting disease gene variants in populations resulting from recent admixture between two genetically distinct populations.[17–22] Admixture mapping takes advantage of the relatively long-range linkage disequilibrium (LD) in recently admixed populations such as African Americans. This long range LD is seen between markers that have high allele frequency differences between ancestral populations. Because larger blocks of the genome are in LD in recently admixed populations, whole-genome association mapping can be performed with about1% of the markers that are typically required for a genome-wide association study.[23] This method has been used to localize regions of the genome with variants predisposing to increased risk of several common diseases. [24–27]
We first assessed whether individual proportion of African ancestry, as estimated from a set of ancestry-informative markers, was associated with AAI using data from two large U.S. population-based African-American cohorts; the Health, Aging, and Body Composition (Health ABC-AA) Study, and the Cardiovascular Health Study (CHS-AA). We then used a larger set of genome-wide ancestry-informative markers to perform an admixture mapping study to search for genomic loci for low AAI among subjects in Health ABC-AA, and validated these results using the genotypes at two loci in participants in Health ABC-AA and CHS-AA.
METHODS
Study Population 1: Health ABC-AA (primary cohort)
Study participants were enrolled in the Health ABC Study between April 1997 and June 1998. Subjects were well-functioning men and women aged 70–79 sampled from Medicare beneficiaries living in or around Alleghany County, PA and Memphis, TN. Inclusion and exclusion criteria have been described in detail elsewhere.[27] Of a sample of 3075 subjects, 1281 were self-reported African Americans. Participants without AAI data (n=125) or genetic data (n=125) or with AAI above 1.5 (n=2) were excluded from the analysis, leaving a study population of 1040 for most of our analyses (443 men, 597 women). The Health ABC study has been approved by the institutional review boards of the University of Pittsburgh, the University of Tennessee, Memphis and the University of California, San Francisco.
Study Population 2: CHS-AA (replication cohort)
CHS is a prospective study of risk factors for cardiovascular disease comprised of 5,888 men and women aged 65 and older. Participants were recruited from Forsyth County, NC, Sacramento County, CA, Washington County, MD, and Pittsburgh, PA. The original cohort of 5,201 participants (including 254 African-Americans) was recruited in 1989–1990, and an additional cohort of 687 African Americans was enrolled in 1992. CHS methods have been described in greater detail elsewhere.[28] The current study population of 753 African Americans (284 men, 469 women) was derived from the 941 African Americans who were recruited into CHS-AA. Subjects without AAI, those who did not consent for DNA testing, or those whose genetic data was unusable were excluded from this analysis.
All participants in both studies have given informed consent.
Ankle-arm index measurement
AAI was assessed at the baseline Health ABC-AA visit (1997–1998). Subjects were recumbent for a rest period of 5 minutes, at which point blood pressure cuffs were placed on the right arm and right and left ankles. Arterial systolic blood pressure was assessed by an 8 Megahertz Doppler probe (Huntleigh Healthcare Inc., Eatontown, NJ) and mercury syphygmomanometer. Two measurements were taken at each of the three arterial sites. The lower of the two posterior tibial pressures was used for the calculation of AAI. Peripheral arterial disease was defined as a self-report of history of peripheral angioplasty or bypass procedure or an AAI of <0.9 in either leg. AAI and PAD were assessed similarly in CHS-AA. The methods have been described previously.[15]
Baseline participant characteristics
All covariates were assessed at the baseline exams of Health ABC-AA and CHS-AA. Smoking status was coded as current, former or never smoker by self-report. Diabetes mellitus was defined as self-report of a physician’s diagnosis of diabetes, self-report of using insulin or oral hypoglycemic medications, or fasting plasma glucose of greater than 125mg/dL. Coronary heart disease was defined as physician’s diagnosis of myocardial infarction or angina or history of coronary artery bypass surgery or percutaneous coronary angioplasty. Cerebrovascular disease was defined as history of transient ischemic attack, cerebrovascular accident, or carotid endarterectomy. Cardiovascular disease was defined as self-reported history of angina, myocardial infarct, stroke, transient ischemic attack, congestive heart failure, claudication, cardiovascular re-vascularization or bypass surgery. Hypertension was defined as seated systolic blood pressure greater than 140 mmHg or seated diastolic blood pressure greater than 90 mmHg or self-reported history of hypertension and self-report of taking an antihypertensive medication. Total serum cholesterol and high-density lipoprotein (HDL) were analyzed enzymatically.
Genotyping
Genotypes were analyzed in 1184 self-reported African-American Health ABC-AA participants. The Broad Institute of Harvard and MIT carried out the genotyping across 1536 single nucleotide polymorphisms (SNPs) using whole-genome amplified DNA samples with an Illumina BeadLab platform.[29] These SNPs were previously identified as being ancestry informative, with particular genotypes occurring with a significantly higher frequency in African populations.[26] Quality control criteria for genotype data included an 85% genotype success rate and consistent clustering of calls for each SNP, and not being in linkage disequilibrium in African population samples. [30,31] 201 SNPs not meeting these criteria were removed from the panel. Individual genome-wide percentage of European ancestry was calculated from a subset of 35 ancestry-informative SNPs in Health ABC-AA and 24 ancestry-informative SNPs in CHS-AA. [32] The ancestry-informative SNPs were chosen based upon their high degree of allele frequency difference among ancestral populations as well as relatively even spacing across the genome, so that each SNP provided unique information.
Additional genotyping of rs9665943 was performed in 753 African American CHS-AA participants. Additionally, one of the 24 ancestry-informative markers previously typed in CHS-AA (rs1042602) happened to be located within the chromosome 11 locus identified in Health ABC-AA and was subsequently typed in Health ABC-AA for further analysis. Both additional SNPs (rs9665943 in CHS and rs1042602 in Health ABC-AA) were genotyped following an identical protocol.[32]
Association between individual ancestry and AAI
To assess the relationship between individual ancestry and AAI in each cohort, we first assessed the correlation between AAI and genome-wide percentage of European ancestry and then performed linear regressions of genome-wide percentage of European ancestry on AAI, adjusting for age, study, site, and gender. The relationship between continuous variation in genome-wide percentage of European ancestry and AAI was additionally evaluated by multivariate regression, which was adjusted for gender, age, study site (and study in combined models), socio-economic status, total serum cholesterol, HDL cholesterol, weight, height, smoking status, diabetes status, hypertension status, and prevalent cardiovascular disease.
Admixture mapping in Health ABC-AA
Admixture scans were conducted using three different analytic platforms, STRUCTURE[18], ADMIXMAP[33] and ANCESTRYMAP[21]. Briefly, each of these methods uses a Hidden Markov Model approach to determining ancestry at each locus. All three programs also use Markov Chain Monte Carlo (MCMC) algorithms to calculate genome-wide associations.
STRUCTURE 2.2 was used to obtain multi-locus estimates of ancestry within each genomic region, under the assumption of a 2 population model with African and European ancestry. In addition to the locus-specific ancestry estimate, STRUCTURE calculates an overall percentage of European and African ancestry for each participant. These models were run across 2,500 iterations of the MCMC algorithm, with a 2,000 iteration burn in period. Linear regression was performed to test for association between ancestry at each locus and AAI, adjusting for individual percentage of African ancestry, gender, age and study site. Results of the regression models are presented as t-statistics and p-values.
ADMIXMAP v.3.5.3 simultaneously estimates locus-specific ancestry and tests for association between locus-specific ancestry and a quantitative trait by computing probability distributions conditional on genotype, phenotype (in this instance, AAI), and prior ancestral genotype frequencies. These models also ran 2,500 MCMC iterations across a 2000 iteration burn-in period and were adjusted for the same covariates as the STRUCTURE regressions.
For the ANCESTRYMAP analyses, subjects were dichotomized into two homogenous groups with the lowest 20% of AAI values (AAI<0.89) as cases and the highest 20% (AAI>1.16 and AAI<=1.5) as controls. There were 416 individuals split equally between the two groups, matched on criteria of age, gender and study site. A range of risk estimates was tested, and using Bayesian model averaging, the strength of evidence for association with group status at each locus was summarized as a Bayes factor. The log(base 10) of the Bayes Factor represents the locus-specific LOD score. As previously described, a locus-specific LOD score >5 is considered approximately genome-wide significant. [26, 34] ANCESTRYMAP was run for a burn-in period of 200 iterations with a follow-on of an additional 500 iterations. 95% confidence intervals based on a genome-wide LOD 2.2 decrease of the association were calculated.
Combined analysis of rs9665943 and rs1042602 in Health ABC-AA and CHS-AA
Because rs9665943 showed consistent association with low AAI across all three analysis platforms, more detailed logistic regression analysis of the relationship between genotype at rs9665943 and AAI of less than 0.9 was performed in both Health ABC-AA and CHS-AA. The outcome variable of interest in these analyses was an AAI of less than 0.9. By chance, another ancestry-informative SNP located in the putative AAI region on chromosome 11 (rs1042602) had been typed previously in CHS-AA. Therefore, rs1042602 was typed in Health ABC-AA as well and genotype at this SNP was tested for its relationship to low AAI. These models were run for each individual study population separately and in a single model combining the data from both studies. These models were adjusted for gender, age, study site (and study in combined models), total serum cholesterol, HDL cholesterol, weight, smoking status, diabetes status, hypertension status, prevalent cardiovascular disease, and estimated percentage of African ancestry. Results are shown for the homozygous African genotype. Subjects with the homozygous European genotype and heterozygotes comprised the reference group for calculating odds ratios, and a dominant genetic model was used. Subjects with AAI greater than 1.4 were excluded from these analyses.
RESULTS
Participant characteristics
Table 1 shows the baseline demographic information in both the primary and replication cohorts. The mean age of subjects in the Health ABC-AA cohort was 73.4 (range 68–80), the mean AAI was 1.02 (range 0.44–1.50), and the mean percentage of African ancestry was 77.2% (range 11.2–99.9). More than half of subjects (55.1%) had a history of smoking, 26.6% were diabetic, 19.8% had a history of coronary heart disease, and 10.6% had a history of peripheral arterial disease. The baseline characteristics of the CHS-AA population were similar to those of the Health ABC-AA participants, although fewer had clinical PAD. The mean genome-wide percentage of African ancestry in CHS-AA was 73.8% and the mean AAI was 1.02.
Table 1.
Baseline characteristics of study participants
| Health ABC-AA | Men n=443 | Women n=597 | Total n=1040 | CHS-AA | Men n=284 | Women n=469 | Total N=753 | |
|---|---|---|---|---|---|---|---|---|
| Age* | 73.5 (2.8) | 73.3 (3.0) | 73.4 (2.9) | 72.5 (5.5) | 72.9 (5.5) | 72.7 (5.5) | ||
| Systolic BP (mmHg)* | 137.7 (22.1) | 138.4 (21.7) | 138.1 (21.9) | 138.3 (20.4) | 143.1 (23.3) | 141.3 (22.3) | ||
| Diastolic BP (mmHg)* | 75.1 (12.1) | 72.1 (12.4) | 73.3 (12.3) | 75.9 (11.5) | 74.4 (10.8) | 74.9 (11.1) | ||
| Total Cholesterol (mg/dL)* | 193.7 (36.7) | 213.6 (40.5) | 205.1 (40.1) | 198.2 (35.2) | 215.6 (39.1) | 209.0 (38.6) | ||
| Weight (Kg) | 81.6 (15.0) | 75.6 (16.1) | 78.2 (15.9) | 80.1 (14.0) | 75.6 (15.6) | 77.3 (15.2) | ||
| Ever Smoker (%) | 69.8 | 44.2 | 55.1 | 69.0 | 41.9 | 51.5 | ||
| Diabetes (%) | 28.6 | 25.1 | 26.6 | 37.3 | 37.7 | 37.6 | ||
| Peripheral Arterial Disease (%) | 13.2 | 7.9 | 10.2 | 5.6 | 2.4 | 3.6 | ||
| Coronary Heart Disease (%) | 23.0 | 17.4 | 19.8 | 17.3 | 20.7 | 19.4 | ||
| Cerebrovascular Disease (%) | 7.7 | 8.2 | 8.0 | 10.9 | 7.0 | 8.5 | ||
| Ankle-Arm Index* | 1.04 (0.18) | 1.01 (0.18) | 1.02 (0.18) | 1.04 (0.22) | 1.01 (0.19) | 1.02 (0.20) | ||
| Percent African Ancestry* | 78.3 (14.8) | 76.4 (15.9) | 77.2 (15.5) | 73.7 (20.6) | 73.9 (19.8) | 73.8 (20.1) |
Mean (Standard Deviation)
Association between individual ancestry and AAI
We first assessed the relationship between genome-wide percentage of European ancestry and AAI separately in Health ABC-AA and CHS-AA, and obtained similar results in each cohort. The combined results are shown in Figure 1. Overall, there was a statistically significant association between increasing percentage of European ancestry and increasing AAI in the combined analysis group of Health ABC-AA and CHS-AA participants (p = 9.6*10−5, r2 0.68, Figure 1). This association was not attenuated in multivariate regression models that adjusted for gender, age, study site, height, weight, socio-economic status, total serum cholesterol, high-density lipoprotein (HDL) cholesterol, weight, height, smoking status (never, former or current smoker), diabetes status, hypertension status, and prevalent cardiovascular disease.
Figure 1.
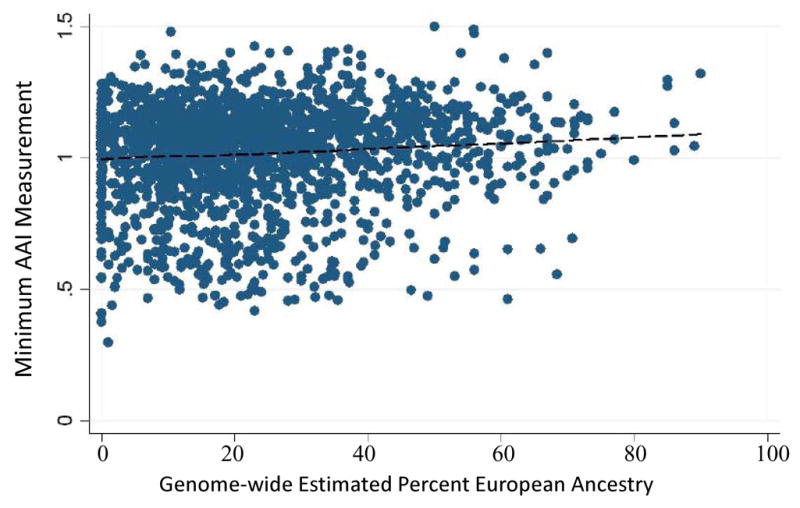
Increasing minimum AAI measurement is associated with increasing percent ancestry from genome-wide estimates. p, trend = 9.6*10−5, r2=0.68
Admixture mapping of AAI in Health ABC-AA
By modeling AAI as a continuous trait and using the locus-specific ancestry estimates from either STRUCTURE or ADMIXMAP, the strongest evidence for ancestry association with AAI was on chromosome 11, including rs12289502 at 80Mb and rs9665943 at 82Mb (Table 2). The admixture mapping results for all SNPs genotyped in Health ABC-AA between 11–76 and 11–88 are shown in table 2. Following Bonferroni correction for multiple testing, the association between rs9665943 and AAI retained borderline statistical significance (p=0.08). The association between locus-specific ancestry at chromosome 11 and AAI eliminated the association between individual genome-wide ancestry and AAI in our STRUCTURE based analyses and regression models.
Table 2.
Admixture scan results for chromosome 11 locus using Health ABC-AA Population
| SNP | Locus* | T, STRUCTURE | p, STRUCTURE | p, ADMIXMAP Ancestry Association | LOD Score, ANCESTRYMAP |
|---|---|---|---|---|---|
| rs3740767 | 11–76 | −3.22 | 0.001 | 0.0007 | 3.8 |
| rs3740677 | 11–78 | −3.37 | 0.001 | 0.001 | 3.9 |
| rs12289502 | 11–80 | −3.80 | 0.00015 | 0.002 | 4.1 |
| rs9665943 | 11–82 | −3.76 | 0.00018 | 0.008 | 3.9 |
| rs7928811 | 11–83 | −3.59 | 0.00035 | 0.02 | 3.7 |
| rs905646 | 11–88 | −3.24 | 0.001 | 0.18 | 2.8 |
Chromosome-Mb
All regressions adjusted for genome-wide African ancestry, gender, age, and study site.
The absolute values for the ANCESTRYMAP association scores are displayed in Figure 2. The 4 SNPs with the strongest ancestry association (LOD ~4) were all between 76 and 83 Mb on chromosome 11 (Table 2). Figure 3 shows the distribution of LOD scores along chromosome 11. The 95% confidence interval calculated for association with SNPs on chromosome 11q is estimated at 79Mb to 104Mb, and includes rs9665943 (LOD=3.9). The highest LOD score found with the ANCESTRYMAP analysis was 4.1, at rs12289502. Associations were nearly identical when ANCESTRYMAP was re-run with differential risk parameters exchanged between the two phenotypic groups.
Figure 2.
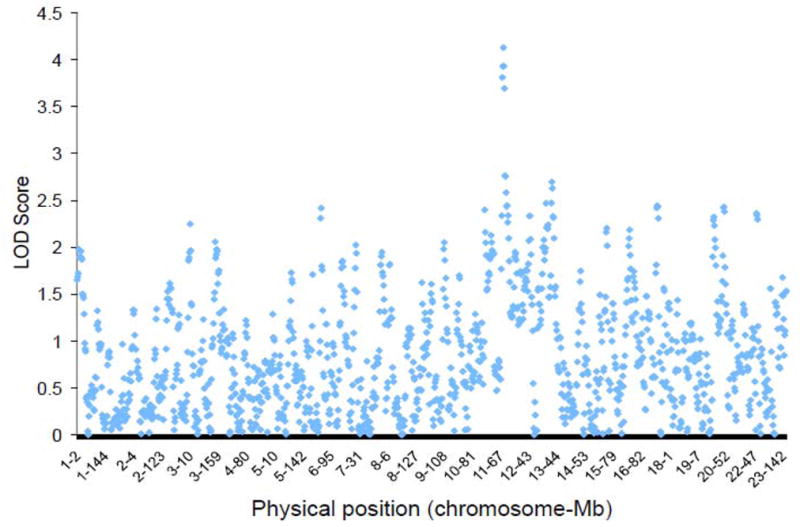
LOD scores of ANCESTRYMAP analyses describing associations between SNP’s and dichotomized AAI phenotype. Each point represents a singe SNP. LOD score of 3.2 is estimate of minimum threshold for suggested association in this analysis.
Figure 3.
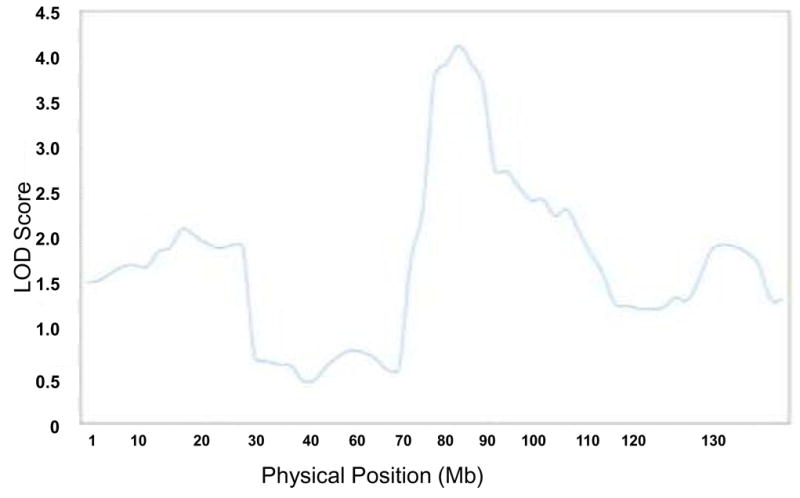
Detailed admixture map of chromosome 11
Logistic regression in Health ABC-AA and CHS-AA
To confirm the association of genotype at rs9665943 with low AAI, we performed multivariate-adjusted logistic regression of genotype at rs9665943 on low AAI in Health ABC-AA and CHS-AA. All regressions were run for each individual study population separately and in a single model combining the data from both studies. Identical models were run for genotype at rs1042602, a SNP in the same region, as well. In all models, homozygous African genotype at both rs9665943 and rs1042602 was significantly associated with low AAI when compared to a reference group of subjects who were carriers of at least one European allele (Table 3). Excluding varied strata of high AAI participants from these models did not attenuate either of the SNP associations’ significance. We also tested our significant SNPs for association with coronary heart disease and cerebrovascular disease, and found no association with either (data not shown).
Table 3.
Odds ratios, 95% confidence intervals coefficients for effects of homozygous African genotype at rs9665943 and rs1042602 for odds of AAI<0.9*
| SNP | Population | OR (95% CI) | p-value |
|---|---|---|---|
| rs9665943 | Health ABC-AA | 1.53 (1.16–2.02) | 0.009 |
| CHS-AA | 1.59 (1.12–2.27) | 0.003 | |
| Combined | 1.56 (1.26–1.93) | <0.001 | |
| rs1042602 | Health ABC-AA | 1.72 (1.08–2.73) | 0.022 |
| CHS-AA | 2.27 (1.13–4.54) | 0.02 | |
| Combined | 1.89 (1.29–2.76) | 0.001 |
Adjusted for gender, age, study site (and study in combined models), total serum cholesterol, HDL cholesterol, weight, smoking status, diabetes status, hypertension status, prevalent cardiovascular disease, and estimated percentage of African ancestry. Genotype AA at rs9665943 represents African homozygote, GG European homozygote. Genotype CC at rs1042602 represents African homozygote, AA European homozygote.
Reference group for calculating odds ratios was subjects with at least one European allele.
In order to assess the degree of linkage disequilibrium in this region, we calculated the correlation coefficients between rs1042602 and the six SNPs listed in Table 2. All of the coefficients were greater than 0.8, indicating a high degree of linkage disequilibrium in this region.
DISCUSSION
Using admixture mapping, we have identified an area of chromosome 11 reaching its peak between 80 and 82 Mb that is potentially associated with a low AAI in African Americans. Although genotype at none of the SNPs in this region met criteria for genome-wide statistical significance following adjustment for multiple testing and genome-wide European ancestry, genotype at rs9665943 had borderline statistical significance. Even though none of the SNPs in this region met the pre-specified threshold for genome-wide significance (LOD of 5) in the ANCESTRYMAP analysis, the fact that several SNPs in this region nearly met criteria for statistical significance across the three different admixture-mapping platforms and showed a strong association with AAI in logistic regression in two different populations suggest that this area might hold a candidate gene associated with PAD and is a promising region for future fine mapping.
Rs9665943 is part of hypothetical protein MGC33846, which has no known function. Another candidate SNP in this area, rs1042602, is a nonsynonymous polymorphism within the coding region of the tyrosinase gene. Tyrosinase is a copper-containing protein that catalyzes several reactions necessary for melanin biosynthesis.[35] Mutations in the gene which lead to deficiencies in the activity of tyrosinase are associated with occulocutaneous albinism, Type IA, although there are no published accounts of patients with OCA1 having increased incidence of PAD or any other vascular disease.[36,37] A search of NCBI Genome View, build 36.2 revealed several potential candidate genes in this region including thyroid hormone responsive gene, which encodes a protein involved in lipogenesis,[38] and angiotensinase C, which encodes an enzyme that regulates both the renin-angiotensin system and the kallikrein-kinin system and has been implicated in preeclampsia in hypertensive women.[39] Angiotensinase C is only 100Kb downstream from rs9665943.
Although we only report results of regression analysis for rs9665943 and rs1042602, there were several other SNPs in the admixture map in the same region of chromosome 11 which showed a potential association with AAI. Because the primary goal of this study was to find a region of the genome for future fine mapping for potential candidate gene discovery, we chose to focus on these two particular SNPs as representative of the region rather than report the outcomes of extensive analysis on all of the SNPs in the region. The results of similar regression analysis for rs12289502, a SNP 2Mb away from rs9665943 yielded similar results (data available upon request).
There are two published reports of whole genome linkage studies for PAD.[7,12] Neither study identified the region on chromosome 11, and only one study included African-Americans in the study population. However, linkage and admixture mapping would not necessarily be expected to map the same loci. Linkage analysis depends on the contribution of a locus to heritability of the trait and is independent of the allele frequency distribution at the locus while admixture mapping examines how loci contribute to a trait. Thus, the power of admixture mapping is dependent upon the allele frequency difference between ancestral populations at a particular locus.
A potential limitation to our study was our use of AAI as a continuous outcome in our ADMIXMAP and STRUCTURE analyses even though prior work has shown that the relationship between AAI and mortality is, in fact, non-linear[16, 40]. However, this locus remained associated with low AAI when AAI was analyzed as a dichotomous trait.
A challenge in the use of AAI as a phenotype is the treatment of subjects with high AAI, which might reflect some degree of arterial calcification, a different etiology than the plaques and stenoses associated with low AAI,[16] Although one study has suggested that subjects with AAI>1.4 have similar risk factor and cardiovascular outcome profiles to subjects with low AAI and increased mortality rates [7,16], the cut-off for the upper limit of what constitutes a normal AAI is not known. Because the pathophysiology associated with having an abnormally high AAI differs from that associated with having a low AAI (PAD), subjects with AAI between 1.4 and 1.5 were not excluded from the admixture mapping. When we excluded these subjects from the regression analysis, the associated between the locus on chromosome 11 and AAI was still present. While more work must be done to clarify what constitutes an abnormally high AAI, we do not believe that our analysis was confounded by our inclusion of subjects with AAI between 1.4 and 1.5 in the admixture mapping. When we excluded these subjects, the results we obtained were similar to the results reported (data available upon request.) Studies with larger populations or studies focused on arterial calcification may help to determine if the genes associated with PAD and high AAI are the same as those for PAD with low AAI. Further, we were unable to exclude subjects with “incompressible” arteries, so this may represent an additional source of confounding.
Another issue is the potential confounding of our association by socioeconomic status, given the complex nature of the relationship between genetics, population structure, and demographics.[32] In order to address this, we performed a post-hoc analysis adjusting for education status and estimated family income, commonly-used indicators of socioeconomic status, and the significance of the AAI-genotype relationship was not mitigated (data available upon request.) In addition, while socioeconomic factors and access to appropriate care and prevention could confound the association between overall individual ancestry and PAD or AAI, it should not affect the association between any locus and PAD or AAI. Thus, locus specific associations with PAD or AAI that are significant after adjustment for ancestry are unlikely to be confounded by non-genetic factors. Finally, the power of admixture mapping is limited by the allele frequency of causative marker. We can only detect causative loci which have high ancestral allele frequency differences between African and European populations.
In summary, we have used an admixture mapping approach to identify a genetic locus on chromosome 11 associated with AAI. Future work will focus on fine mapping of this region to identify the gene and genetic variants underlying this association.
Acknowledgments
The authors wish to acknowledge Drs. Nick Patterson and David Reich for their role in developing the set of admixture markers used in this article. The research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant number U01 HL080295 from the National Heart, Lung, and Blood Institute, as well as the Intramural Research Program of the National Institute on Aging, contracts N01-AG-6-2101, N01-AG-6-2103, and N01-AG-6-2106 with additional contribution from the National Institute of Neurological Disorders and Stroke. DR was supported by a Burroughs Wellcome Career Development Award in the Biomedical Sciences. EZ’s effort was supported by a career development award from the NCI (K22 CA109351), the dept of defense breast cancer research program (BC030551) and the NIH/NIA (U19 AG23122). A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm.
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd to permit this article (if accepted) to be published in Journal of Medical Genetics and any other BMJPGL products and sublicences such use and exploit all subsidiary rights, as set out in our licence (http://JMG.bmj.com/misc/ifora/licenceform.shtml).
References
- 1.Selvin E, Erlinger TP. Prevalence of and risk factors for peripheral arterial disease in the United States: results from the National Health and Nutrition Examination Survey, 1999–2000. Circulation. 2004;110:738–743. doi: 10.1161/01.CIR.0000137913.26087.F0. [DOI] [PubMed] [Google Scholar]
- 2.Newman AB, Sutton-Tyrrell K, Vogt MT, Kuller LH. Morbidity and mortality in hypertensive adults with a low ankle/arm blood pressure index. Jama. 1993;270:487–489. [PubMed] [Google Scholar]
- 3.McDermott MM, Fried L, Simonsick E, Ling S, Guralnik JM. Asymptomatic peripheral arterial disease is independently associated with impaired lower extremity functioning: the women’s health and aging study. Circulation. 2000;101:1007–1012. doi: 10.1161/01.cir.101.9.1007. [DOI] [PubMed] [Google Scholar]
- 4.Aboyans V, Criqui MH, McClelland RL, Allison MA, McDermott MM, Goff DC, Manolio TA. Intrinsic contribution of gender and ethnicity to normal ankle-brachial index values: the Multi-Ethnic Study of Atherosclerosis (MESA) J Vasc Surg. 2007;45:319–327. doi: 10.1016/j.jvs.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 5.Allison MA, Ho E, Denenberg JO, Langer RD, Newman AB, Fabsitz RR, Criqui MH. Ethnic-specific prevalence of peripheral arterial disease in the United States. Am J Prev Med. 2007;32:328–333. doi: 10.1016/j.amepre.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Carmelli D, Fabsitz RR, Swan GE, Reed T, Miller B, Wolf PA. Contribution of genetic and environmental influences to ankle-brachial blood pressure index in the NHLBI Twin Study. National Heart, Lung, and Blood Institute. Am J Epidemiol. 2000;151:452–458. doi: 10.1093/oxfordjournals.aje.a010230. [DOI] [PubMed] [Google Scholar]
- 7.Kullo IJ, Turner ST, Kardia SL, Mosley TH, Jr, Boerwinkle E, deAndrade M. A genome-wide linkage scan for ankle-brachial index in African American and non-Hispanic white subjects participating in the GENOA study. Atherosclerosis. 2006;187:433–438. doi: 10.1016/j.atherosclerosis.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Danielsson P, Truedsson L, Eriksson KF, Norgren L. Inflammatory markers and IL-6 polymorphism in peripheral arterial disease with and without diabetes mellitus. Vasc Med. 2005;10:191–198. doi: 10.1191/1358863x05vm617oa. [DOI] [PubMed] [Google Scholar]
- 9.Fowkes FG, Connor JM, Smith FB, Wood J, Donnan PT, Lowe GD. Fibrinogen genotype and risk of peripheral atherosclerosis. Lancet. 1992;339:693–696. doi: 10.1016/0140-6736(92)90596-u. [DOI] [PubMed] [Google Scholar]
- 10.Fowkes FG, Lee AJ, Hau CM, Cooke A, Connor JM, Lowe GD. Methylene tetrahydrofolate reductase (MTHFR) and nitric oxide synthase (ecNOS) genes and risks of peripheral arterial disease and coronary heart disease: Edinburgh Artery Study. Atherosclerosis. 2000;150:179–185. doi: 10.1016/s0021-9150(99)00366-4. [DOI] [PubMed] [Google Scholar]
- 11.Fontana P, Gaussem P, Aiach M, Fiessinger JN, Emmerich J, Reny JL. P2Y12 H2 haplotype is associated with peripheral arterial disease: a case-control study. Circulation. 2003;108:2971–2973. doi: 10.1161/01.CIR.0000106904.80795.35. [DOI] [PubMed] [Google Scholar]
- 12.Gudmundsson G, Matthiasson SE, Arason H, Johannsson H, Runarsson F, Bjarnason H, Helgadottir K, Thorisdottir S, Ingadottir G, Lindpaintner K, Sainz J, Gudnason V, Frigge ML, Kong A, Gulcher JR, Stefansson K. Localization of a gene for peripheral arterial occlusive disease to chromosome 1p31. Am J Hum Genet. 2002;70:586–592. doi: 10.1086/339251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowkes FG. The measurement of atherosclerotic peripheral arterial disease in epidemiological surveys. Int J Epidemiol. 1988;17:248–254. doi: 10.1093/ije/17.2.248. [DOI] [PubMed] [Google Scholar]
- 14.Newman AB, Shemanski L, Manolio TA, Cushman M, Mittelmark M, Polak JF, Powe NR, Siscovick D. Ankle-arm index as a predictor of cardiovascular disease and mortality in the Cardiovascular Health Study. The Cardiovascular Health Study Group. Arterioscler Thromb Vasc Biol. 1999;19:538–545. doi: 10.1161/01.atv.19.3.538. [DOI] [PubMed] [Google Scholar]
- 15.Newman AB, Siscovick DS, Manolio TA, Polak J, Fried LP, Borhani NO, Wolfson SK. Ankle-arm index as a marker of atherosclerosis in the Cardiovascular Health Study. Cardiovascular Heart Study (CHS) Collaborative Research Group. Circulation. 1993;88:837–845. doi: 10.1161/01.cir.88.3.837. [DOI] [PubMed] [Google Scholar]
- 16.O’Hare AM, Katz R, Shlipak MG, Cushman M, Newman AB. Mortality and cardiovascular risk across the ankle-arm index spectrum: results from the Cardiovascular Health Study. Circulation. 2006;113:388–393. doi: 10.1161/CIRCULATIONAHA.105.570903. [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci U S A. 1998;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKeigue PM. Mapping genes underlying ethnic differences in disease risk by linkage disequilibrium in recently admixed populations. Am J Hum Genet. 1997;60:188–196. [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens JC, Briscoe D, O’Brien SJ. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 24.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 25.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, Oakley-Girvan I, Whittemore AS, Cooney KA, Ingles SA, Altshuler D, Henderson BE, Reich D. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nalls MA, Wilson JG, Patterson NJ, Tandon A, Zmuda JM, Huntsman S, Garcia M, Hu D, Li R, Beamer BA, Patel KV, Akylbekova EL, Files JC, Hardy CL, Buxbaum SG, Taylor HA, Reich D, Harris TB, Ziv E. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wassel Fyr CL, Kanaya AM, Cummings SR, Reich D, Hsueh WC, Reiner AP, Harris TB, Moffett S, Li R, Ding J, Miljkovic-Gacic I, Ziv E. Genetic admixture, adipocytokines, and adiposity in Black Americans: the Health, Aging, and Body Composition study. Hum Genet. 2007;121:615–624. doi: 10.1007/s00439-007-0353-z. [DOI] [PubMed] [Google Scholar]
- 28.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 29.Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 30.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 31.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM, Kwok PY. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 34.Reich D, Patterson N. Will admixture mapping work to find disease genes? Philos Trans R Soc Lond B Biol Sci. 2005;360:1605–1607. doi: 10.1098/rstb.2005.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tripathi RK, Hearing VJ, Urabe K, Aroca P, Spritz RA. Mutational mapping of the catalytic activities of human tyrosinase. J Biol Chem. 1992;267:23707–23712. [PubMed] [Google Scholar]
- 36.Opitz S, Kasmann-Kellner B, Kaufmann M, Schwinger E, Zuhlke C. Detection of 53 novel DNA variations within the tyrosinase gene and accumulation of mutations in 17 patients with albinism. Hum Mutat. 2004;23:630–631. doi: 10.1002/humu.9248. [DOI] [PubMed] [Google Scholar]
- 37.King RA, Pietsch J, Fryer JP, Savage S, Brott MJ, Russell-Eggitt I, Summers CG, Oetting WS. Tyrosinase gene mutations in oculocutaneous albinism 1 (OCA1): definition of the phenotype. Hum Genet. 2003;113:502–513. doi: 10.1007/s00439-003-0998-1. [DOI] [PubMed] [Google Scholar]
- 38.Kinlaw WB, Church JL, Harmon J, Mariash CN. Direct evidence for a role of the “spot 14” protein in the regulation of lipid synthesis. J Biol Chem. 1995;270:16615–16618. doi: 10.1074/jbc.270.28.16615. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Feng Y, Zhang Y, Zhou H, Jiang S, Niu T, Wei LJ, Xu X, Xu X, Wang X. Prolylcarboxypeptidase gene, chronic hypertension, and risk of preeclampsia. Am J Obstet Gynecol. 2006;195:162–171. doi: 10.1016/j.ajog.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 40.Resnick HE, Lindsay RS, McDermott MM, Devereux RB, Jones KL, Fabsitz RR, Howard BV. Relationship of high and low ankle brachial index to all-cause and cardiovascular disease mortality: the Strong Heart Study. Circulation. 2004;109:733–739. doi: 10.1161/01.CIR.0000112642.63927.54. [DOI] [PubMed] [Google Scholar]


