Abstract
An important aspect of catalysis by cholesterol oxidase (3β-hydroxysteroid oxidase) is the nature of its association with the lipid bilayer that contains the sterol substrate. Efficient catalytic turnover is affected by the association of the protein with the membrane as well as the solubility of the substrate in the lipid bilayer. In this review, the binding of cholesterol oxidase to the lipid bilayer, its turnover of substrates presented in different physical environments, and how these conditions affect substrate specificity are discussed. The physiological functions of the enzyme in bacterial metabolism, pathogenesis, and macrolide biosynthesis are reviewed in this context.
Keywords: Cholesterol oxidase, interfacial enzyme, lipid phase, bilayer, membrane, sterol, catabolism, GMC oxidoreductase, virulence, macrolide biosynthesis
3β-Hydroxysteroid:oxygen oxidoreductase (EC 1.1.3.6), commonly known as cholesterol oxidase (ChOx), is a flavoenzyme that catalyzes the oxidation and isomerization of cholesterol to cholest-4-en-3-one and has been well characterized structurally and chemically (see minireview 1). The enzyme is extracellular and occurs in a secreted form and/or cell surface-associated form depending on the producer microorganism and growth conditions. They are both products of the same gene. The secreted form is a soluble, globular protein, and the X-ray crystal structures [1–3] revealed that it is essentially two fused domains: the flavin-binding domain and the substrate-binding domain. An important aspect of catalysis by this enzyme is the nature of its association with the lipid bilayer that contains the sterol substrate. Efficient catalytic turnover is affected by the association of the protein with the membrane as well as the solubility of the substrate in the lipid bilayer. In this review, we discuss the binding of ChOx to the lipid bilayer, its turnover of substrates presented in different physical environments, and how these conditions affect substrate specificity. Defining substrate specificity with respect to these parameters is important for understanding the physiological functions of the enzyme in bacterial metabolism and perhaps pathogenesis. We will begin with an overview of interfacial membrane kinetics and how they pertain to the reaction catalyzed by ChOx and to understanding the implications for investigating substrate specificity.
Interfacial kinetics
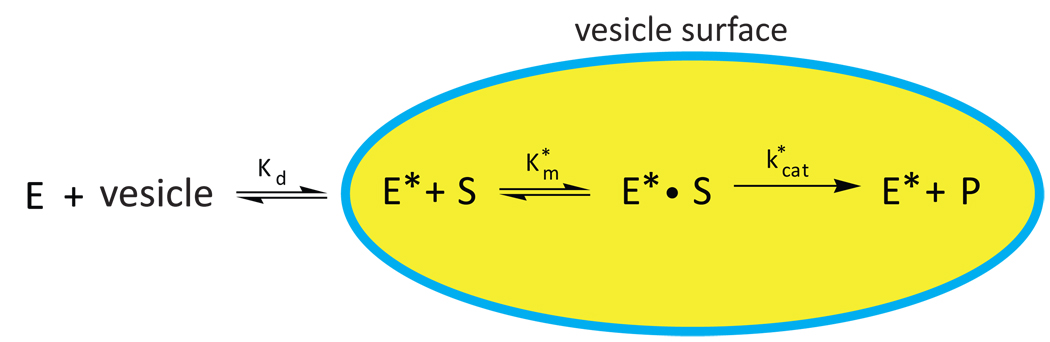
An interfacial enzyme is a protein that binds transiently to the membrane surface during catalysis. Perhaps, the best-studied family of interfacial enzymes is the phospholipase A2 family and extensive development of the kinetic model derives from its study [4]. The two key components of the interfacial kinetic model are that the enzyme freely dissociates from the membrane surface, and that the important measure of substrate quantity is its mole fraction in the membrane, not bulk solution concentration (Scheme 1). The initial velocity dependence on mole fraction of substrate may be determined analogous to what is done with soluble substrates as long as the fraction of enzyme bound to the surface is known. Typically, the initial rate dependence on mole fraction is hyperbolic and is fit to the Michaelis-Menten equation. Km* is the mole fraction of substrate at which the initial velocity is half-maximal and kcat* is the first order rate constant for turnover when all enzyme molecules are bound to the membrane and saturated with substrate. The paradigm for understanding phospholipase interfacial kinetics is applicable to ChOxs. Although cholesterol desorbs from membranes much faster than phospholipids, the rate of cholesterol desorption is about 5 orders of magnitude slower than the catalytic turnover rate of ChOx measured with a variety of membranes. Therefore, the enzyme does not capture free cholesterol from aqueous solution. Rather, ChOx associates with lipid bilayers, and binds cholesterol from the membrane.
Scheme 1.
Model for interfacial Michaelis-Menten kinetics at a membrane surface. E, free enzyme; E*, membrane-bound enzyme; S, substrate in the membrane; P, product in the membrane; kcat*, interfacial first-order rate constant in min−1; Km*, interfacial Michaelis constant in units of mole fraction.
Cholesterol mixed into membranes does not behave as an ideal solute. In the ideal case, as more cholesterol is added to a single phospholipid component membrane (binary mixture), the chemical activity of the cholesterol increases in proportion to mole fraction. However, the activity can change non-linearly depending on the structure of the lipids mixed with cholesterol and the liquid phase present. In other words, the chemical activity of the cholesterol substrate is not only dependent on its mole fraction; it also depends on the probability that cholesterol will leave the membrane [5].
The rate at which cholesterol is desorbed from the membrane is affected as intermolecular packing changes within the membrane. The liquid membrane is essentially a solvent for the substrate and changing the nature of the solvent, for example, the structures of the lipid acyl chains, results in a change in desorption rate. The molecular interactions between lipids and cholesterol determine the free energy of the cholesterol in the lipid. Consequently, the equilibrium between substrate binding to the enzyme and substrate residing in the membrane changes as the structure of the lipid membrane changes. Lipids with saturated acyl chains have a greater affinity for cholesterol than lipids with unsaturated acyl chains. Head groups affect the affinity of the membrane for cholesterol in the order sphingomyelin > phosphatidylserine > phosphatidylcholine > phosphatidylethanolamine (reviewed in [6]). Thus, the enzyme’s apparent specificity for different sterols can depend on the molecular interactions between lipids and substrates as much as it depends on interactions between substrate and enzymes. This relationship was recognized early and forms the basis of using ChOx to probe the physiological partitioning of cholesterol in biological membranes [7–13]. Moreover, ChOx has insecticidal properties against Coeloptera larvae, agricultural pests (covered in minireview 3), and was being developed for use in agricultural crop treatments [14]. The efficacy of the treatment depends on the relative specificity of the enzyme for pest membranes versus plant membranes [15].
Membrane effects on cholesterol oxidase activity
Generally, lipid bilayers exist in a gel phase or a liquid phase in the absence of cholesterol. Lipids above their melting temperature have greater lateral mobility within the lipid bilayer and hence behave more like liquids than solids. In membranes composed of lipids with saturated acyl chains, e.g., dipalmitoylphosphatidylcholine, introduction of cholesterol into the liquid phase results in an increase in membrane order to form a phase that is still liquid, that is, the lipids still have lateral mobility, but there is a higher degree of order. The cholesterol constrains the saturated lipid acyl chains to an S-trans conformation that limits disorder in the center of the lipid bilayer. This liquid-ordered state separates from the cholesterol-free liquid-disordered state, and the two phases can coexist [16–18]. In addition, due to multibody molecular interactions that occur at critical cholesterol mole fractions, ChOx activity may be decreased or increased non-proportionately with mole fraction within a single-phase region [6, 19–21]. Thus, increasing the mole fraction of cholesterol can perturb the physical state of the membrane and alter the affinity of the membrane for cholesterol. Therefore, the enzyme’s apparent substrate specificity will depend on the lipid composition and substrate mole fraction at which the enzyme activity is determined.
Further complicating the determination of ChOx substrate specificity, many studies are performed in detergent micelles [22, 23]. Early studies demonstrated that the rate of cholesterol oxidation was highest with nonionic detergent micelles, e.g., Triton X-100 (polyethylene glycol octylphenyl ether) or Thesit (polyethylene glycol monododecyl ether), containing cholesterol [24, 25]. The elements of the kinetic model are the same with detergent micelles. The enzyme must associate with the surface and the mole fraction of cholesterol is the important element. The free energy of interaction of cholesterol with the detergent can have a large effect on the apparent catalytic activity of the enzyme. For example, no turnover is detected with cetyltrimethylammonium bromide/cholesterol micelles, whereas at the same concentration, pH, and temperature, Triton X-100/cholesterol micelles are oxidized readily [24]. In addition, the Km values widely reported using Triton X-100/cholesterol micelles are actually apparent Km values that include a term for binding to the micellar surface. The consequence is that many changes to active site binding have no apparent effect on the Km [26–28] because this kinetic term is dominated by the micelle binding.
The corollary is that comparing different substrates in detergent micelles is only useful for understanding substrate specificity in the presence of detergent. The free energy of association between substrates and detergent can dictate the apparent preference of the enzyme for different steroids. For example, ChOx is 2–4-fold more specific for cholesterol over sitosterol and stigmasterol in detergent micelles, but shows equal specificity for all three sterols in dioleoylphosphatidylcholine/sterol vesicles [29]. For biotechnology applications like measuring concentrations of sterols (see review 3 of this miniseries), the use of detergent micelles makes sense. However, for understanding physiological function or the potential role of ChOx in pathogenesis, the specificity must be studied in the context of lipid membranes.
Cholesterol oxidase-membrane interactions
The association mechanism of interfacial catalysis (Scheme 1) requires that the substrate-binding site is oriented toward the lipid bilayer and is in contact with it. X-ray crystal structures [2, 3, 30] of the soluble enzyme in the absence of lipid identified the face that must be oriented towards the membrane containing the substrate. The Streptomyces and Rhodococcus equi (misclassified as Brevibacterium sterolicum) enzymes have been the focus of these studies because they are amenable to expression, mutagenesis, and high-resolution crystallography. Their mechanism and structures are nearly identical [31]. On this face, the substrate-binding cavity is protected from solvent by a protein loop comprising residues 79–83.
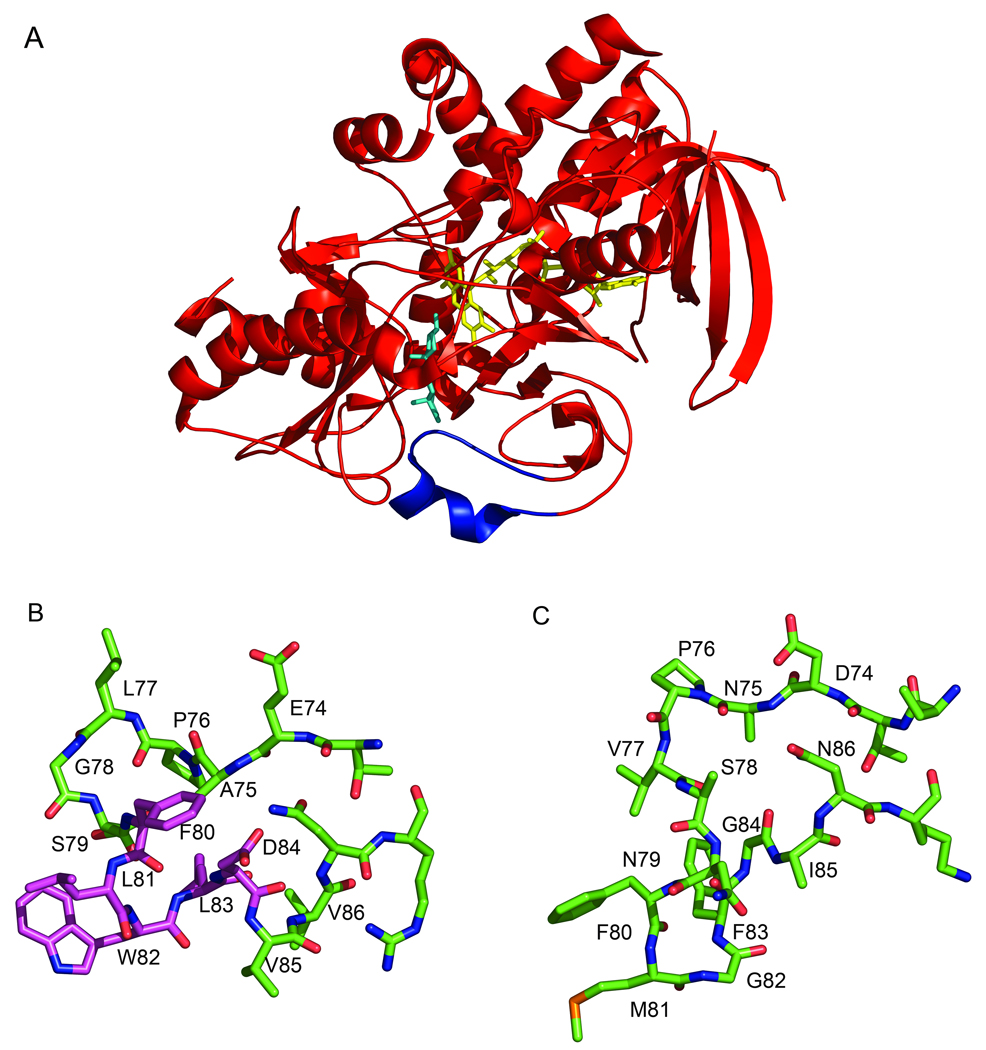
The role of this surface loop was investigated by deletion of the 5 amino acid residues, Ser 79, Phe 80, Leu 81, Trp 82 and Leu 83 at the tip of the loop [32] (Fig. 1). The Kd for binding to phosphatidylcholine-cholesterol vesicles was not affected in the mutant. However, k*cat/Km* was reduced nearly 3000-fold with 1:1 phosphatidylcholine-cholesterol vesicles as substrate. These experiments were interpreted to suggest that the loop is required for cholesterol to bind to the enzyme, but not for binding to the membrane. The loop is amphipathic and the four tip residues deleted are hydrophobic groups that must pack with the eight carbons of the cholesterol side chain in the loop open form (Fig. 1). In the Streptomyces enzyme, the region comprising residues 78–87 adopts a small amphipathic helical turn with hydrophobic residues directed toward the active site cavity and hydrophilic residues directed toward the external surface of the molecule [33]. In this conformation, the active site is covered and thus aggregation of the protein at its active site is prevented. Upon substrate binding, hydrophobic interactions between the hydrophobic residues and cholesterol minimize energy loss.
Fig. 1.
Three-dimensional structure of ChOx illuminating the active site loop. (a) Ribbon cartoon of Streptomyces ChOx (1MXT [3]) with epiandrosterone modeled into the active site (cyan). The FAD cofactor is shown in yellow. The active site loop that must move to allow substrate binding is shown in blue. (b) Stick atomic representation of Streptomyces ChOx active site loop from (a) in the same orientation. (c) Stick atomic representation of active site loop from Rhodococcus equi (formerly B. sterolicum) ChOx (1COY) [2]. The entire Rhodococcous ChOx structure was overlaid with the Streptomyces ChOx structure and the loops in (b) and (c) are depicted in the same enzyme orientations. Side-chains for which there is no electron density were modeled as alanines. The residues that were deleted in [32] are shown with a magenta carbon backbone. This figure was made with Pymol [103].
From inspection of the X-ray crystal structures, conformational changes must accompany substrate binding. The position of the active site loop upon binding to model membranes (lipid vesicles) was determined using fluorescence quenching [34]. In this study, a cysteine was introduced into the loop at position 81 of ChOx from R. equi (Brevibacterium sterolicum) and labeled with acrylodan, an environmentally sensitive fluorescence probe. Modeling the acrylodan-labeled cysteine as an extended chain of the loop revealed that the backbone of this loop does not penetrate into the lipid bilayer but interacts with the head groups of the lipid bilayer. This experiment suggests that the enzyme sits on the membrane surface. Slotte has also demonstrated that tetramethylrhodamine-labeled ChOx associates with cholesterol/dimyristoylphosphatidylcholine monolayers [35]. This surface binding is consistent with vesicle lysis studies that demonstrated binding of the enzyme to membrane surfaces does not disrupt the membrane [36]. ChOx’s association with lipid bilayers together with partitioning of cholesterol into its active site does not alter the bilayer sufficiently to allow pore formation and consequent leakage of dye encapsulated in vesicles. However, conversion of cholesterol to cholest-4-en-3-one does increase membrane permeability by expansion (actually decondensation) of the lipid bilayer.
Using the fluorescently-labeled enzyme, binding to a variety of lipid vesicle types was monitored. Enzyme binding to the membrane is insensitive to charge and appears to be driven by hydrophobic interactions [34]. Compared to phospholipase A2 binding to anionic membranes, the binding affinity of ChOx is very weak which means that many studies have been performed without saturating the membrane surface with enzyme and differences between substrate preparations may simply reflect differences in binding affinity.
Enzyme substrate specificity
After enzyme binding to the membrane surface, sterol must bind in the enzyme active site for catalysis to occur. Studies of different sterols in lipid bilayer environments are few. An absolute requirement for activity is the presence of the 3β-hydroxy group on the steroid framework. Modification of the cholesterol side chain ranging from no side chain, 5-androstene-3β-ol, to a branched side chain, sitosterol, has little effect on substrate specificity when the steroids are monolayers [37] or in dioleoylphosphatidylcholine/sterol unilamellar vesicles [29]. This lack of specificity is in distinct contrast to substrate specificity studies in detergent micelles or with propan-2-ol co-solvent that demonstrate a specificity for cholesterol over sitosterol, androsten-3β-ol and related steroid structures (see review 3 of this miniseries) [38, 39].
Two types of model membrane have been used to look at ChOx specificity for cholesterol in different membranes: unilamellar vesicles and monolayers. The specificities observed follow the same trend. In both cases, the activities of the enzyme correlate with the chemical activity of cholesterol in the lipid membrane. The more favorable the packing interactions between cholesterol and lipid, the lower the catalytic activity of the enzyme. For example, the kcat*/Km* with cholesterol mixed with dioleoylphosphatidylcholine is two-fold higher than the kcat*/Km* with cholesterol mixed with dipalmitoylphosphatidylcholine in similar mole fraction regimes [40]. When the dipalmitoylphosphatidylcholine is substituted with sphingomyelin, the decrease in kcat*/Km* is 40-fold [41, 42]. In addition, it is not possible to saturate the enzyme with cholesterol. That is, the initial velocity dependence on cholesterol mole fraction is linear throughout all experimentally achievable mole fractions [40]. Thus, the true maximal rate for cholesterol in membranes has never been measured. Because microbial ChOxs are active with various natural sterols, it is necessary to test the mole fraction dependence with a more extensive variety of membranes composed of a variety of sterols to determine which substrate is the best.
Moreover, the kcat*/Km* varies depending on the multibody interactions in the membrane. When the mole fraction of cholesterol exceeds a sustainable packing ratio [6, 19–21], the excess cholesterol is a better substrate [7, 25, 40, 43]. Restated, cholesterol can exist in membranes as free cholesterol clusters. These clusters appear above cholesterol/phospholipid ratios that depend on the precise lipid and method of preparing the membranes. However, they are typically detected above stoichiometries of 1/2 or 1/1 [6]. All of the results in these studies are consistent with a catalytic model in which ChOx sits on the surface of the membrane and binds sterol by passive partitioning from the membrane into the active site.
Thus, the question arises, what is the relevant cholesterol matrix mixture to study? If the desired use of ChOx is a technical application, e.g., serum cholesterol assays in the clinic, then micelles or other non-native mixtures of cholesterol are the best form of substrate to use in order to obtain maximal activity. In contrast, if the aim is to understand physiological function, then cholesterol must be assayed with lipid mixtures that reflect the native environment of the enzyme. There are three major physiological functions of ChOxs that have been studied to varying degrees. The first function is in a catabolic pathway for nutrition, the second function is a proposed role in virulence. More recently, ChOx has been implicated as a biosensor for macrolide biosynthesis.
Cholesterol as a nutritional source
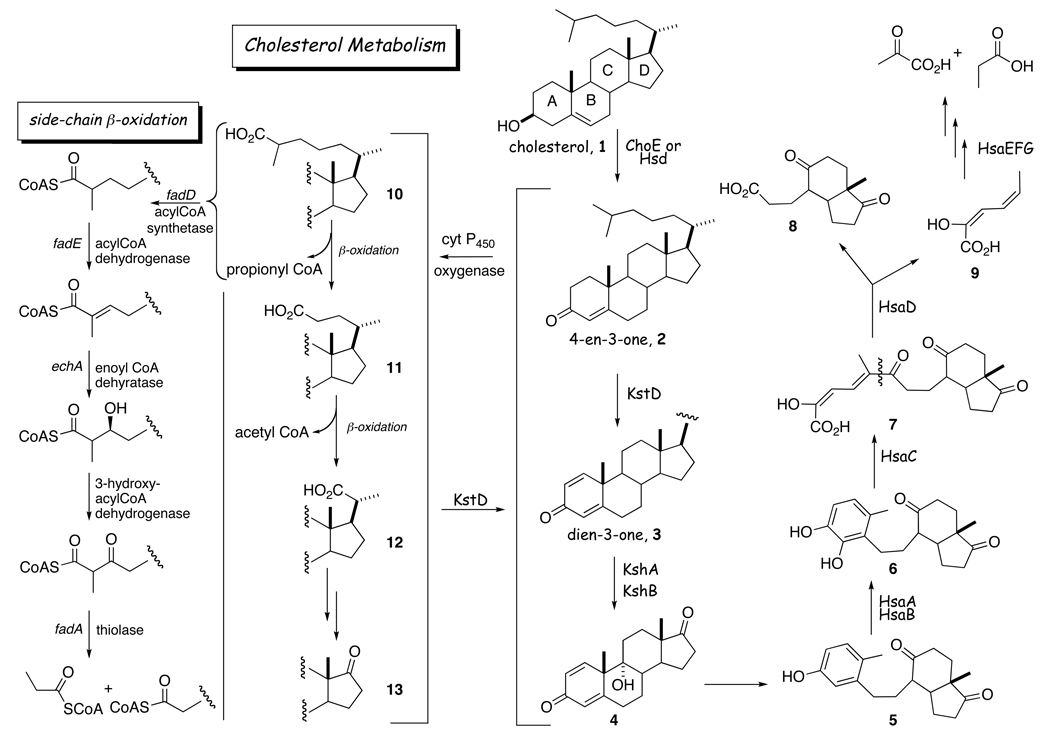
Conversion of a 3β-hydroxy-5-ene steroid to the corresponding 4-en-3-one product is the first and compulsory step in bacterial sterol catabolic pathways. Following this step, sterol-catabolizing microorganisms proceed to degrade the steroid nucleus and the sterol side chain simultaneously, but independently, at different rates. Some species cleave the side chain before C-1(2) dehydrogenation and/or 9α-hydroxylation of the steroid skeleton (Scheme 2, reviewed in [44]). Moreover, an enzyme requiring O2 (ChOx) is always involved in the sterol 3β-hydroxy-5-ene conversion by actinomycete genera, Corynebacterium, Gordona, Proactinomyces and Rhodococcus; this conversion is carried out by a dehydrogenase/isomerase enzyme that utilizes NAD+ or NADP+ in a Pseudomonas sp. [45], Comomonas testosteroni (formerly Pseudomonas testosteroni [46], Nocardia [47, 48] and proteobacteria [49, 50]. In mycobacteria, the existing evidence suggests that this step is catalyzed by a dehydrogenase [51], although an oxidase has been suggested to perform this function [52] (vide infra). Species of the genera Rhodococcus, Mycobacterium and Gordona are widespread in nature, where they play major roles in the degradation of organic wastes that include sterols, and thus have evolved the ability to use them as sources of carbon and energy. Thus, in the ChOx-producing species one role of this enzyme is nutritional. The precise matrix in which the substrate is presented to the enzyme is unclear. In nature, phytosterols are common in wood pulp waste streams, and thus are likely to be found in membranous form. Similarly, cholesterol, an animal product, is presumably presented as decaying membranous material to soil bacteria.
Scheme 2.
Canonical cholesterol catabolism pathway [44] based on studies in Rhodococcus [100], C. testosteroni [46, 101] and fast-growing mycobacteria [102]. It is believed that the cholesterol derivatives 2 and 3 are both substrates for the C26 hydroxylation enzyme. The cholesterol side chain β-oxidation intermediates are potential substrates for KstD and KshA/B. The preferred substrates have not been established.
Rhodococcal ChOx is an induced enzyme; its biosynthesis requires the presence of cholesterol or plant sterols (a detailed description of ChOx production level in different strains is reported in review 3). ChOx induction in Rhodococcus sp. GK1 is independent of the steroid 3β-hydroxy-5-ene, since cholest-4-en-3-one was demonstrated to be inducer [53, 54]. Moreover, androstenedione or testosterone, intermediates in cholesterol catabolism (Scheme 2), completely repressed ChOx synthesis by this strain. Thus, enzyme induction is dependent on the presence of the sterol side chain. This regulation is consistent with the preferred substrates being cholesterol and phytosterols, e.g., sitosterol and stigmasterol.
The exact taxonomy of some species of genera recognized to catabolize cholesterol has changed over time. For example, Nocardia restrictus ATCC 14887, a strain extensively used by Sih et al. [55, 56] in their studies of cholesterol catabolism, is now recognized as R. equi ATCC 14887 [57]. A second example is the ChOx producing B. sterolicum ATCC 21387 that is actually an R. equi strain [58]. The original term, nocardioform bacteria, encompasses the genera Corynebacterium, Gordona (or Gordonia), Mycobacterium, Nocardia, Rhodococcus and Tsukamurella. Bacteria belonging to these genera all contain mycolic acid in their capsule and they have been known as mycolate-containing nocardioform actinomycetes. The term, nocardioform describes the morphology and refers to mycelial growth with fragmentation into rod-shaped and/or coccoid elements. These actinomycetes form a distinct suprageneric group; they are ubiquitous in nature and have the ability to catabolize different natural substances including cholesterol and plant sterols. The group encompasses pathogenic species that are generally opportunistic. New species are still being found, for example, a Gordona actinomycete was recently isolated from sewage sludge and found to catabolize cholesterol [59].
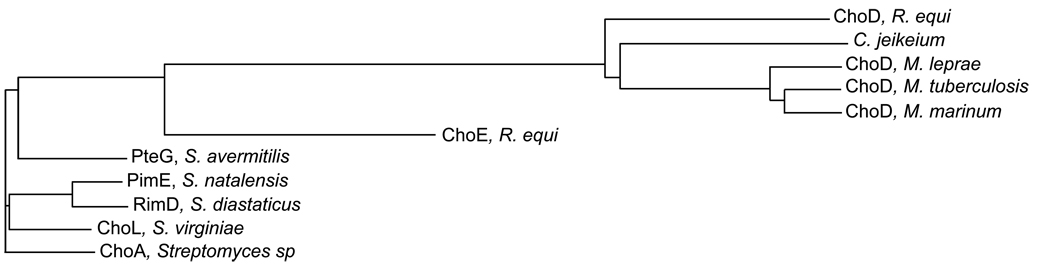
Recent interest has focused on cholesterol oxidation in mycobacteria. There are many reports that mycobacteria oxidize sterols [60–65]. However, there is no definitive evidence that mycobacteria produce a ChOx. The taxonomy of Mycobacterium sp. used in the studies of Stadtman and collaborators [66, 67] has been revised, first to Nocardia cholesterolicum and finally to R. rhodochrous [68, 69]. The enzyme isolated by Stadtman et al. [66] was a ChOx [68]. Sequencing of whole genomes has allowed a bioinformatics approach to gene identification. No orthologs of the biochemically-verified streptomycete (choA) and rhodococcal (choE) ChOx genes are present. However, a putative ChOx was identified in the mycobacterial genomes and annotated as choD. The choD gene is also present in the R. equi genome [70]. Phylogenetic analysis of streptomycete, rhodococcal, and mycobacterial annotated ChOxs reveals that R. equi ChoE is 56% identical to the ChOxs from Streptomyces species (Fig. 2 and [71]). The R. equi and Streptomyces proteins have signal peptide sequences and their corresponding ChOxs are, in most cases, extracellular. Although Rhodococcus is genetically more closely related to Mycobacterium than to Streptomyces, the identity of ChoE with the putative cholesterol oxidase ChoD of M. tuberculosis or M. leprae is low, around 25%. In addition, ChoD lacks a signal peptide sequence suggesting it is localized inside the bacterium. Importantly, M. tuberculosis has a 3β-hydroxysteroid dehydrogenase (Rv1106c, hsd) that has been expressed and purified. This enzyme converts NAD+ and cholesterol to NADH and cholest-4-en-3-one [51]. Disruption of the hsd gene in M. tuberculosis abrogates cholesterol conversion to cholest-4-en-3-one as determined by HPLC analysis. There is a report that expression of ChoD in M. smegmatis lysates increases cholesterol oxidation activity [52]. However, this activity was measured colorimetrically and conversion of cholesterol to cholest-4-en-3-one was not verified. Taking into consideration these observations, we support the opinion of Navas and collaborators [71] who suggested that ChoDs may be proteins without ChOx activity, but belonging to the GMC oxidoreductase group.
Fig. 2.
Unrooted phylogenetic tree for functionally characterized and putative cholesterol oxidase protein sequences from Streptomyces, Rhodococcus and Mycobacterium. The length of the horizontal lines corresponds to relative evolutionary distance. The tree was generated using a ClustalW2 alignment [104] with the neighbor-joining method [105]. Proteins are identified by Genbank id and gene id if assigned: ORF1948, R. equi ChoD; CAC44897, R. equi ChoE; CAI36788, Corynebacterium jeikeium; CAR70482, M. leprae ChoD; CAB01014, M. tuberculosis H37Rv ChoD; ACC39597, M. marinum ChoD; CAC20926, S. natalensis PimE; AAR16516, S. diastaticus RimD; ABS32193, S. virginiae ChoL; BAB69314, S. avermitilis PteG; AAA26719, Streptomyces sp. ChoA.
Cholesterol oxidase and virulence
Related to the high activity of ChOx with membranes containing clusters of free cholesterol, treatment of cell membranes with sphingomyelinase before ChOx addition results in higher activity of the enzyme [72]. Most commonly, this activity is reported as hemolysis because red blood cells are used as the source of cell membranes. This effect is seen both upon addition of purified enzymes, or upon addition of bacterial strains that secrete sphingomyelin-specific phospholipase D and ChOx. The hemolysis activity of R. equi ChOx was confirmed through molecular genetic experiments [71]. ChoE-negative R. equi mutants lose cooperative hemolysis (CAMP reaction) that occurs with sphingomyelinase-producing Listeria ivanovii. The CAMP-reaction was also observed for L. monocytogenes and R. equi [73]. In other cases, the hydrolases may be secreted by the same strain, e.g., choline phosphohydrolase and sphingomyelinase C are produced by R. equi that also produces ChOx [74]. These observations are consistent with a model in which ceramide formation displaces cholesterol from liquid-ordered regions [75] making the cholesterol more accessible to ChOx.
These studies defined the parameters required for cell lysis and suggested a possible role that ChOx may play in pathogenesis. All the ChOxs isolated to date are extracellular, either secreted or cell associated. Rhodococcus equi is primarily a horse pathogen. However, it is an emerging, opportunistic human infection, especially in immune compromised individuals, for example, those infected with HIV [76]. These bacteria infect and multiply inside the macrophage, a potentially rich source of cholesterol.
An in vitro study suggested that during bacterial invasion of the host cell, membrane lysis is facilitated by the induction of extracellular ChOx [77]. Oxidation of macrophage membrane cholesterol by R. equi (ATCC 33701) was studied under infection-mimicking conditions [78]. In this study, uptake of R. equi cells by cultured mouse macrophages (ATCC PD388D1) was accompanied by intracellular survival of the bacterium and enzymatic oxidation of macrophage cholesterol. Cholesterol oxidation was significantly increased when the strain was co-phagocytosed with Corynebacterium pseudotuberculosis, a sphingomyelinase-producing bacterium and cooperative partner of R. equi in in vitro hemolysis of sheep erythrocytes [72]. Synergistic actions of cytotoxic enzymes may also take place in vivo, since pathogens and/or ubiquitous commensal organisms can exist at the same time in infected hosts, especially in immunocompromised individuals. Moreover, intracellular survival of the bacterium in the host macrophage is enhanced by the induction of oxidative enzymes, catalase (EC 1.11.1.6) and superoxide dismutase (EC 1.15.1.1) [77]. Both enzymes reduce oxidizing agents, H2O2 and free radicals and, thus, contribute to pathogen protection from oxidative stress effects. These studies did not address whether it is the lytic or the nutritional function that contributes to bacterial survival in the macrophage.
A mutant of R. equi, originally isolated from foals with pneumonia, in which choE (ChOx) was disrupted was constructed by allelic exchange. This mutant was devoid of ChOx activity. The mutant was assessed for in vivo virulence in mice or foals and in vitro cytotoxicity to macrophages [79, 80]. The virulence of the mutant strain was not attenuated and mutation did not reduce cytotoxicity in infected macrophages. Based on the rates of multiplication of the mutant and the parent strain in the infected animals or in the macrophages, and their similar cytotoxicities to macrophages, it was concluded that ChOx is not a virulence factor and its role may be limited to the catabolism of cholesterol as a carbon and energy source of the infecting bacterium. Furthermore, a partial deletion mutant in the supAB genes, which encode for the permease subunits of the cholesterol uptake transporter (mce4) [81], was used to infect macrophages in vitro and the mutant was tested for growth on cholesterol as a sole carbon source [82]. Although disruption of the permease genes blocks cholesterol catabolism, cholesterol uptake and catabolism are not essential for survival of R. equi in the macrophage. We note that in Rhodococcus, ChOx is either secreted and/or cell-surface-linked (see for example, [54, 83–85]). Thus, cholesterol conversion into cholest-4-en-3-one occurs outside the bacterial cell and the supAB/mce4 transporter system in rhodococci probably transports cholest-4-en-3-one rather than cholesterol.
These studies were all performed for short time courses (2 to 4 weeks in foals) and it is possible that the cholesterol catabolism and/or cell lysis by ChOx may be required at advanced stages of infection. Consistent with a catabolic role, the orthologous cholesterol transporter in M. tuberculosis is required for bacterial persistence at the chronic stage of mouse lung infection, but not in the initial stages of infection [86]. In addition, the transporter is only required for growth within interferon-γ activated macrophages and its mutation has no effect on infection of resting macrophages. Both mycobacteria and rhodococci catabolize cholesterol and the pathways share many similarities [87–89]. However, in these two genera, the conversion of cholesterol to cholest-4-en-3-one is catalyzed by different enzymes, a dehydrogenase [51] and an oxidase, which are intracellular and extracellular (secreted and/or cell-surface bound), respectively. These differences may be a consequence of additional functions that are distinct in the two genera. The primary question at this point in time, is whether cholesterol oxidation plays only a nutritional role in pathogenesis, or has additional consequences in microbial infection. In the host, cholesterol mixed with phospholipid or sphingomyelin is the presumed form of the substrate in vivo and is the matrix that should be studied in assessments of cholesterol oxidation in pathogenesis.
Cholesterol oxidase and polyene macrolide biosynthesis
Streptomyces natalensis produces the polyene macrolide, pimaricin. This macrolide is used in the food industry to prevent mold contamination of cheese and non-sterile food, and also for treatment of keratitis. The mechanism of pimaricin antifungal activity relies on its interaction with sterols, primarily ergosterol, in the cell membrane of molds, thus causing alteration of the membranes and lysis of mold cells.
Aparicio and collaborators [90, 91] identified a gene cluster involved in pimaricin biosynthesis. In the center of this gene cluster is a gene, pimE, which encodes for a cholesterol oxidase. A transcriptional activator gene, pimR, is located at the 5’-end of the cluster. Disruption of pimR results in total abrogation of pimE transcription as well as a significant reduction in transcription of biosynthetic genes, thus blocking pimaricin production completely [92].
PimE shares high amino acid identity with other known ChOxs that are in the GMC oxidoreductase family, including the active site residues and the enzyme is a catalytically active ChOx [93]. The location of pimE in the middle of the pimaricin gene cluster is intriguing, since the biosynthesis of this macrolide does not require cholesterol oxidation. Moreover, the pimE gene is required for production of pimaricin by S. natalensis [93]. Complementation of the ΔpimE mutant restores macrolide production. Unexpectedly, when purified enzyme is added to the growth media of the ΔpimE mutant, pimaricin production is recovered. ChOxs from other microbial sources also restore pimaricin production in the ΔpimE mutant, provided that they belong to the GMC oxidoreductase family, i.e., are type I cholesterol oxidases (see review 1 of this miniseries). It is hypothesized that the S. natalensis ChOx acts as a signaling protein for the macrolide biosynthesis pathway [94, 95].
The regulatory model of pimaricin biosynthesis in S. natalensis cells is an attractive paradigm because ChOx genes are present in additional antifungal polyketide biosynthetic gene clusters in Streptomyces [96, 97]. The precise mechanism of signaling is unclear. One possible mechanism is that ChOx acts as a fungal sensor via oxidation of ergosterol, or another unknown mold sterol. Alternatively, the enzyme itself may act as a ligand for a receptor signaling system.
Early studies with extracellular ChOxs concluded that ergosterol is a poor substrate for the enzyme from Streptomyces [98] or from Rhodococcus sp. [85]. However, to our knowledge, substrate specificity studies with ergosterol have only been performed with detergent micelles. For these studies, the relevant form of the substrate is ergosterol mixed with fungal lipids. Alternatively, the ergosterol may bind to the enzyme without undergoing oxidation and induce a conformational change in PimE. This activated complex would then interact with a receptor signaling system to promote pimaricin biosynthesis.
Concluding comments
The present minireview considers the interaction of microbial ChOx with biological membrane surfaces. In order to sequester its substrate, the ChOx molecule binds to a membrane surface through hydrophobic interactions. However, the exact mechanism of binding, sterol sequestration and 4-en-3-one release is not well understood. The questions that remain for future investigation are: does the contact surface extend beyond the entrance to the substrate-binding site, or does any part of the protein insert more deeply than another? In its interfacial mechanism, ChOx is like the well-studied family of phospholipases.
From the accumulated studies of ChOx action on model membranes, both monolayers and bilayers, it is known that binding and substrate sequestration are the parameters that limit enzyme activity. Kinetically, the rate dependence on substrate mole fraction (k*cat/K*m, Scheme 1), rather than bulk substrate concentration, is the important rate constant to consider for determining substrate specificity.
The interfacial characteristics of ChOx are linked to its physiological roles as the enzyme that initiates sterol catabolism mainly in species of the actinomycetal genera, Corynebacterium, Gordona, and Rhodococcus. However, in the case of Mycobacterium and Nocardia, sterol conversion to the corresponding 4-en-3-one may be carried out by a dehydrogenase/isomerase system requiring NAD+ or NADP+. Future studies that consider taxonomically well-determined mycobacterial members are needed to understand the role of cholesterol oxidation in these microorganisms.
The actinomycetes encompass species that are generally opportunistic pathogens. A possible role of ChOx in rhodococcal virulence has been proposed to be a consequence of the enzyme’s membrane disruption characteristics, which were determined with model membranes and erythrocyte and macrophage cells. The question as to whether ChOx plays a role in virulence or not remains open.
The third possible function of ChOx (PimE) is as a possible regulator in pimaricin biosynthesis by S. natalensis. ChOx may act as a signaling protein via catalysis of mold ergosterol and/or other sterols, or the enzyme itself may act as ligand for a receptor signaling system, since PimE is extracellular [99]. The receptor activator might be a PimE reaction product or PimE itself could play the role of an activating ligand. In either case, elucidation of the precise mechanism by which ChOx promotes production of this macrolide is an interesting new avenue of research for an old enzyme.
Acknowledgements
The work in the authors’ laboratories was supported by the National Institutes of Health (AI065251, HL53306, N.S.S), the American Heart Association (0725861T, N.S.S.) and NATO (Collaborative Linkage Grant LST.CLG.980121).
Abbreviations
- ChOx
cholesterol oxidase
- M.
Mycobacterium
- R.
Rhodococcus
- S.
Streptomyces
References
- 1.Vrielink A, Lloyd LF, Blow DM. Crystal structure of cholesterol oxidase from Brevibacterium sterolicum refined at 1.8 Å resolution. J Mol Biol. 1991;219:533–554. doi: 10.1016/0022-2836(91)90192-9. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Vrielink A, Brick P, Blow DM. Crystal structure of cholesterol oxidase complexed with a steroid substrate: implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry. 1993;32:11507–11515. [PubMed] [Google Scholar]
- 3.Lario PI, Sampson N, Vrielink A. Sub-atomic resolution crystal structure of cholesterol oxidase: What atomic resolution crystallography reveals about enzyme mechanism and the role of the FAD cofactor in redox activity. J Mol Biol. 2003;326:1635–1650. doi: 10.1016/s0022-2836(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 4.Berg OG, Gelb MH, Tsai MD, Jain MK. Interfacial enzymology: the secreted phospholipase A(2)-paradigm. Chem Rev. 2001;101:2613–2654. doi: 10.1021/cr990139w. [DOI] [PubMed] [Google Scholar]
- 5.Radhakrishnan A, McConnell HM. Chemical activity of cholesterol in membranes. Biochemistry. 2000;39:8119–8124. doi: 10.1021/bi0005097. [DOI] [PubMed] [Google Scholar]
- 6.Silvius J. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim Biophys Acta. 2003;1610:174–183. doi: 10.1016/s0005-2736(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 7.Barenholz Y, Patzer EJ, Moore NF, Wagner RR. Cholesterol oxidase as a probe for studying membrane composition and organization. Adv Exp Med Biol. 1978;101:45–56. doi: 10.1007/978-1-4615-9071-2_5. [DOI] [PubMed] [Google Scholar]
- 8.Lange Y, Matthies H, Steck TL. Cholesterol oxidase susceptibility of the red cell membrane. Biochim Biophys Acta. 1984;769:551–562. doi: 10.1016/0005-2736(84)90053-1. [DOI] [PubMed] [Google Scholar]
- 9.Lange Y. Tracking cell cholesterol with cholesterol oxidase. J Lipid Res. 1992;33:315–321. [PubMed] [Google Scholar]
- 10.El Yandouzi EH, Le Grimellec C. Effect of cholesterol oxidase treatment on physical state of renal brush border membranes: Evidence for a cholesterol pool interacting weakly with membrane lipids. Biochemistry. 1993;32:2047–2052. doi: 10.1021/bi00059a023. [DOI] [PubMed] [Google Scholar]
- 11.Zager RA, Burkhart KM, Johnson A. Sphingomyelinase and membrane sphingomyelin content: determinants of proximal tubule cell susceptibility to injury. J Am Soc Nephrol. 2000;11:894–902. doi: 10.1681/ASN.V115894. [DOI] [PubMed] [Google Scholar]
- 12.Lange Y, Ye J, Steck TL. Scrambling of phospholipids activates red cell membrane cholesterol. Biochemistry. 2007;46:2233–2238. doi: 10.1021/bi6023397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange Y, Steck TL. Cholesterol homeostasis and the escape tendency (activity) of plasma membrane cholesterol. Prog Lipid Res. 2008;47:319–332. doi: 10.1016/j.plipres.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbin DR, Greenplate JT, Purcell JP. The identification and development of proteins for control of insects in genetically modified crops. Hortscience. 1998;33:614–617. [Google Scholar]
- 15.Corbin DR, Grebenok RJ, Ohnmeiss TE, Greenplate JT, Purcell JP. Expression and chloroplast targeting of cholesterol oxidase in transgenic tobacco plants. Plant Physiol. 2001;126:1116–1128. doi: 10.1104/pp.126.3.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ipsen JH, Karlström G, Mouritsen OG, Wennerström H, Zuckermann MJ. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim Biophys Acta. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 17.Ipsen JH, Mouritsen OG, Zuckermann MJ. Theory of thermal anomalies in the specific heat of lipid bilayers containing cholesterol. Biophys J. 1989;56:661–667. doi: 10.1016/S0006-3495(89)82713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sankaram MB, Thompson TE. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry. 1990;29:10670–10675. doi: 10.1021/bi00499a014. [DOI] [PubMed] [Google Scholar]
- 19.McConnell HM, Radhakrishnan A. Condensed complexes of cholesterol and phospholipids. Biochim Biophys Acta. 2003;1610:159–173. doi: 10.1016/s0005-2736(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Buboltz JT, Feigenson GW. Maximum solubility of cholesterol in phosphatidylcholine and phosphatidylethanolamine bilayers. Biochim Biophys Acta. 1999;1417:89–100. doi: 10.1016/s0005-2736(98)00260-0. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Feigenson GW. A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers. Biophys J. 1999;76:2142–2157. doi: 10.1016/S0006-3495(99)77369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uwajima T, Yagi H, Terada O. Properties of crystalline 3β-hydroxysteroid oxidase of Brevibacterium sterolicum. Agr Biol Chem. 1974;38:1149–1156. [Google Scholar]
- 23.Kreit J, Lefebvre G, Germain P. Membrane-bound cholesterol oxidase from Rhodococcus sp. cells. Production and extraction. J Biotechnol. 1994;33:271–282. [Google Scholar]
- 24.De Martinez SG, Green C. The action of cholesterol oxidase on cholesterol in vesicles and micelles [proceedings] Biochem Soc Trans. 1979;7:978–979. doi: 10.1042/bst0070978. [DOI] [PubMed] [Google Scholar]
- 25.Peynet J, Delattre J, Canal J, Rousselet F, Girard ML. Effect of the phosphatidylcholine/cholesterol molar ratio of liposomes on the reactivity of cholesterol with cholesterol:oxygen oxydoreductase. Biochimie. 1979;61:487–494. doi: 10.1016/s0300-9084(79)80205-9. [DOI] [PubMed] [Google Scholar]
- 26.Sampson NS, Kass IJ. Isomerization, but not oxidation, is suppressed by a single point mutation, E361Q, in the reaction catalyzed by cholesterol oxidase. J Am Chem Soc. 1997;119:855–862. [Google Scholar]
- 27.Kass IJ, Sampson NS. Evaluation of the role of His447 in the reaction catalyzed by cholesterol oxidase. Biochemistry. 1998;37:17990–18000. doi: 10.1021/bi982115+. [DOI] [PubMed] [Google Scholar]
- 28.Kass IJ, Sampson NS. The importance of Glu361 position in the reaction catalyzed by cholesterol oxidase. Bioorg Med Chem Lett. 1998;8:2663–2668. doi: 10.1016/s0960-894x(98)00478-8. [DOI] [PubMed] [Google Scholar]
- 29.Xiang J, Sampson NS. Library screening studies to investigate substrate specificity in the reaction catalyzed by cholesterol oxidase. Prot Eng Design & Select. 2004;17:341–348. doi: 10.1093/protein/gzh041. [DOI] [PubMed] [Google Scholar]
- 30.Vrielink A, Li J, Brick P, Blow D. Structure and mechanism of cholesterol oxidase. Spec Publ (R Soc Chem) 1992;111:83–94. [Google Scholar]
- 31.Sampson NS, Vrielink A. Cholesterol oxidases: A study of nature’s approach to protein design. Acc Chem Res. 2003;36:713–722. doi: 10.1021/ar9800587. [DOI] [PubMed] [Google Scholar]
- 32.Sampson NS, Kass IJ, Ghoshroy KB. Assessment of the role of an Ω loop of cholesterol oxidase: a truncated loop mutant has altered substrate specificity. Biochemistry. 1998;37:5770–5778. doi: 10.1021/bi973067g. [DOI] [PubMed] [Google Scholar]
- 33.Yue QK, Kass IJ, Sampson NS, Vrielink A. Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants. Biochemistry. 1999;38:4277–4286. doi: 10.1021/bi982497j. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Wolfgang DE, Sampson NS. Use of the parallax-quench method to determine the position of the active-site loop of cholesterol oxidase in lipid bilayers. Biochemistry. 2000;39:13383–13389. doi: 10.1021/bi001407j. [DOI] [PubMed] [Google Scholar]
- 35.Slotte JP. Direct observation of the action of cholesterol oxidase in monolayers. Biochim Biophys Acta. 1995;1259:180–186. doi: 10.1016/0005-2760(95)00161-5. [DOI] [PubMed] [Google Scholar]
- 36.Ghoshroy KB, Zhu W, Sampson NS. Investigation of membrane disruption in the reaction catalyzed by cholesterol oxidase. Biochemistry. 1997;36:6133–6140. doi: 10.1021/bi962190p. [DOI] [PubMed] [Google Scholar]
- 37.Slotte JP. Cholesterol oxidase susceptibility of cholesterol and 5-androsten-3β-ol in pure sterol monolayers and in mixedmonolayers containing 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine. Biochim Biophys Acta. 1992;1124:23–28. doi: 10.1016/0005-2760(92)90121-b. [DOI] [PubMed] [Google Scholar]
- 38.Brooks CJW, Smith AG. Cholesterol oxidase: Further studies of substrate specificity in relation to the analytical characterisation of steroids. J Chromatog. 1975;112:499–511. doi: 10.1016/s0021-9673(00)99979-5. [DOI] [PubMed] [Google Scholar]
- 39.Smith AG, Brooks CJW. The substrate specificity and stereochemistry, reversiblility and inhibition of the 3-oxo steroid Δ4– Δ5 isomerase component of cholesterol oxidase. Biochem J. 1977;167:121–129. doi: 10.1042/bj1670121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn KW, Sampson NS. Cholesterol oxidase senses subtle changes in lipid bilayer structure. Biochemistry. 2004;43:827–836. doi: 10.1021/bi035697q. [DOI] [PubMed] [Google Scholar]
- 41.Grönberg L, Slotte JP. Cholesterol oxidase catalyzed oxidation of cholesterol in mixed lipid monolayers: effects of surface pressure and phospholipid composition on catalytic activity. Biochemistry. 1990;29:3173–3178. doi: 10.1021/bi00465a003. [DOI] [PubMed] [Google Scholar]
- 42.Sampson NS, Kwak S. Catalysis at the membrane interface: Cholesterol oxidase as a case study. In: Kettner C, Hicks MG, editors. Proceedings of 3rd International symposium on experimental standard conditions of enzyme characterizations (ESCEC) Germany: Beilstein-Institut, Rüdesheim/Rhein; 2008. pp. 13–22. [Google Scholar]
- 43.Slotte JP. Enzyme-catalyzed oxidation of cholesterol in mixed phospholipid monolayers reveals the stoichiometry at which free cholesterol clusters disappear. Biochemistry. 1992;31:5472–5477. doi: 10.1021/bi00139a008. [DOI] [PubMed] [Google Scholar]
- 44.Donova MV. Transformation of steroids by actinobacteria: a review. App Biochem Microbiol. 2007;43:1–14. [PubMed] [Google Scholar]
- 45.Owen RW, Mason AN, Bilton RF. The degradation of cholesterol by Pseudomonas sp. NCIB 10590 under aerobic conditions. J Lipid Res. 1983;24:1500–1511. [PubMed] [Google Scholar]
- 46.Horinouchi M, Kurita T, Yamamoto T, Hatori E, Hayashi T, Kudo T. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem Biophys Res Commun. 2004;324:597–604. doi: 10.1016/j.bbrc.2004.09.096. [DOI] [PubMed] [Google Scholar]
- 47.Horinouchi S, Ishizuka H, Beppu T. Cloning, nucleotide sequence, and transcriptional analysis of the NAD(P)-dependent cholesterol dehydrogenase gene from a Nocardia sp. and its hyperexpression in Streptomyces spp. Appl Environ Microbiol. 1991;57:1386–1393. doi: 10.1128/aem.57.5.1386-1393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kishi K, Watazu Y, Katayama Y, Okabe H. The characteristics and applications of recombinant cholesterol dehydrogenase. Biosci Biotechnol Biochem. 2000;64:1352–1358. doi: 10.1271/bbb.64.1352. [DOI] [PubMed] [Google Scholar]
- 49.Chiang YR, Ismail W, Müller M, Fuchs G. Initial steps in the anoxic metabolism of cholesterol by the denitrifying Sterolibacterium denitrificans. J Biol Chem. 2007;282:13240–13249. doi: 10.1074/jbc.M610963200. [DOI] [PubMed] [Google Scholar]
- 50.Chiang YR, Ismail W, Heintz D, Schaeffer C, Van Dorsselaer A, Fuchs G. Study of anoxic and oxic cholesterol metabolism by Sterolibacterium denitrificans. J Bacteriol. 2008;190:905–914. doi: 10.1128/JB.01525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Dubnau E, Smith I, Sampson NS. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry. 2007;46:9058–9067. doi: 10.1021/bi700688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brzostek A, Dziadek B, Rumijowska-Galewicz A, Pawelczyk J, Dziadek J. Cholesterol oxidase is required for virulence of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2007;275:106–112. doi: 10.1111/j.1574-6968.2007.00865.x. [DOI] [PubMed] [Google Scholar]
- 53.Kreit J, Germain P. Induction and stability of cholesterol oxidase from cells of a Rhodoccus. In Stability and stabilization of biocatalysts. In: Ballesteros A, Plou FJ, Iborra JL, Halling PJ, editors. Progress in Biotechnology. Vol 15. Amsterdam: Elsevier Science; 1998. pp. 639–644. [Google Scholar]
- 54.Elalami A, Baessler K, Kong F, Sampson N, Kreit J. Subcellular forms of cholesterol oxidase from Rhodococcus sp. CIP 105 335: induction, solubilization and characterization. In: Mendez-Vilas A, editor. Current Research Topics in Applied Microbiology and Microbial Biotechnology. London: World Scientific Publishing Co. Pte. Ltd; 2009. pp. 729–735. [Google Scholar]
- 55.Sih CJ, Wang KC, Tai HH. C22 acid intermediates in the microbiological cleavage of the cholesterol side chain. J Am Chem Soc. 1967;89:1956–1957. doi: 10.1021/ja00984a038. [DOI] [PubMed] [Google Scholar]
- 56.Sih CJ, Tai HH, Tsong YY. The mechanism of microbial conversion of cholesterol into 17-keto steroids. J Am Chem Soc. 1967;89:1957–1958. doi: 10.1021/ja00984a039. [DOI] [PubMed] [Google Scholar]
- 57.Goodfellow M, Alderson G. The actinomycete-genus Rhodococcus: a home for the "rhodochrous" complex. J Gen Microbiol. 1977;100:99–122. doi: 10.1099/00221287-100-1-99. [DOI] [PubMed] [Google Scholar]
- 58.Ladrón N, Fernández M, Agüero J, González Zörn B, Vázquez-Boland JA, Navas J. Rapid identification of Rhodococcus equi by a PCR assay targeting the choE gene. J Clin Microbiol. 2003;41:3241–3245. doi: 10.1128/JCM.41.7.3241-3245.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drzyzga O, Navarro Llorens JM, Fernández de Las Heras L, García Fernández E, Perera J. Gordonia cholesterolivorans sp. nov., a cholesterol-degrading actinomycete isolated from sewage sludge. Int J Syst Evol Microbiol. 2009;59:1011–1015. doi: 10.1099/ijs.0.005777-0. [DOI] [PubMed] [Google Scholar]
- 60.Arima K, Nagasawa M, Bae M, Tamura G. Microbial transformation of sterols Part I. Decomposition of cholesterol by microorganisms. Agr Biol Chem. 1969;33:1636–1643. [Google Scholar]
- 61.Marsheck WJ, Kraychy S, Muir RD. Microbial degradation of sterols. Appl Microbiol. 1972;23:72–77. doi: 10.1128/am.23.1.72-77.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wovcha MG, Antosz FJ, Knight JC, Kominek LA, Pyke TR. Bioconversion of sitosterol to useful steroidal intermediates by mutants of Mycobacterium fortuitum. Biochim Biophys Acta. 1978;531:308–321. doi: 10.1016/0005-2760(78)90213-8. [DOI] [PubMed] [Google Scholar]
- 63.Schoemer U, Martin CKA. Microbial transformation of sterols. Biotechnol Bioeng. 1980;22 Suppl 1:11–25. [Google Scholar]
- 64.Martin CKA. Sterols. In: Rehm HJ, Reed G, editors. Biotechnology, a Comprehensive Treatise in Eight Volumes: Biotransformations, Vol 6a (Kieslich K, vol ed) Weinheim, Germany: Wiley-VCH Verlag GmbH; 1984. pp. 79–95. [Google Scholar]
- 65.Lee CY, Liu WH. Production of androsta-1,4-diene-3,17-dione from cholesterol using immobilized growing cells of Mycobacterium sp. NRRL B-3683 adsorbed on solid carriers. Appl Microbiol Biotechnol. 1992;36:598–603. doi: 10.1007/BF00183235. [DOI] [PubMed] [Google Scholar]
- 66.Stadtman TC, Cherkes A, Anfinsen CB. Studies on the microbiological degradation of cholesterol. J Biol Chem. 1954;206:511–523. [PubMed] [Google Scholar]
- 67.Stadtman TC. Cholesterol dehydrogenase from a Mycobacterium. Methods Enzymol. 1955;1:678–681. [Google Scholar]
- 68.Bell KS, Philp JC, Aw DWJ, Christofi N. The genus Rhodococcus. J Appl Microbiol. 1998;85:195–210. doi: 10.1046/j.1365-2672.1998.00525.x. [DOI] [PubMed] [Google Scholar]
- 69.Halpern M. Industrial enzymes from microbial sources. (Chem Tech Rev no 186) NJ, USA: Noyes Data Corp; 1981. Cholesterol oxidase from bacteria; pp. 3–22. [Google Scholar]
- 70.Rahman MT, Herron LL, Kapur V, Meijer WG, Byrne BA, Ren J, Nicholson VM, Prescott JF. Partial genome sequencing of Rhodococcus equi ATCC 33701. Vet Microbiol. 2003;94:143–158. doi: 10.1016/s0378-1135(03)00100-7. [DOI] [PubMed] [Google Scholar]
- 71.Navas J, Gonález-Zorn B, Ladrón N, Garrido P, Vázquez-Boland JA. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J Bacteriol. 2001;183:4796–4805. doi: 10.1128/JB.183.16.4796-4805.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linder R, Bernheimer AW. Enzymatic oxidation of membrane cholesterol in relation to lysis of sheep erythrocytes by corynebacterial enzymes. Arch Biochem Biophys. 1982;213:395–404. doi: 10.1016/0003-9861(82)90565-3. [DOI] [PubMed] [Google Scholar]
- 73.Fernández-Garayzábal JF, Delgado C, Blanco MM, Domínguez L. Cholesterol oxidase from Rhodococcus equi is likely the major factor involved in the cooperative lytic process (CAMP reaction) with Listeria monocytogenes. Lett App Microbiol. 1996;22:249–252. doi: 10.1111/j.1472-765x.1996.tb01153.x. [DOI] [PubMed] [Google Scholar]
- 74.Machang'u RS, Prescott JF. Purification and properties of cholesterol oxidase and choline phosphohydrolase from Rhodococcus equi. Can J Vet Res. 1991;55:332–340. [PMC free article] [PubMed] [Google Scholar]
- 75.Megha, London E. Ceramide selectively displaces cholesterol from ordered lipid domains (rafts): implications for lipid raft structure and function. J Biol Chem. 2004;279:9997–10004. doi: 10.1074/jbc.M309992200. [DOI] [PubMed] [Google Scholar]
- 76.Weinstock DM, Brown AE. Rhodococcus equi: an emerging pathogen. Clin Infect Dis. 2002;34:1379–1385. doi: 10.1086/340259. [DOI] [PubMed] [Google Scholar]
- 77.Fuhrmann H, Dobeleit G, Bellair S, Gück T. Cholesterol oxidase and resistance of Rhodococcus equi to peroxidative stress in vitro in the presence of cholesterol. J Vet Med B. 2002;49:310–311. doi: 10.1046/j.1439-0450.2002.00555.x. [DOI] [PubMed] [Google Scholar]
- 78.Linder R, Bernheimer AW. Oxidation of macrophage membrane cholesterol by intracellular Rhodococcus equi. Vet Microbiol. 1997;56:269–276. doi: 10.1016/s0378-1135(97)00095-3. [DOI] [PubMed] [Google Scholar]
- 79.Pei Y, Dupont C, Sydor T, Haas A, Prescott JF. Cholesterol oxidase (ChoE) is not important in the virulence of Rhodococcus equi. Vet Microbiol. 2006;118:240–246. doi: 10.1016/j.vetmic.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 80.Pei Y, Nicholson V, Woods K, Prescott JF. Immunization by intrabronchial administration to 1-week-old foals of an unmarked double gene disruption strain of Rhodococcus equi strain 103+ Vet Microbiol. 2007;125:100–110. doi: 10.1016/j.vetmic.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 81.Mohn WW, van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. The actinobacterial Mce4 locus encodes a steroid transporter. J Biol Chem. 2008;283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Geize R, de Jong W, Hessels GI, Grommen AWF, Jacobs AAC, Dijkhuizen L. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucl Acids Res. 2008;36:e151. doi: 10.1093/nar/gkn811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Atrat PG, Wagner B, Wagner M, Schumann G. Localization of the cholesterol oxidase in Rhodococcus erythropolis IMET 7185 studied by immunoelectron microscopy. J Steroid Biochem Molec Biol. 1992;42:193–200. doi: 10.1016/0960-0760(92)90028-h. [DOI] [PubMed] [Google Scholar]
- 84.Sojo M, Bru R, LopezMolina D, GarciaCarmona F, Argüelles JC. Cell-linked and extracellular cholesterol oxidase activities from Rhodococcus erythropolis. Isolation and physiological characterization. App Microbiol Biotechnol. 1997;47:583–589. doi: 10.1007/s002530050977. [DOI] [PubMed] [Google Scholar]
- 85.Kreit J, Germain P, Lefebvre G. Extracellular cholesterol oxidase from Rhodococcus sp. cells. J Biotechnol. 1992;24:177–188. [Google Scholar]
- 86.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yam KC, D'Angelo I, Kalscheuer R, Zhu H, Wang JX, Snieckus V, Ly LH, Converse PJ, Jacobs WRJ, Strynadka N, Eltis LD. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000344. doi: 10.1371/journal.ppat.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Capyk JK, D'Angelo I, Strynadka NC, Eltis LD. Characterization of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J Biol Chem. 2009;284:9937–9946. doi: 10.1074/jbc.M900719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aparicio JF, Fouces R, Mendes MV, Olivera N, Martín JF. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem Biol. 2000;7:895–905. doi: 10.1016/s1074-5521(00)00038-7. [DOI] [PubMed] [Google Scholar]
- 91.Aparicio JF, Mendes MV, Antón N, Recio E, Martín JF. Polyene macrolide antibiotic biosynthesis. Curr Med Chem. 2004;11:1645–1656. doi: 10.2174/0929867043365044. [DOI] [PubMed] [Google Scholar]
- 92.Antón N, Mendes MV, Martín JF, Aparicio JF. Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J Bacteriol. 2004;186:2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mendes MV, Recio E, Antón N, Guerra SM, Santos-Aberturas J, Martín JF, Aparicio JF. Cholesterol oxidases act as signaling proteins for the biosynthesis of the polyene macrolide pimaricin. Chem Biol. 2007;14:279–290. doi: 10.1016/j.chembiol.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 94.Nesbitt NM, Sampson NS. Antifungal tradecraft by cholesterol oxidase. Chem Biol. 2007;14:238–241. doi: 10.1016/j.chembiol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aparicio JF, Martín JF. Microbial cholesterol oxidases: bioconversion enzymes or signal proteins? Mol Biosyst. 2008;4:804–809. doi: 10.1039/b717500k. [DOI] [PubMed] [Google Scholar]
- 96.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seco EM, Pérez-Zúñiga FJ, Rolón MS, Malpartida F. Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem Biol. 2004;11:357–366. doi: 10.1016/j.chembiol.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 98.Kamei T, Takiguchi Y, Suzuki H, Matsuzaki M, Nakamura S. Purification of 3β-hydroxysteroid oxidase of Streptomyces violascens origin by affinity chromatography on cholesterol. Chem Pharm Bull (Tokyo) 1978;26:2799–2804. doi: 10.1248/cpb.26.2799. [DOI] [PubMed] [Google Scholar]
- 99.Martín JF, Aparicio JF. Enzymology of the polyenes pimaricin and candicidin biosynthesis. Methods Enzymol. 2009;459:215–242. doi: 10.1016/S0076-6879(09)04610-2. [DOI] [PubMed] [Google Scholar]
- 100.van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol Microbiol. 2002;45:1007–1018. doi: 10.1046/j.1365-2958.2002.03069.x. [DOI] [PubMed] [Google Scholar]
- 101.Horinouchi M, Hayashi T, Koshino H, Kurita T, Kudo T. Identification of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2,4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441. Appl Environ Microbiol. 2005;71:5275–5281. doi: 10.1128/AEM.71.9.5275-5281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Andor A, Jekkel A, Hopwood DA, Jeanplong F, Ilköy E, Kónya A, Kurucz I, Ambrus G. Generation of useful insertionally blocked sterol degradation pathway mutants of fast-growing mycobacteria and cloning, characterization, and expression of the terminal oxygenase of the 3-ketosteroid 9α-hydroxylase in Mycobacterium smegmatis mc(2)155. Appl Environ Microbiol. 2006;72:6554–6559. doi: 10.1128/AEM.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.DeLano WL. San Carlos, CA, USA: DeLano Scientific; 2006. The PyMOL Molecular Graphics System. http://www.pymol.org. [Google Scholar]
- 104.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 105.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]