Abstract
The innate immune response of insects includes induced expression of genes encoding a variety of antimicrobial peptides. The signaling pathways that stimulate this gene expression have been well characterized by genetic analysis in Drosophila melanogaster, but are not well understood in most other insect species. One such pathway involves proteolytic activation of a cytokine called Spätzle, which functions in dorsal-ventral patterning in early embryonic development and in the antimicrobial immune response in larvae and adults. We have investigated the function of Spätzle in a lepidopteran insect, Manduca sexta, in which hemolymph proteinases activated during immune responses have been characterized biochemically. Two cDNA isoforms for M. sexta Spätzle-1 differ due to alternative splicing, resulting in a 10 amino acid residue insertion in the pro-region of proSpätzle-1B that is not present in proSpätzle-1A. The proSpätzle-1A cDNA encodes a 32.7 kDa polypeptide that is 23% and 44% identical to D. melanogaster and Bombyx mori Spätzle-1, respectively. Recombinant proSpätzle-1A was a disulfide-linked homodimer. M. sexta hemolymph proteinase 8 (HP8) cleaved proSpätzle-1A to release Spätzle-C108, a dimer of the carboxyl-terminal 108-residue cystine-knot domain. Injection of Spätzle-C108, but not proSpätzle-1A, into larvae stimulated expression of several antimicrobial peptides and proteins, including attacin-1, cecropin-6, moricin, lysozyme, and the immunoglobulin domain protein hemolin, but did not significantly affect expression of two bacteria-inducible pattern recognition proteins, immulectin-2 and β-1,3-glucan recognition protein-2. Results from this paper and other recent studies support a model for a pathway in which the clip-domain proteinase proHP6 becomes activated in plasma upon exposure to Gram-negative or Gram-positive bacteria or to β-1,3-glucan. HP6 then activates proHP8, which in turn activates Spätzle-1. The resulting Spätzle-C108 dimer is likely to function as a ligand to activate a Toll pathway in M. sexta as a response to a wide variety of microbial challenges, stimulating a broad response to infection.
Keywords: Spätzle, M. sexta, proteolytic activation, antimicrobial peptides, innate immunity
Introduction
A prominent feature of the innate immune systems of insects is the activation of serine proteinase cascade pathways in hemolymph, which function to activate plasma proteins that perform immune functions. This mechanism leads to activation of phenoloxidase, which oxidizes catechols leading to formation of toxic quinones and melanin [1,2], and to activation of cytokines that stimulate hemocyte adhesion [3] or synthesis of antimicrobial peptides [4]. These antimicrobial peptides from several families reach high concentrations in the hemolymph and efficiently kill invading microorganisms [4,5].
The signaling mechanisms that elicit expression of antimicrobial peptides are best understood in Drosophila melanogaster. In this species, the Toll pathway operates by transmitting an extracellular signal initiated by recognition of microbial surface polysaccharides, leading to activation of serine proteinases to produce an active Toll ligand called Spätzle [4,6]. The Spätzle ligand and Toll receptor also establish the dorsal-ventral axis in Drosophila embryo, although this activation of proSpätzle is carried out by a different set of proteinases [7].
ProSpätzle is secreted as an inactive precursor, consisting of an unstructured pro-domain [8–10] and a carboxyl terminal fragment that adopts a cystine knot structure similar to that of mammalian neurotrophins such as nerve growth factor (NGF) [7]. This cystine knot motif contains three intramolecular disulfide linkages and an intermolecular disulfide bond, which joins two subunits to form a homodimer [7]. The proSpätzle precursor requires proteolytic processing at a specific site, 106 amino acid residues from the carboxyl terminus to produce an active ligand, termed C106 [7,11]. In the cascade for dorsal-ventral development, the clip-domain serine proteinase [12] Easter cleaves proSpätzle to yield active C106 [7,13]. C106 then binds to the ectodomain of the transmembrane receptor Toll and thereby initiates a cytoplasmic signaling pathway resulting in release of are l-family transcription factor Dorsal from the inhibitor protein Cactus to activate genes involved in dorsal-ventral differentiation [9,14,15]. The proteinases acting upstream of Spätzle during the immune response are distinct from those mediating Toll activation during embryonic development [16]. A clip-domain proteinase called Spätzle processing enzyme (SPE) converts proSpätzle in hemolymph to active C106 [11,17].
In addition to Spätzle-1, the D. melanogaster genome encodes five additional Spätzle homologues (Spz2-6) [18], although functions for these have not yet been identified. Orthologs of all six D. melanogaster Spätzle genes have been identified in the genomes of the mosquitoes Anopheles gambiae and Aedes aegypti [19,20], but only two Spätzle homologs are present in genomes of the honeybee Apis mellifera and the red flour beetle Tribolium castaneum [21,22]. A probable ortholog of Spätzle-1 has been studied in the silkworm, Bombyx mori [23]. A. aegypti Spätzle-1 was demonstrated by RNA interference experiments to function in antifungal immunity [20], while injection of the active form of B. mori and T. castaneum Spätzle-1 into insects has been shown to induce antimicrobial peptide-expression [23–25].
A serine proteinase that activates proSpätzle-1 in immune responses has been identified in a beetle, Tenebrio molitor. The T. molitor clip-do main SPE has been demonstrated to be activated by a proteinase cascade stimulated by peptidoglycan or β-1,3-glucan and to convert Tribolium castneum proSpätzle to its active form [24,25]. Jang et al. [11] described a B. mori clip-domain proteinase called BAEEase as a candidate proSpätzle-1 activator, because it is activated by upstream serine proteinase cascade components in the presence of peptidoglycan and β-1,3-glucan, and has sequence similarity to Easter.
The tobacco hornworm, Manduca sexta, has been a useful model system for biochemical investigations of innate immunity, including the function of hemolymph proteinase cascades and antimicrobial peptides [26–28]. In M. sexta larvae, hemolymph antimicrobial activity is strongly induced by both Gram-negative and Gram-positive bacteria [29], and thirty hemolymph proteins whose synthesis is induced by microbial exposure have been studied [30].
A proteinase pathway activated by exposure to bacteria or β-1,3-glucan was shown to contain M. sexta proteinase HP6, most similar in sequence to the D. melanogaster clip-domain proteinase Persephone. HP6 activates clip-domain proteinase HP8, which is most similar to Drosophila SPE and Easter [31]. Injection of either of these M. sexta proteinases into larvae stimulated expression of antimicrobial peptide genes, suggesting that they might function in activation of a Toll pathway [31]. We present here results characterizing M. sexta Spätzle-1, identifying HP8 as its activating proteinase, and demonstrating that processed Spätzle-1 functions to stimulate expression of several antimicrobial peptides in M. sexta.
Results
Isolation and analysis of M. sexta proSpätzle-1 cDNAs
We identified a 130-bp fragment in a M. sexta fat body and hemocyte EST collection [32], which encoded a polypeptide sequence with 46% identity to B. mori Spätzle-1 [23]. We carried out 3′ and 5′ RACE to obtain the missing ends of the cDNA and then used primers encompassing the start and stop codons with larval fat body cDNA as template to obtain eight individual clones containing the complete coding sequence. Two variants of the full-length proSpätzle-1 cDNA sequence were identified. The shorter proSpätzle-1A cDNA contains 2,532 nucleotides, with a 181-bp 5′-noncoding sequence, an 888-bp open reading frame, and a 1463-bp 3′-noncoding sequence, including a poly(A) tail (Fig. 1A). The 3′-noncoding region contains two putative polyadenylation signals just upstream of the poly(A) tail. The longer variant, proSpätzle-1B contained a 30 bp insertion in the open reading frame, beginning at nucleotide 516. This results in insertion of a 10 amino acid residue segment (TREIDYPETI) and one substitution (Ser→Gly) at the carboxyl-terminal end of the insertion (Fig. 1B).
Fig. 1.
(A) cDNA and deduced amino acid sequences of M. sexta proSpätzle. The one-letter code for each amino acid residue is aligned with the second nucleotide of the corresponding codon. The stop codon is marked with “*”. The predicted secretion signal peptide is underlined. The proteolytic activation site is indicated with “‖”. The amino terminal sequence, determined by Edman degradation, of the activated form of Spätzle (C108) after cleavage by HP8, is shown in bold. Putative N- and O- linked glycosylation sites are shaded. AATAAA sequences (double-underlined) near the end of the 3′ untranslated region are potential polydenylation signals. Intron positions identified within the ORF are indicated by “◊”, with a filled symbol “♦” showing the position of an intron conserved in the orthologous Spätzle genes from D. melanogaster, B. mori, and T. castaneum. (B) The alternative splicing boundaries leading to two proSpätzle isoforms (accession numbers GQ249944, GQ249945).
To examine the origin of the two proSpätzle-1 variants, we used primers designed from the cDNA sequence to amplify overlapping genomic DNA fragments corresponding to nearly the complete open reading frame (Figure S1). Four introns were identified in the proSpätzle-1 gene, all of which are conserved in the B. mori proSpätzle-1 gene (data not shown). We were not able to amplify a genomic sequence containing the first ~300 bp of the M. sexta proSpätzle-1 mRNA, perhaps because of a large intron in this region, as occurs in the B. mori proSpätzle-1 gene [23]. One intron is at a conserved position in proSpätzle-1 genes of D. melanogaster [18], B. mori [23], and T. castaneum [22] (Fig. 1A and Fig. S1). The two M. sexta proSpätzle-1 variants apparently arise from use of alternative 3’ splicing sites for the first intron in the genomic region that was sequenced (Fig. 1B and Fig. S1). RT-PCR analysis, using primers flanking the alternative splice sites to produce different sized products for the two variants (Table S1), indicated that both isoforms are expressed, with proSpätzle-1B more abundant than proSpätzle-1A (Fig. S3).
The conceptual proteins deduced from nucleotide sequences of proSpätzle-1A and proSpätzle-1B cDNA consisted of 295 and 305 amino acid residues, respectively, both including a predicted 18-residue secretion signal peptide. The calculated mass and isoelectric point of mature proSpätzle-1A are 31,861 Da and 6.97, whereas those of proSpätzle-1B are 33,050 Da and 6.08. There is one potential N-linked glycosylation site at Asn75 and one potential O-linked glycosylation site (Thr109 in proSpätzle-1A and Thr119 in proSpätzle-1B). The putative activation cleavage site, identified by alignment with D. melanogaster and B. mori proSpätzle (Fig. 2), is located after IAQR169 in proSpätzle-1A (IAQR179 in proSpätzle-1B), suggesting that an activating proteinase would cleave after this specific Arg residue.
Fig. 2.
Alignment of full length of M. sexta proSpätzle-1A (Ms_Spz), B. mori Spätzle-1 (Bm_Spz) and D. melanogater Spätzle (Dm_Spz). Completely conserved amino acid residues are indicated by “*”, and conservative substitutions by “:” below the sequences. The P1 residue at the activation cleavage site is shown in bold, and the scissile bond is indicated with an arrow.. Absolutely conserved cysteine residues are shaded and numbered. The paired numbers (1-1, 2-2, 3-3) indicate the intrachain disulfide linkage in Dm_Spz [10]. Cys-4 forms an intermolecular disulfide bond with its counterpart in another subunit. The GenBank accession numbers are: Ms_Spz, GQ249944; Bm_Spz, NM_001114594; Dm_Spz, NM_079802.
In preliminary experiments to express proSpätzle-1B, we found that it was cleaved at Arg-95, within the alternatively spliced insertion, by a proteinase activity produced by both the D. melanogaster S2 cell line and the Spodoptera frugiperda Sf9 cell line (data not shown), but this was not the case for proSpätzle-1A, which lacks this residue. For this reason, we focused further experiments on proSpätzle-1A to avoid complications of this artifact.
Sequence comparisons and phylogenetic analysis
Database searches and sequence alignment indicated that M. sexta proSpätzle-1A is most similar in amino acid sequence to B. mori proSpätzle-1, with 44% identity. Of the six D. melanogaster Spätzle paralogs, the sequence of one Spätzle-1 splice variant (accession number NM_079802) is the most similar to M. sexta proSpätzle-1A (23% identity). In the genome of a beetle, T. castaneum, putative proSpätzle GLEAN01054 is most similar to M. sexta proSpätzle-1A (22% identity). The putative active domain at the carboxyl terminus is generally more conserved among different species (26–42% identity) than the amino-terminal pro-region (14–23% identity). An exception is the B. mori sequence, in which the pro-region is 40% identical to that of M. sexta. Seven Cys residues in the putative carboxyl-terminal active cystine knot domain of M. sexta proSpätzle-1 are conserved with those found in D. melanogaster and B. mori Spätzle-1 (Fig. 2) and in nearly all known Spätzle cystine knot domains (Fig. S2). In D. melanogaster Spätzle, six of these Cys residues form intramolecular disulfide bridges, and the seventh makes an intermolecular linkage with its counterpart in another subunit to form a disulfide-linked homodimer [10].
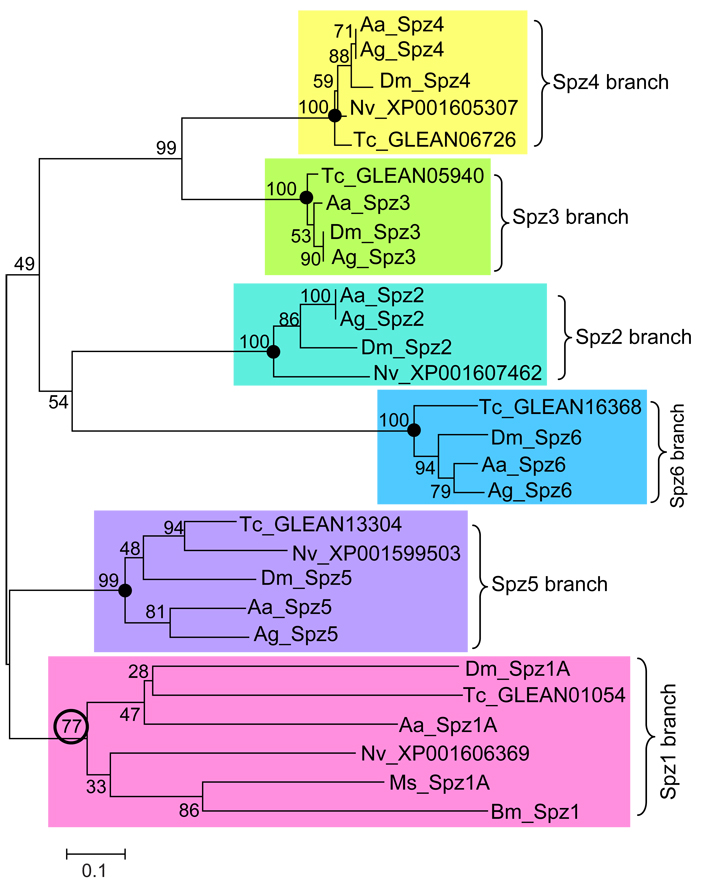
To assess the relationship between M. sexta proSpätzle-1 and other insect Spätzle proteins, we performed a phylogenetic analysis by aligning the homologous cystine knot domain sequences from D. melanogaster, A. aegypti, A. gambiae, B. mori, M. sexta, Nasonia vitripennis, and T. castaneum. We could not include A. gambiae Spätzle-1 in the analysis, because the partial sequence of Spätzle-1 available for this species is, as yet, missing the cystine knot domain. The phylogenetic tree (Fig. 3) suggests that all Spätzle homologues can be assigned to a 1:1 orthologous group with one of the Drosophila Spätzle gene products (Spz1 to Spz6). The inclusion of M. sexta proSpätzle-1A in the same branch as Drosophila Spätzle-1, with a bootstrap value of 77, suggests that M. sexta proSpätzle-1A is an ortholog of this gene. In the clade including Spätzle-1, the branch lengths are noticeably longer and the bootstrap values are lower than in the other clades containing Spätzle-2 through Spätzle-6, indicating a lower degree of sequence conservation in Spätzle-1.
Fig. 3.
Phylogenetic analysis of the cystine-knot domains in Spätzle from M. sexta and other insect species. The tree was derived from an alignment that can be found in supplementary Figure S2. Numbers at the nodes are bootstrap values as percentage. The nodes signifying Spz2, Spz3, Spz4, Spz5, and Spz6 specific branches are denoted by “•”. The circled bootstrap value indicates that M. sexta Spätzle-1A probably belongs to the Spz1 group. The scale bar indicates the number of substitutions per site. Abbreviations used are: Aa, Aedes aegypti; Ag, Anopheles gambiae; Bm, Bombyx mori; Dm, Drosophila melanogaster; Ms, Manduca sexta; Nv, Nasonia vitripennis; Tc, Tribolium castaneum.
M. sexta Spätzle-1 gene expression
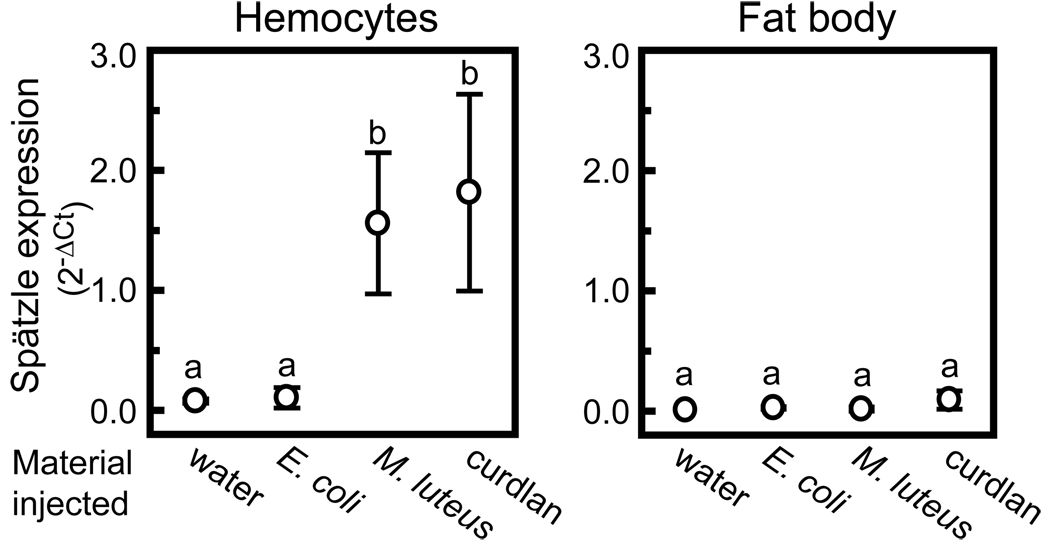
To test whether M. sexta Spätzle-1 mRNA level changes after exposure to microbial elicitors, we analyzed the Spätzle-1 transcript level in hemocytes and fat body after larvae were injected with killed E. coli, M. luteus, curdlan (insoluble β-1,3-glucan), or water as a control. An approximately 20-fold increase in Spätzle-1 transcript level was observed in hemocytes at 24 h after injection of M.luteus or curdlan , but not after injection of killed E. coli (Fig. 4). Spätzle-1 mRNA was also detected the fat body, although at a much lower level than in hemocytes. No significant change was observed in fat body after injection of microbial elicitors. We attempted to investigate concentration of Spätzle-1 in hemolymph by immunoblot analysis, but failed to detect the protein in hemolymph samples. Based on the detection limit of our antibody with purified recombinant Spätzle-1, we estimate the concentration of Spätzle-1 in plasma to be less than 10 µg/ml.
Fig. 4.
M. sexta Spätzle gene expression is up-regulated after injection of microbial elicitors. Quantitative RT-PCR was used to assess the transcript level of Spätzle-1, with ribosomal protein S3 (rpS3) as in internal standard to indicate consistent total mRNA amount. Day 2, fifth instar larvae were injected with water, E. coli, M. luteus, or curdlan. RNA was extracted from hemocytes and fat body collected 24 h after injection. The bars represent mean ± S.D. (n=3). Bars labeled with different letters are significantly different (one-way ANOVA followed by the Newman-Keuls test, P < 0.05).
Recombinant Spätzle-1A is a disulfide-linked dimmer
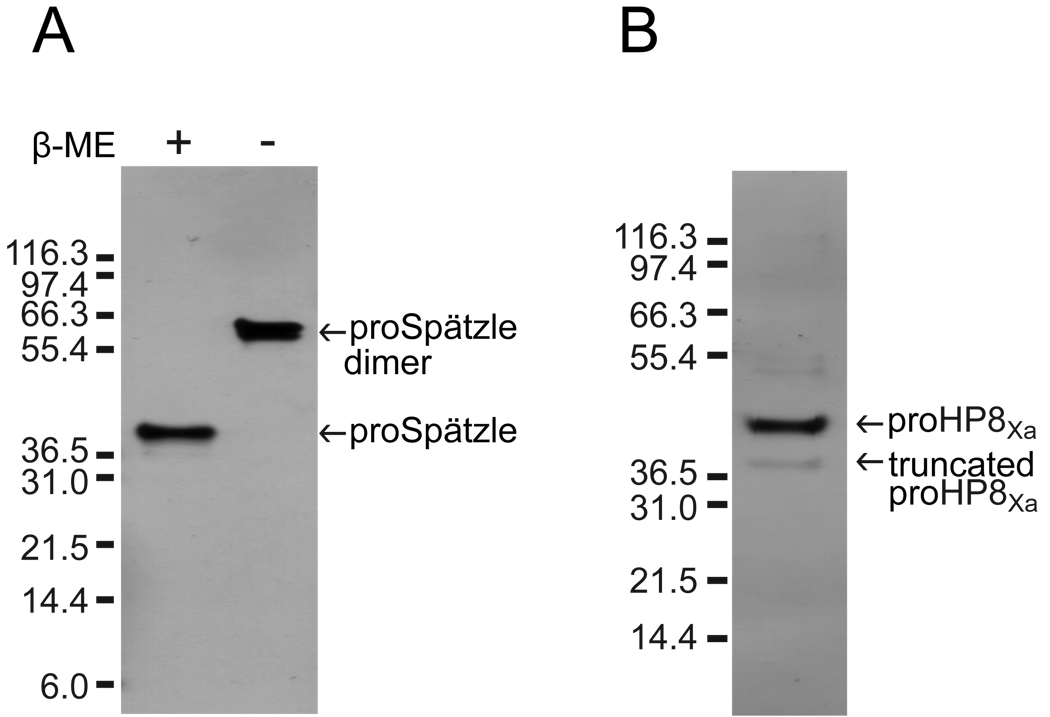
To investigate potential immune functions of M. sexta Spätzle, we expressed proSpätzle-1A with six histidine residues at its carboxyl terminus, using a baculovirus system and Sf9 insect cells. ProSpätzle-1A was secreted into the cell culture medium under control of its own signal peptide and was purified by nickel-affinity chromatography, followed by anion exchange chromatography. SDS-PAGE analysis in the presence of β-mercapoethanol indicated that the purified spätzle had an apparent molecular mass of 38 kDa, which is slightly larger than that predicted from its cDNA sequence plus 6×His-tag (32.7 kDa) (Fig. 5A). Recombinant proSpätzle-1A bound to Concanavalin A (data not shown), indicating that N-linked glycosylation may account for the increased mass. In the absence of β-mercapoethanol, proSpätzle-1A migrated to a position around 64 kDa (Fig. 5A), suggesting that the recombinant protein is a disulfide-linked dimer.
Fig. 5.
SDS-PAGE analysis of purified recombinant proteins. (A) Purified proSpätzle-1A (0.1 µg) was treated with SDS sample buffer in the absence or presence of 0.14 M β-mercaptoethanol (β-ME) at 95°C for 5 min and separated by SDS-PAGE followed by silver staining. (B) Purified proHP8Xa (75 ng) was analyzed by SDS-PAGE under reducing conditions followed by silver staining. The sizes and positions of the molecular weight markers are indicated on the left side of each gel.
ProSpätzle-1A is activated by proteinase HP8Xa
In other insect species, proSpätzle is activated through proteolysis by a clip-domain serine proteinase. The similarity of M. sexta clip-domain proteinase HP8 to D. melanogaster SPE and Easter, which cleave D. melanogaster proSpätzle to produce the active form (C106) [7,11,17], and to Tenebrio molitor SPE [24] led us to predict that HP8 is an activating proteinase for M. sexta proSpätzle [31]. To test this hypothesis, we prepared a recombinant form of proHP8 (proHP8Xa), mutated to permit its activation by commercially available bovine Factor Xa.
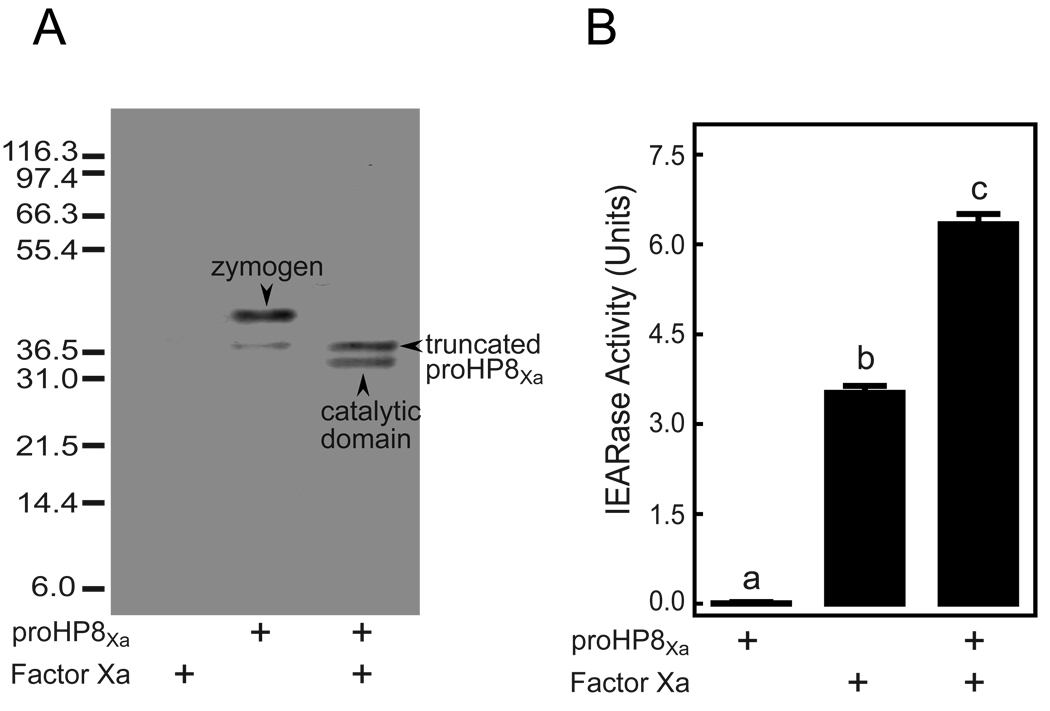
Recombinant proHP8Xa secreted from Drosophila S2 cells was purified by sequential chromatography steps of Blue Gel affinity (to remove contaminating fetal bovine serum albumin), Concanavalin A (ConA) affinity, Q-Sepharose anion exchange, and Sephacryl S-300 HR gel permeation. SDS-PAGE analysis indicated that proHP8Xa was essentially pure, but had in addition to the predominant band at 42 kDa corresponding to the proHP8Xa zymogen [31] a minor band with an apparent molecular mass of 37 kDa (Fig. 5B). This band, which was also detected by antibody to HP8 (Fig. 6A), was shown by amino-terminal sequencing by Edman degradation to be identical to the proHP8 sequence beginning at Gly60 (GAFGNDQG), indicating that it is a truncated form of proHP8, cleaved after Arg59. As the activation site of proHP8 is at Arg90 [31], this truncated form of proHP8Xa is not expected to be active. Incubation of proHP8Xa with Factor Xa, resulted in appearance of a 34-kDa band corresponding to the catalytic domain (Fig. 6A), as previously observed when wild-type proHP8 was activated by M. sexta HP6 [31]. To confirm the activation of proHP8Xa by Factor Xa, we tested whether activated HP8Xa can hydrolyze the HP8 substrate Ile-Glu-Ala-Arg-p-nitroanilide (IEARpNA) [31]. ProHP8Xa lacked IEARase activity, but after the zymogen was activated by Factor Xa, IEARase activity increased significantly above that of Factor Xa alone, which can also hydrolyze the substrate (Fig. 6B). These results indicate that Factor Xa cleaved and activated proHP8Xa.
Fig. 6.
Activation of purified recombinant proHP8Xa by Factor Xa. (A) Purified recombinant proHP8Xa (50 ng) and Factor Xa (100 ng) were incubated separately or mixed together in the presence of 0.005% Tween-20 at 95°C for 5 min, and the mixtures were separated by SDS-PAGE, followed by immunoblot analysis using antiserum against M. sexta HP8. Bands representing the proHP8Xa zymogen, a truncated form of proHP8Xa, and the catalytic domain of active HP8 are marked by arrowheads. The size and position of molecular weight standards are indicated on the left. (B) Catalytic activity of activated HP8Xa (50 ng) was detected by spectrophotometric assay using IEARpNA as a substrate, as described in Experimental Procedures. The bars represent mean ± S.D. (n=3). Bars labeled with different letters are significantly different (one-way ANOVA followed by the Newman-Keuls test, P < 0.05).
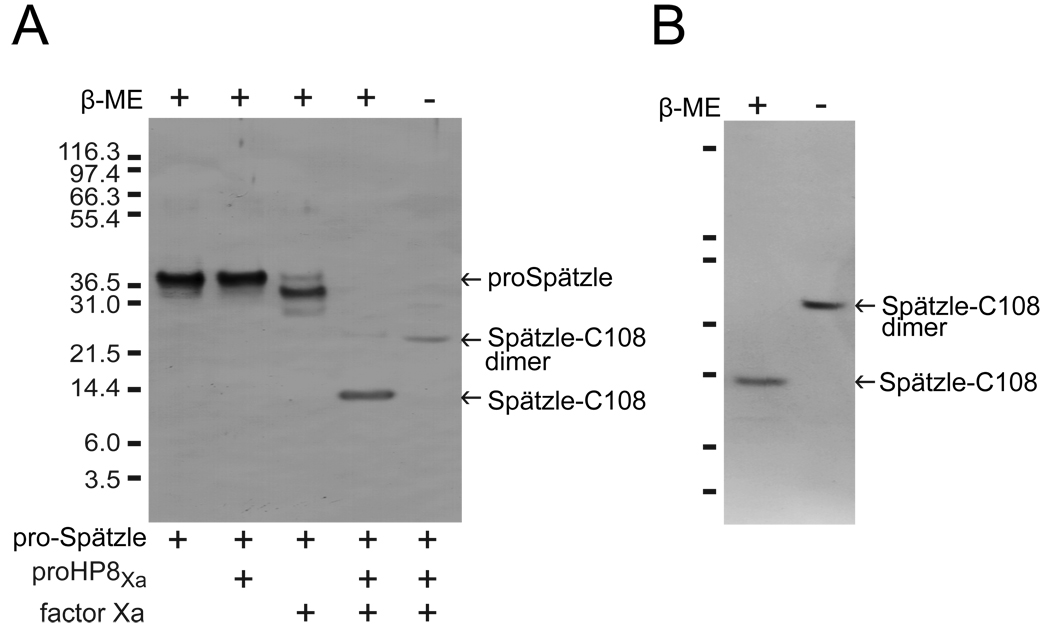
When activated HP8Xa was mixed with proSpätzle-1A, the 38 kDa pro-Spätzle band disappeared, and a 12 kDa product was produced (Fig. 7A). Amino-terminal sequencing of the 12-kDa polypeptide indicated that it corresponds to the carboxyl terminal cystine-knot domain of Spätzle, beginning at Leu170 (LGPQEDNM). This is the expected proteolytic activation site, after Arg169, based on the alignment with D. melanogaster and B. mori proSpätzle sequences (Fig. 2). This product of proSpätzle-1A cleaved by HP8 was named Spätzle-C108, as it consists of the carboxyl-terminal 108 residues. This band did not appear after treatment of proSpätzle-1A with Factor Xa alone or with proHP8Xa zymogen (Fig. 7A), indicating that the observed cleavage of pro-Spätzle was a result of HP8Xa proteolytic activity. We did not observe any cleavage of proSpätzle-1A after incubation with M. sexta clip-domain serine proteinases HP6 or pro PO-activating proteinase-1 (data not shown). In the absence of β-mercapoethanol, Spätzle-C108 migrated to a position around 23 kDa on SDS-PAGE (Fig.7A), indicating that it is a disulfide-linked dimer. Spätzle-C108 was purified after cleavage by HP8 by binding of its carboxyl-terminal 6 × His tag to a nickel-affinity column. SDS-PAGE followed by silver staining demonstrated that this step effectively separated Spätzle-C108 from its pro-domain and the activating proteinases and that it remained as a disulfide linked homodimer (Fig. 7B).
Fig. 7.
(A) Proteolytic activation of proSpätzle by HP8Xa. ProHP8Xa (25 ng) was activated by bovine Factor Xa (50 ng) and then incubated with proSpätzle (100 ng) at 37°C for 1 h. The mixtures were subjected to SDS-PAGE and immunoblotting using Spätzle antibodies. The sizes and positions of molecular weight standards are indicated on the left. Bands representing proSpätzle precursors, cysteine-knot domain (Spätzle-C108), and Spätzle-C108 dimer are marked by arrows. (B) SDS-PAGE analysis of Spätzle-C108. Spätzle-C108 (40 ng) purified after activation by HP8Xa, was treated with SDS sample buffer in the absence or presence of 0.14 M β-mercaptoethanol (β-ME) at 95°C for 5 min and separated by SDS-PAGE followed by silver staining. Sizes and positions of the molecular weight markers are indicated on the left.
M. sexta Spätzle-1 stimulates antimicrobial peptide gene expression
To investigate whether Spätzle-1 plays a role in stimulating the expression of AMP genes in M. sexta, we injected purified Spätzle-C108, proSpätzle-1A, or buffer into fifth instar larvae and 20 h later collected hemolymph to measure antimicrobial activity and protein levels, and we isolated RNA from fat body to measure antimicrobial peptide transcript levels.
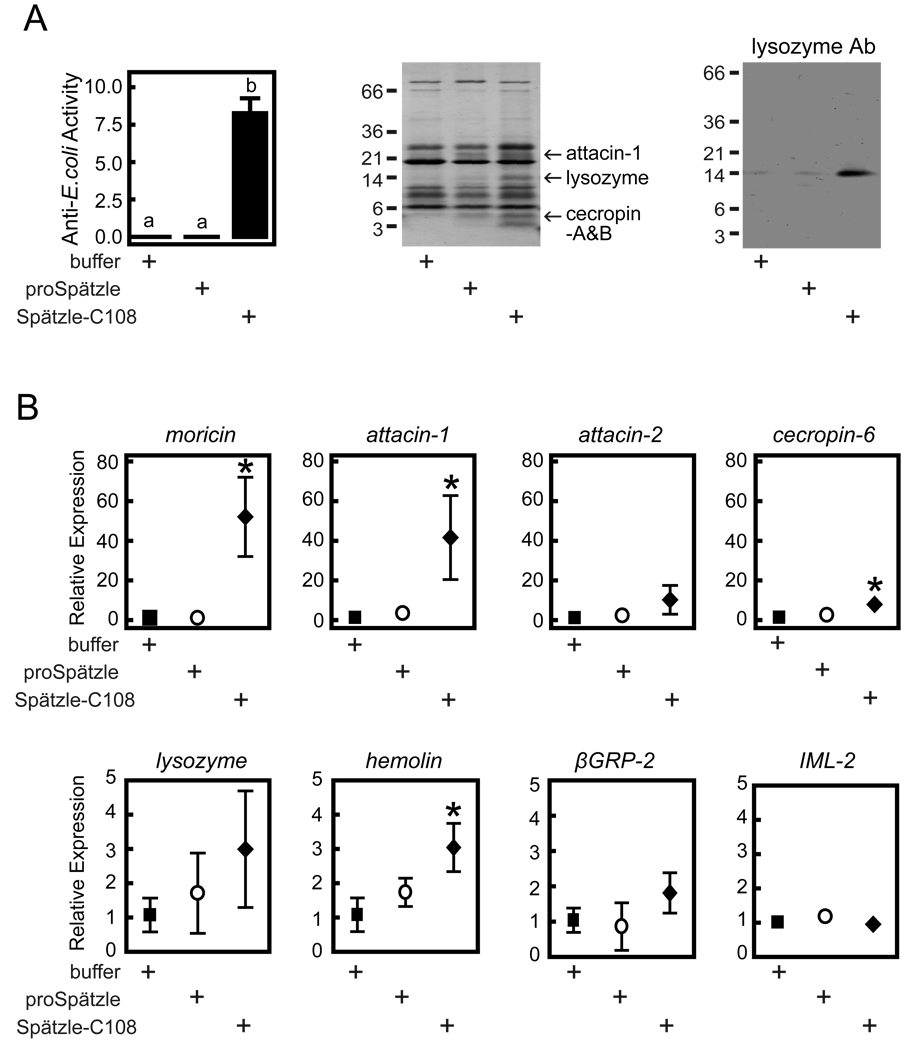
Plasma antimicrobial activity against E. coli was not detected after injection of buffer or proSpätzle-1A but increased significantly after injection with Spätzle-C108 (Fig. 8A). We analyzed heat-stable polypeptides in plasma by SDS-PAGE and identified protein bands that consistently had higher intensities after injection of Spätzle-C108 (Fig. 8A). Analysis of tryptic peptides from these bands by MS/MS and Mascot software identified them as attacin-1, lysozyme, and cecropin-A/B (Table S3). Immunoblot analysis with antibody to M. sexta lysozyme confirmed the elevated level of lysozyme in plasma after injection of Spätzle-C108 (Fig. 8A).
Fig. 8.
Effects of Spätzle injection on the humoral immune response. Fifth instar, day 0 larvae were injected with buffer, proSpätzle-1A, or activated Spätzle-C108. Twenty h later, hemolymph was collected, and fat body RNA samples were prepared from each insect. (A) Antimicrobial activity of plasma assayed against E. coli and identification of antimicrobial plasma proteins by SDS-PAGE and peptide mass fingerprinting or immunoblotting. Sizes and positions of molecular weight standards are indicated on the left. (B) Relative transcript levels for indicated genes were assayed by quantitative RT-PCR as described in Experimental Procedures. Symbols represent mean ± S.D. (n=3). Lack of error bars indicates that S.D. was smaller than the size of the symbol. Asterisks indicate means that are significantly different from the buffer-injected control (one-way ANOVA followed by the Newman-Keuls test, P < 0.05).
Quantitative real-time PCR results revealed increased levels of mRNAs for moricin (50-fold), attacin-1 (40-fold), and cecropin-6 (10-fold) after the injection of Spätzle-C108 compared to the control injections with buffer or proSpätzle-1A (Fig. 8B). Levels of attacin-2 and lysozyme mRNA were higher after Spätzle-C108 injection but did not reach a statistically significant level in this experiment. These results indicate that proSpätzle-1A is not itself active, but that its proteolytic cleavage by HP8 produces Spätzle-C108, which acts as a cytokine to stimulate expression of a set of genes whose products have antimicrobial activity.
Transcript levels for immulectin-2 [33]and β-1,3-glucan recognition protein-2, pattern recognition proteins that are upregulated after injection of bacteria [33,34] were not affected by injection of proSpätzle-1A or Spätzle-C108 (Fig. 8B), and mRNA for hemolin, the most abundant M. sexta plasma protein induced after injection of bacteria [35], increased only 3-fold after injection of Spätzle-C108. Hemolymph concentrations of hemolin and β-1,3-glucan recognition protein-2 were not significantly affected by injection of Spätzle-C108, as detected by immunoblotting (data not shown). Therefore, it appears that Spätzle-C108 signaling may stimulate expression of a sub-set of the genes whose expression is induced by microbial exposure in M. sexta.
Discussion
Progress in understanding the biochemical pathways that operate in innate immune systems requires investigation of molecular function in diverse taxa. We have identified a key cytokine, Spätzle-1, in a lepidopteran insect. The sequence of this protein is weakly conserved in the insects from which it has been characterized (Fig. S2), but it retains a common function in stimulating expression of antimicrobial peptides. It also controls dorsal/ventral patterning in the D. melanogaster embryo, but this role has apparently not been studied yet in other insect species.
Although it is clear that Drosophila Spätzle acts as the ligand of the Toll receptor in two important physiological pathways [16,36], the functions of other homologues, Spz2-6, are still unknown. Phylogenetic analysis indicated that the M. sexta cDNAs isolated in this study belong to the Spätzle-1 clade. The sequences for Spätzle-1 orthologs from different insect species are much less conserved than those of the other groups, with longer branches and lower bootstrap values for the Spätzle-1 clade. It appears that the Spätzle-1 orthologs, which are predicted to have immune functions may, like other genes of the immune system, be subject to positive natural selection, with higher rates of adaptive evolution than most other genes in the genome. For example, persephone, spirit, Toll and necrotic in the Toll pathway, and Imd, Dredd and Relish in the Imd pathway have evolved faster than non-immunity genes [37–39].
We identified two proSpätzle-1 isoforms in M. sexta larval cDNA, which resulted in a ten amino acid residue insertion present in proSpätzle-1B but not in proSpätzle-1A caused by use of two alternate 3’ splice sites (Fig. 1). In the currently available B. mori proSpätzle-1 cDNA and EST sequences, the splicing site is equivalent to that in M. sexta proSpätzle-1, with no evidence for a longer form. Ten splicing isoforms of Drosophila Spätzle occur in the precellular blastoderm embryo [8]. One pair of splicing isoforms appears as D. melanogaster Spätzle 11.7 and 11.15, with amino acid sequences identical except for a nine-residue segment present in 11.7 but not in 11.15 caused by use of an alternative 3’ splice site [8], although at a different position within the pro-region than observed in the M. sexta splicing isoforms. Spätzle 11.15 was as active as isoform 11.7 in restoring ventrolateral pattern elements [8]. How these sequences, which differ in the pro-region rather than the active signaling molecule might differ in function, remains to be explored further.
An unusual 3′ splice site sequence (TG) exists at the end of the alternately spliced intron in pro-Spätzle-1A rather than the consensus (AG), which does occur at the 3’ end of the intron for pro-Spätzle-1B, following the GT-AG splicing rule [40]. Both splicing isoforms were present in RNA samples tested, with proSpätzle-1B more abundant than proSpätzle-1A, indicated that the unusal splicing city may be less preferred. The GT-TG exon-intron boundary is less common but has also been reported in other genes, such as human and Drosophila Gsα (the α subunit of the guanine nucleotide-binding protein) isoforms [41,42].
ProSpätzle-1 mRNA was detected in hemocytes and fat body of M. sexta larvae, with much higher level of expression in hemocytes. We can not exclude the possibility that the signal detected in fat body may have been from contaminating hemocytes. Expression of D. melanogaster Spätzle in hemocytes but not fat body has been reported [43]. In B. mori, Spätzle-1 transcript was observed in fat body and midgut samples, but hemocytes were not tested [23]. M. sexta ProSpätzle-1 expression in hemocytes increased approximately 20-fold by 24 h after injection of larvae with a Gram-positive bacterium or β-1,3-glucan (a component of fungal cell walls) but no significant change was observed after injection of a Gram-negative bacterium. Microarray analysis in D. melanogaster has shown increased Spätzle expression after inoculation with a mixture of M. luteus and E. coli [43, 44], and genetic analysis indicated that induced Spätzle expression was regulated by the Toll but not the imd pathway [44], an indication that Spätzle gene expression may not be stimulated by Gram negative bacteria in D. melanogaster. The enhanced expression of proSpätzle during an infection may lead to increased ability to stimulate production of antimicrobial peptides during an infection, as a type of feed-forward positive regulation.
We previously found that proteinase HP8, which is most similar to D. melanogaster proSpätzle activating proteinases Easter and SPE, could stimulate expression of antimicrobial peptide genes when injected into M. sexta larvae [31]. These observations led us to test a hypothesis that HP8 can process proSpätzle-1 to release a carboxyl-terminal fragment that forms the active cystine knot cytokine. HP8 cleaved proSpätzle-1 with specificity at the expected position, based on sequence alignment with other proSpätzle-1 sequences, to release the Spätzle-C108 disulfide-linked homodimer. The sequence around the activation cleavage site of proSpätzle-1 from different species is relatively well conserved, suggesting that this may be required to allow specific recognition by the activating proteinase. Demonstration that the Spätzle-C108 fragment produced by HP8 is active as a cytokine to stimulate expression of antimicrobial peptide genes, along with previous results showing that the Persephone ortholog HP6 can activate proHP8 [31], leads to the following model for an extracelluar immune-stimulatory pathway in M. sexta. ProHP6 is activated in hemolymph upon exposure to Gram-positive or Gram-negative bacteria or β-1,3-glucan [31]. HP6 then cleaves and activates proHP8, which in turn cleaves and activates proSpätzle-1. The Spätzle-C108 dimer then binds to a Toll receptor in fat body cytoplasmic membrane to trigger an intracellular signal transduction pathway leading to activation of rel-family transcription factors that stimulate transcription of antimicrobial peptide genes. A Toll cDNA from M. sexta has been identified [45], and role of this protein as a Spätzle-1 receptor needs to be examined. Upstream of the components characterized to date, the proteinase that activates proHP6 is still undiscovered, and pattern recognition proteins that may trigger this pathway have not yet been identified.
Even though the activation and function of M. sexta proSpätzle-1 have similarities with the pathways characterized in D. melanogaster and T. molitor, there are also some notable differences. Exposure to β-1,3-glucan or to dead E. coli or M. luteus leads to proHP6 activation and antimicrobial peptide synthesis in M. sexta, suggesting the existence of endogenous pattern recognition factors and a proHP6-activating proteinase in plasma. However, D. melanogaster Persephone, a putative ortholog of M. sexta HP6 [31], activates the Toll pathway after it is cleaved by fungal or bacterial proteinases [46,47]. In T. molitor, a three-component pathway generates active SPE that can activate both proSpätzle and prophenoloxidase [24,25]. In contrast, in M. sexta, activation proSpätzle and prophenoloxidase is carried out by different clip-domain proteinases, which are activated in separate cascade pathways [28,31].
In conclusion, the results presented here support a conclusion that the function of the cytokine Spätzle-1 is conserved in the immune system of a lepidopteran insect, suggesting that a cytokine-activated Toll pathway is an ancient feature of innate immunity in insects. Although the families of extracellular molecules involved in this pathway are conserved between Diptera, Coleoptera, and Lepidoptera, there are interesting variations in how the pathways are initiated by recognition of microbial patterns and by microbial proteinases. Further biochemical and genetic research is required for more complete understanding of the extracellular reactions of plasma proteins that regulate innate immune responses.
Experimental procedures
Insect rearing
M. sexta eggs were originally purchased from Carolina Biological Supplies. The larvae were reared on an artificial diet under conditions described previously [48].
DNA sequencing
DNA sequences were determined using an Applied Biosystems 3730 DNA Analyzer in the DNA Sequencing and Genotyping Facility at Kansas State University.
Cloning of proSpätzle cDNAs
A 130-bp expressed sequence tag (contig 4514) obtained through pyrosequencing of M. sexta fat body and hemocyte cDNA [32] encoded a protein fragment 46% identical to B. mori Spätzle-1. Based on this fragment, primers (Table S1) were synthesized for rapid amplification of cDNA ends (RACE), which was carried out using a GeneRacer kit (Invitrogen) with cDNA from the fat body of fifth instar M. sexta larvae collected 24 h after injection with 100 µl of Micrococcus luteus (1 µg/µl). The resulting products were cloned into TOPO PCR 4.0 T-vector, and their sequences were determined. cDNAs encompassing the entire reading frame of M. sexta Spätzle-1 were amplified from larval fat body cDNA by using primers encoding the start and stop codon regions (Table S1). The products were cloned into TOPO PCR 4.0 T-vector, and the nucleotide sequences were confirmed by DNA sequencing.
Amplification and sequencing of M. sexta genomic DNA
Primer pairs designed from the Spätzle-1 cDNA sequence were used to amplify corresponding fragments of M. sexta genomic DNA (Table S1). These were cloned into TOPO PCR 4.0 T-vector and sequenced.
Sequence analysis
The program Splign [49] was used to assign intron/exon boundaries by comparison of the genomic and cDNA sequences. Analysis of the amino acid sequences deduced from the cDNA, including prediction of signal peptide and glycosylation sites, was carried out in the ExPASy (Expert Protein Analysis System) proteomics server (http://www.expasy.org).
The deduced amino acid sequence of M. sexta Spätzle-1 was used to search the non-redundant database from NCBI and sequences from the Human Genome Sequencing Center at Baylor College of Medicine by using the TBLASTN program [50]. Similar protein sequences retrieved from GenBank or deduced from the assembled contigs from insect genomes were aligned with M. sexta Spätzle-1 sequence using Clustal W program. Phylogenetic trees were constructed by the neighbor-joining method with a Poisson correction model using the MEGA version 3.1 software [51]. For neighbor-joining method, gaps were treated as characters and statistical analysis was performed by the bootstrap method using 1,000 repetitions.
Quantitative RT-PCR analysis of Spätzle-1 mRNA level
Fifth instar day 2 larvae were injected with formalin-killed Escherichia coli XL1-Blue, Micrococcus luteus, curdlan, or with water as a control as described previously [31]. At 24 h after injection, total RNA was prepared from fat body and hemocytes, and first-strand cDNA was synthesized as described previously [31]. Each cDNA sample (diluted 1:500 for hemocyte cDNA or 1:25 for fat body cDNA) was used as template for quantitative RT-PCR analysis. The M. sexta ribosomal protein S3 (rpS3) mRNA was used as an internal standard to normalize the amount of RNA template. The primer pairs used are listed in Table S2. The thermal cycling conditions and calculations were as described previously [31].
Antiserum preparation
A cDNA fragment encoding the last 108 amino acid residues of M. sexta proSpätzle-1 (Spätzle-C108) was amplified by PCR using cDNA from day 2 fifth instar larval hemocytes (collected 24 h after injection of 100 µg curdlan) and primers listed in Table S1. The forward primer, corresponding to nucleotides 774–786 of the cDNA, also contained an NcoI restriction site. The reverse primer, corresponding to nucleotides 1084–1099 of the cDNA, included codons for six histidine residues, followed by a stop codon and an XhoI site. The PCR product was cloned into plasmid TOPO PCR 4.0 T-vector (Invitrogen) and then digested with NcoI and XhoI. The cDNA fragment was subcloned into the same restriction sites in the expression vector pET-28a (Novagen). After sequence verification, the resulting plasmid was used to transform E. coli strain BL21(DE3). For recombinant protein expression, these bacteria were grown at 30°C in LB medium containing 50 µg/ml of kanamycin. When A600 of the culture reached 1.0, D-sorbitol was added to the culture to a final concentration of 100 mM. Thirty min later, isopropyl β-D-thiogalactoside was added at 0.5 mM, and the recombinant protein was expressed for 6 h at 30°C. The bacteria were harvested by centrifugation and resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 8.0). Cells were lysed by sonication, and a cleared lysate was obtained by centrifugation. The soluble Spätzle-C108 in the supernatant was purified by nickel-nitrilotriacetic acid (NTA) agarose chromatography as described by Jiang et al. [52]. Spätzle-C108 was further purified by preparative 12 % SDS-polyacrylamide gel electrophoresis (PAGE) and used as antigen for the production of a rabbit polyclonal antiserum (Cocalico Biologicals).
SDS-PAGE and immunoblot analysis
Protein samples were treated with 6 × SDS sample buffer with or without β-mercapoethanol at 95°C for 5 min and then separated by SDS-PAGE using 4–12% NuPAGE Bis-Tris gels (Invitrogen). Gels were stained with silver nitrate [53]. Immunoblot analysis was carried out using 1:1000 diluted antiserum to M. sexta Spätzle-C108 as the primary antibody. Antibody binding was visualized using alkaline phosphate-conjugated goat anti-rabbit IgG and alkaline phosphate substrate kit (Bio-Rad).
Expression and purification of recombinant proSpätzle-1A
The entire M. sexta proSpätzle-1A coding region including the signal peptide was amplified by PCR using forward and reverse primers described in Table S1, to include an SpeI site at the 5’ end and codons for an in-frame six-histidine sequence followed by a stop codon and an XhoI site at the 3’ end. The PCR product was recovered by agarose gel electrophoresis, digested with SpeI and XhoI and then inserted into the same restriction sites in the vector pFastBac1 (Invitrogen). The resulting plasmid, after sequence verification, was used to generate a recombinant baculovirus according to the manufacturer’s instructions (Invitrogen).
To express proSpätzle, Sf9 cells (2 × 106 cells/ml) in 800 ml of Sf-900 II serum-free medium (Invitrogen) were infected with the recombinant baculovirus at multiplicity of infection of 2 and were incubated at 28°C with shaking at 150 rpm. The culture was harvested at 48 h post infection, and cells were removed by centrifugation at 5000×g for 20 min at 4°C. The cell-free medium was incubated for one h at 4°C with 2 ml of Ni-NTA agarose (Qiagen) equilibrated with initial buffer (20 mM Tris-HCl, 200 mM NaCl, pH 8.0). Ni-NTA agarose was then packed into a column (1.5 cm × 1 cm), which was washed with buffer (20 mM Tris-HCl, 200 mM NaCl, 20 mM imidazole, pH 8.0) until the A280 of the effluent was near 0. The bound proteins were sequentially eluted with one ml aliquots of the initial buffer containing 50 mM, 100 mM, or 250 mM imidazole. Fractions containing recombinant proSpätzle were pooled and dialyzed against buffer (20 mM Tris-HCl, 20 mM NaCl, pH 8.0), and then applied to a pre-equilibrated Q-Sepharose Fast Flow column (1 cm × 2.5 cm). The column was washed with the dialysis buffer until the absorbance at 280 nm was near 0 and then eluted with a linear gradient of NaCl (20–500 mM, 40 ml total) in 20 mM Tris-HCl, pH 8.0, at 1 ml/min. Fractions of one ml were collected and analyzed by SDS-PAGE.
Production, purification and activation of a mutant of M. sexta hemolymph proteinase-8 (HP8Xa)
The entire coding region of proHP8 inserted into plasmid vector pMT/V5-His A (Invitrogen) [31] was used as a template to produce mutant proHP8 (proHP8Xa) plasmid according to the instructions of Quick Change® Multi Site-Directed Mutagenesis Kit (Stratagene). The cleavage activation site of proHP8, NNDR90 was replaced with IEGR90, the preferred cleavage site for bovine Factor Xa, by using the mutagenic oligonucleotide primer (5′-TGCGGCATTCAAATCGAGGGCAGAATTGTTGGAGG-3′, sequence encoding IEGR underlined). After DNA sequence verification, the plasmid was used to transfect Drosophila S2 cells and produce the mutant protein, proHP8Xa, from a stable cell line, following the manufacturer’s instructions (Invitrogen).
The S2 culture medium was collected at 48 h after induction with CuSO4 at a final concentration of 500 µM. ProHP8Xa was secreted into cell culture medium under control of its own signal peptide. The secreted recombinant proHP8Xa was purified following the method described previously [31]. To determine the amino-terminal sequence of a truncated band visible after SDS-PAGE under reducing conditions, the protein was transferred onto a polyvinylidene difluoride (PVDF) membrane and stained with 40% methanol containing 0.025% Coomassie Brilliant Blue R-250. After de-staining with 50% methanol, the band corresponding to the truncated proHP8Xa was excised and subjected to Edman degradation sequencing at the W. M. Keck Facility at Yale University.
To test whether proHP8Xa could be activated by Factor Xa, 50 ng of purified recombinant proHP8Xa was incubated with 100 ng of bovine Factor Xa (New England BioLabs) in the reaction buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween-20) at 37°C for 6 h. The mixtures were separated by SDS-PAGE using NuPAGE 4–12% Bis-Tris gels (Invitrogen) followed by immunblotting with antiserum against M. sexta HP8 (diluted 1:2000) as the primary antibody. The activation of proHP8Xa was confirmed by measuring the amidase activity of activated HP8Xa with 200 µl of 50 µM acetyl-Ile-Glu-Ala-Arg-p-nitroanilide (IEARpNa) in 0.1 M Tris-HCl, pH 8.0, 0.1 M NaCl, 5 mM CaCl2 as colorimetric substrate. The amidase activity was measured by monitoring A405 in a microplate reader (Bio-Tek Instrument, Inc.). One unit of amidase activity was defined as ΔA405/min=0.001.
Activation of recombinant proSpätzle by HP8Xa
To test the ability of HP8Xa to cleave proSpätzle, 25 ng of Factor Xa-activated HP8Xa or 50 ng of Factor Xa alone as a control was incubated with 100 ng of proSpätzle in the presence of 20 mM imidazole at 37°C for 1 h. The reaction mixtures were separated by SDS-PAGE using NuPAGE 4–12% Bis-Tris gel (Invitrogen) followed by immunoblotting with antiserum against Spätzle-C108 as primary antibody. The cleavage site of proSpätzle was determined by Edman sequencing as described above for truncated proHP8Xa.
To obtain active spätzle for injection into larvae to test biological activity, 100 µg of purified proSpätzle-1 A (20 ng/µl) was activated as described above and diluted in 10 ml of 20 mM Tris-HCl, 200 mM NaCl, pH 8.0, then incubated with 100 µl of Ni-NTA agarose at 4°C for 1 h. The Ni-NTA agarose was collected by centrifugation at 500 × g for 5 min at 4°C, and washed twice with 1 ml of 20 mM Tris-HCl, 200 mM NaCl, pH 8.0. The bound Spätzle-C108 was sequentially eluted three times with 200 µl aliquots of the same buffer containing 20 mM, 50 mM, and 100 mM imidazole. The eluted fractions were analyzed by SDS-PAGE followed by silver staining [52].
Effects of Spätzle on antimicrobial peptide gene expression
Day 0 fifth instar larvae were injected with filtered buffer (20 mM Tris-HCl, 200 mM NaCl, 20 mM imidazole, pH 8.0) (100 µl/larva) as negative control, purified proSpätzle (100 µl/larva, 30 ng/µl), or purified Spätzle-C108 (100 µl/larva, 10 ng/µl, 3 larvae) derived from cleavage of proSpätzle-1A by HP8Xa and re-purified by nickel-affinity chromatography as described above. Twenty hours later, fat body and hemolymph samples were collected. Total RNA samples were prepared from fat body and cDNA was prepared as described previously [31]. Cell-free hemolymph samples were heated at 95°C for 5 min to remove most high molecular weight proteins and then centrifuged at 10,000×g for 5 min. The supernatant was stored at −20°C. Quantitative real-time PCR, identification of plasma proteins by mass spectrometry, and assay of antimicrobial activity against E. coli strain XL1-Blue were carried out as described previously [31].
Supplementary Material
Acknowledgements
We thank Dr. Peter Dunn for antiserum to M. sexta lysozyme. This work was supported by National Institutes of Health Grants GM41247 (to MK) and GM58643 (to HJ). This is contribution 10-009-J from the Kansas Agricultural Experiment Station. Protein digestion and mass spectrometry was performed by the Nevada Proteomics Center at the University of Nevada, which is supported by P20 RR-016464 from the INBRE Program of the National Center for Research Resources.
Abbreviations
- Spz
Spätzle
- SPE
Spätzle processing enzyme
- HP6
hemolymph proteinase 6
- HP8
hemolymph proteinase 8
- RT-PCR
reverse transcriptase-polymerase chain reaction
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis.
Footnotes
References
- 1.Kanost MR, Gorman MJ. Phenoloxidase in insect immunity. In: Beckage N, editor. Insect Immunology. San Diego: Academic Press/Elsevier; 2008. pp. 69–96. [Google Scholar]
- 2.Cerenius L, Lee BL, Soderhall K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Prakash O, Kanost MR. Insect ENF family peptides with diverse biological activities display similar core structures in solution. Medical Chem. Res. 2001;10:493–501. [Google Scholar]
- 4.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat.Rev.Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie JP, Kanost MR, Trenczek T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997;42:611–643. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 6.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLotto Y, DeLotto R. Proteolytic processing of the Drosophila Spatzle protein by easter generates a dimeric NGF-like molecule with ventralising activity. Mech. Dev. 1998;72:141–148. doi: 10.1016/s0925-4773(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 8.DeLotto Y, Smith C, DeLotto R. Multiple isoforms of the Drosophila Spatzle protein are encoded by alternatively spliced maternal mRNAs in the precellular blastoderm embryo. Mol. Gen. Genet. 2001;264:643–652. doi: 10.1007/s004380000350. [DOI] [PubMed] [Google Scholar]
- 9.Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat. Immunol. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann A, Funkner A, Neumann P, Juhnke S, Walther M, Schierhorn A, Weininger U, Balbach J, Reuter G, Stubbs MT. Biophysical characterization of refolded Drosophila Spatzle, a cystine knot protein, reveals distinct properties of three isoforms. J. Biol. Chem. 2008;283:32598–32609. doi: 10.1074/jbc.M801815200. [DOI] [PubMed] [Google Scholar]
- 11.Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, Kambris Z, Brun S, Hashimoto C, Ashida M, Brey PT, Lee WJ. A Spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev.Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem. Mol. Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 13.LeMosy EK, Tan YQ, Hashimoto C. Activation of a protease cascade involved in patterning the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5055–5060. doi: 10.1073/pnas.081026598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Yagi Y, Tanji T, Zhou S, Ip YT. Multimerization and interaction of Toll and Spatzle in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9369–9374. doi: 10.1073/pnas.0307062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangloff M, Murali A, Xiong J, Arnot CJ, Weber AN, Sandercock AM, Robinson CV, Sarisky R, Holzenburg A, Kao C, Gay NJ. Structural insight into the mechanism of activation of the Toll receptor by the dimeric ligand Spatzle. J. Biol. Chem. 2008;283:14629–14635. doi: 10.1074/jbc.M800112200. [DOI] [PubMed] [Google Scholar]
- 16.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 17.Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, Lee WJ, Ueda R, Lemaitre B. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr. Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Parker JS, Mizuguchi K, Gay NJ. A family of proteins related to Spatzle, the toll receptor ligand, are encoded in the Drosophila genome. Proteins. 2001;45:71–80. doi: 10.1002/prot.1125. [DOI] [PubMed] [Google Scholar]
- 19.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 20.Shin SW, Bian G, Raikhel AS. A toll receptor and a cytokine, Toll5A and Spz1C, are involved in toll antifungal immune signaling in the mosquito Aedes aegypti. J. Biol. Chem. 2006;281:39388–39395. doi: 10.1074/jbc.M608912200. [DOI] [PubMed] [Google Scholar]
- 21.Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost M, Thompson GJ, Zou Z, Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tribolium Genome Sequencing Consortium. Richards S, Gibbs RA, Weinstock GM, Brown SJ, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Cheng T, Rayaprolu S, Zou Z, Xia Q, Xiang Z, Jiang H. Proteolytic activation of pro-spatzle is required for the induced transcription of antimicrobial peptide genes in lepidopteran insects. Dev. Comp. Immunol. 2007;31:1002–1012. doi: 10.1016/j.dci.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, Yang Y, Park JW, Lee HH, Ha NC, Kang HJ, Nonaka M, Soderhall K, Lee BL. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J. Biol. Chem. 2008;283:7599–7607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- 25.Kan H, Kim CH, Kwon HM, Park JW, Roh KB, Lee H, Park BJ, Zhang R, Zhang J, Soderhall K, Ha NC, Lee BL. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J. Biol. Chem. 2008;283:25316–25323. doi: 10.1074/jbc.M804364200. [DOI] [PubMed] [Google Scholar]
- 26.Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 27.Ragan EJ, An C, Jiang H, Kanost MR. Roles of haemolymph proeins in antimicrobial defences of Manduca sexta. In: Rolff J, Reynolds SE, editors. Insect Infection and Immunity. Oxford University Press; 2009. pp. 34–48. [Google Scholar]
- 28.Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- 29.Kanost MR, Dai W, Dunn PE. Peptidoglycan fragments elicit antibacterial protein synthesis in larvae of Manduca sexta. Arch. Insect Biochem. Physiol. 1988;8:147–164. [Google Scholar]
- 30.Kanost MR, Nardi JB. Innate immune responses of Manduca sexta. In: Goldsmith MR, Marec F, editors. Molecular Biology and Genetic of Lepidoptera. CRC press; 2009. in press. [Google Scholar]
- 31.An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca sexta Hemolymph Proteinases HP6 and HP8 in Two Innate Immune Pathways. J. Biol. Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem. Mol. Biol. 2008;38:677–682. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu XQ, Kanost MR. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 2000;275:37373–37381. doi: 10.1074/jbc.M003021200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Ma C, Lu ZQ, Kanost MR. Beta-1,3-glucan recognition protein-2 (betaGRP-2)from Manduca sexta; an acute-phase protein that binds beta-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem. Mol. Biol. 2004;34:89–100. doi: 10.1016/j.ibmb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Ladendorff NE, Kanost MR. Bacteria-induced protein P4 (hemolin) from Manduca sexta: a member of the immunoglobulin superfamily which can inhibit hemocyte aggregation. Arch. Insect Biochem. Physiol. 1991;18:285–300. doi: 10.1002/arch.940180410. [DOI] [PubMed] [Google Scholar]
- 36.Morisato D, Anderson KV. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 1995;29:371–399. doi: 10.1146/annurev.ge.29.120195.002103. [DOI] [PubMed] [Google Scholar]
- 37.Begun DJ, Whitley P. Adaptive evolution of relish, a Drosophila NF-kappaB/IkappaB protein. Genetics. 2000;154:1231–1238. doi: 10.1093/genetics/154.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiggins FM, Kim KW. A screen for immunity genes evolving under positive selection in Drosophila. J. Evol. Biol. 2007;20:965–970. doi: 10.1111/j.1420-9101.2007.01305.x. [DOI] [PubMed] [Google Scholar]
- 40.Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu. Rev. Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 41.Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Isolation and characterization of the human Gs alpha gene. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2081–2085. doi: 10.1073/pnas.85.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quan F, Forte MA. Two forms of Drosophila melanogaster Gs alpha are produced by alternate splicing involving an unusual splice site. Mol. Cell. Biol. 1990;10:910–917. doi: 10.1128/mcb.10.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell. Microbiol. 2005;7:335–350. doi: 10.1111/j.1462-5822.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 44.De Gregario E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophlia immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ao JQ, Ling E, Yu XQ. A Toll receptor from Manduca sexta is in response to Escherichia coli infection. Mol. Immunol. 2008;45:543–552. doi: 10.1016/j.molimm.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 46.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of 'danger signals' and pathogen-associated molecular patterns defines binary signaling pathways 'upstream' of Toll. Nat. Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl. Acad. Sci. U. S. A. 2009 doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn PE, Drake D. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm, Manduca sexta. J. Invertebr. Pathol. 1983;41:77–85. [Google Scholar]
- 49.Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol. Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 52.Jiang H, Mulnix AB, Kanost MR. Expression and characterization of recombinant Manduca sexta serpin-1B and site-directed mutants that change its inhibitory selectivity. Insect Biochem. Mol. Biol. 1995;25:1093–1100. doi: 10.1016/0965-1748(95)00042-9. [DOI] [PubMed] [Google Scholar]
- 53.Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT. Current protocols in protein science. John Wiley & Sons, Inc.; 1995. pp. 10.5.6–10.5.7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.