Abstract
Type I diabetes (T1D) results from interactions between environmental exposures and genetic susceptibility leading to immune dysfunction and destruction of the insulin-producing β cells of the pancreas. Vitamin D deficiency is likely to be one of the many environmental factors influencing T1D development and diagnosis, and, hence, the hormone receptor gene, VDR, was examined for association with T1D risk. The Type I Diabetes Genetics Consortium genotyped 38 single nucleotide polymorphisms (SNPs) in 1654 T1D nuclear families (6707 individuals, 3399 affected). Genotypes for 38 SNPs were assigned using the Illumina (ILMN) and Sequenom (SQN) technology. The analysis of data release as of July 2008 is reported for both platforms. No evidence of association of VDR SNPs with T1D at P < 0.01 was obtained in the overall sample set, nor in subgroups analyses of the parent-of-origin, sex of offspring and HLA risk once adjusted for multiple testing.
Keywords: autoimmune disease, VDR, genetic susceptibility, vitamin D, single nucleotide polymorphism, type I diabetes
Introduction
Type I diabetes (T1D) is considered to be an autoimmune disease but its precise cause is unknown. Familial clustering and genetic susceptibility have been associated with strong association of the major histocompatibility complex (MHC) class II genotypes with T1D, as well as variants of other MHC genes and many additional susceptibility loci across the genome, including functional candidate genes in immune genes.1,2 Amongst the non-MHC candidate genes, the vitamin D receptor locus (VDR) is a functional candidate, owing to its functionality in the immune system and evidence for a protective function of vitamin D in the experimental non-obese diabetic (NOD) mouse model and in human studies.3–6 As vitamin D deficiency has been described in T1D, seasonal variation as well as vitamin D status at T1D manifestation have been correlated.7–9 This has also supported a general recommendation for vitamin D supplementation.10 VDR knockout mousemodels have been found to differ for key immune pathways that make the vitamin D system attractive for an intervention strategy.11,12
Many association studies of VDR and T1D have been performed with positive or null results.10–17 One case report of T1D occurring in a child with a compound heterozygous VDR mutation that dissociates the promoter and the CYP24 enzyme activation of the receptor illustrates an experiment of nature with the vitamin D system.18 The human VDR gene region is located on chromosome 12q13 spanning over 80 kb. In the Type I Diabetes Genetics Consortium (T1DGC) affected sib-pair (ASP) family collection, a total of 38 SNPs in the VDR region, from the 5′ flanking to 3′ flanking sites19 were evaluated in 1654 ASP families, which were retrieved from the database 2008.07.RR without any further selection.
Results
Illumina (ILMN) genotyping platform
In the T1DGC ASP families that had VDR single nucleotide polymorphisms (SNPs) genotyped using the Illumina GoldenGate assay, an association with T1D (P < 0.10) was observed for rs1859281 (P = 0.0531), rs6580639 (P = 0.05), and rs2239186 (P = 0.07). In the four regional datasets, differences in the transmission rates were observed for rs11608702 in the Asian-Pacific region (paternal transmission, P = 0.03), rs7968585 (P = 0.04) in all Asian-Pacific transmissions, rs6580639 (P = 0.02) in all UK + Sardinian families and in UK + Sardinian maternal transmissions (P = 0.02), rs7975232 in Europe in all transmission (P = 0.03) and in European paternal transmissions (P = 0.04). The rs7975232 SNP also exhibited differences in all North America (P = 0.04) and maternal transmissions (P = 0.02), as well as rs11574113 in all North America transmissions (P = 0.04), rs2189480 in European maternal transmissions (P = 0.04), rs3819545 in UK + Sardinian paternal transmissions (P = 0.05), rs2239186 for maternal transmission in the Asian-Pacific network (P = 0.034), rs2254210 in Europe (P = 0.04), rs11168287 in North America for maternal transmissions (P = 0.03), rs11574026 in Europe for paternal (P = 0.0061), and rs12721377 in Europe (P = 0.04) and in UK + Sardinian (all transmissions, P = 0.04; maternal transmissions, P = 0.0020). These results are summarized in Table 1.
Table 1.
ILMN platform: VDR SNPs showing P < 0.1 in one of the four T1DGC networks
| ID | Marker | Regional datasets |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asian-Pacific |
European |
North American |
UK, Sardinian |
|||||||||||||
|
P-value |
P-value |
P-value |
P-value |
P-value |
||||||||||||
| all | pat. | mat. | all | pat. | mat. | all | pat. | mat. | all | pat. | mat. | all | pat. | mat. | ||
| 1 | rs1859281 | 0.0531 | 0.1302 | 0.1894 | 0.5588 | 0.0653 | 0.4373 | 0.4489 | 0.8383 | 0.3906 | 0.0869 | 0.0837 | 0.4676 | 0.5244 | 0.6963 | 0.2087 |
| 4 | rs11608702 | 0.2950 | 0.4585 | 0.4052 | 0.1674 | 0.0296 | 1.0000 | 0.8045 | 0.6982 | 1.0000 | 0.6283 | 0.9638 | 0.4895 | 0.6362 | 0.9130 | 0.4131 |
| 6 | rs7968585 | 0.2594 | 0.3157 | 0.4914 | 0.0391 | 0.2018 | 0.0760 | 0.8419 | 0.9190 | 0.6998 | 0.7591 | 1.0000 | 0.6526 | 0.2014 | 0.1726 | 0.5798 |
| 7 | rs6580639 | 0.0541 | 0.1973 | 0.1198 | 0.4854 | 0.8149 | 0.3583 | 1.0000 | 0.4577 | 0.4529 | 0.1187 | 0.7062 | 0.0755 | 0.0199 | 0.1881 | 0.0223 |
| 9 | rs7975232 | 0.7926 | 0.5374 | 0.8487 | 0.2674 | 0.5892 | 0.2125 | 0.0319 | 0.0389 | 0.1783 | 0.0135 | 0.1153 | 0.0205 | 0.4522 | 0.4562 | 0.6713 |
| 10 | rs11574113 | 0.2290 | 0.6129 | 0.1970 | 0.4925 | 0.8857 | 0.1981 | 0.9138 | 0.3045 | 0.3637 | 0.0418 | 0.1544 | 0.1218 | 0.6090 | 0.7485 | 0.6673 |
| 22 | rs2189480 | 0.2986 | 0.6356 | 0.2793 | 1.0000 | 0.7473 | 0.7761 | 0.1124 | 0.7472 | 0.0417 | 0.9536 | 0.7899 | 0.8632 | 0.6502 | 0.2818 | 0.7190 |
| 23 | rs3819545 | 0.8840 | 0.9108 | 0.9141 | 0.5060 | 0.6713 | 0.1778 | 0.6546 | 0.7874 | 0.6869 | 0.4596 | 0.2788 | 0.9649 | 0.2005 | 0.0480 | 0.9575 |
| 25 | rs2239186 | 0.0773 | 0.3227 | 0.0945 | 0.2426 | 0.7087 | 0.0343 | 0.1432 | 0.4210 | 0.1557 | 0.6832 | 0.7959 | 0.7238 | 0.0635 | 0.0905 | 0.2584 |
| 27 | rs2254210 | 0.2272 | 0.4539 | 0.2906 | 0.1589 | 0.1187 | 0.5606 | 0.0422 | 0.2900 | 0.0519 | 0.5646 | 0.4529 | 0.8984 | 0.8053 | 0.6233 | 0.9173 |
| 31 | rs11168287 | 0.4162 | 0.8518 | 0.1671 | 0.8990 | 0.8415 | 0.7131 | 0.3720 | 0.4728 | 0.5300 | 0.1640 | 0.8969 | 0.0268 | 0.3816 | 0.6834 | 0.3691 |
| 34 | rs11574026 | 0.7819 | 0.2126 | 0.3888 | 0.8496 | 0.8839 | 0.6756 | 0.2123 | 0.0061 | 0.3469 | 0.1368 | 0.5094 | 0.1170 | 0.2869 | 0.7080 | 0.2338 |
| 37 | rs12721377 | 0.6081 | 0.3150 | 0.0939 | 0.3398 | 0.2458 | 0.7367 | 0.0363 | 0.0970 | 0.1521 | 0.3758 | 0.9055 | 0.1832 | 0.0386 | 1.0000 | 0.0020 |
Abbreviations: ILMN, Illumina; SNP, single nucleotide polymorphism; T1DGC, Type I Diabetes Genetics Consortium; VDR, vitamin D receptor.
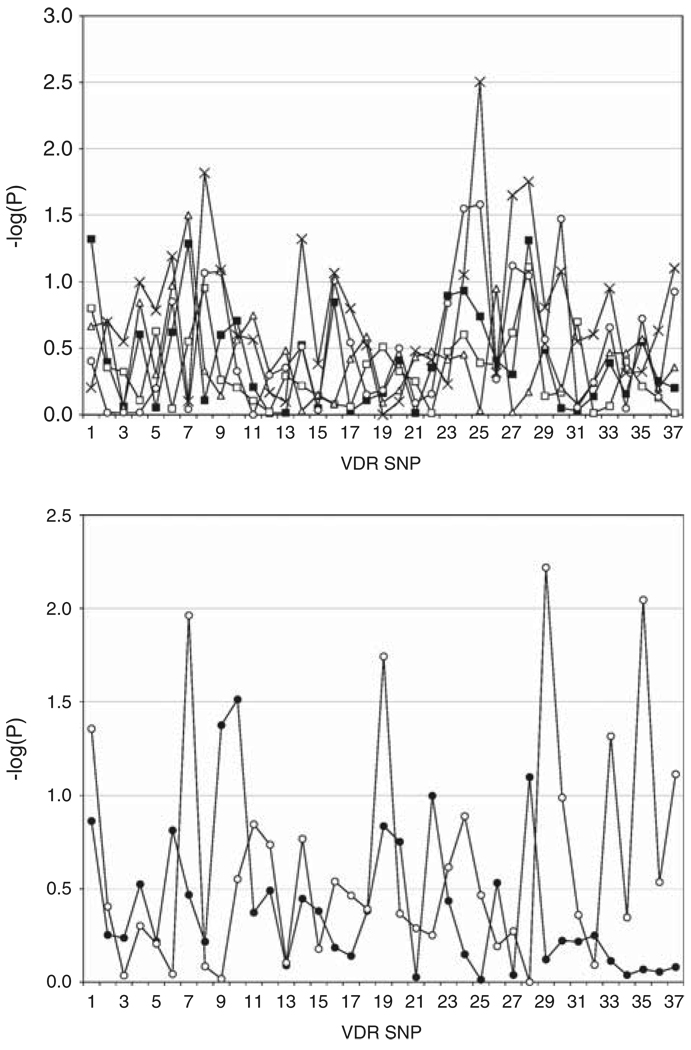
Stratification by HLA risk genotype showed distorted transmission of rs2254210 (−log P = 2.5) in the HLA DR X/X (neither DR3 nor DR4) subgroup. The rs6580639 SNP also had a distorted transmission (−log P = 2.0), as did rs10783219 (−log P = 2.0). All stratified results (−log P-values) for the 38 VDR SNPs are provided in Figure 1. The gender-stratified analyses identified two SNPs, the synonymous BsmI (P = 0.0386) and rs4760648 (P = 0.045) with associations in females; rs6580639 exhibited an association with T1D in males (P = 0.02) (Table 2).
Figure 1.
Sib-pair families were stratified according to the affected sibs’ status at MHC (a) or gender (b). The pedigree disequilibrium test (PDT) results were determined for all 38 SNPs and converted to −log P-values. (a) HLA subgroups: all sibs (filled squares); DR3/4 sibs (open squares); DR3/X sibs (open circles); DR4/X (open triangles) and DRX/X (crosses). (b) Subgroups by gender: females (filled circles), males (open circles).
Table 2.
SQN platform: association analyses of SNPs in families in subgroups of parent-of-origin and by sex of sibling
| ID | Marker | Allele | Freq. % | T% | P-value | Parental sex |
Sibling sex |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Father |
Mother |
Son |
Daughter |
||||||||||||||
| Freq. % | T % | P-value | Freq. | T % | P-value | Freq. % | T % | P-value | Freq. % | T % | P-value | ||||||
| 7 | rs6580639 | C | 3.4 | 44.4 | 0.01208 | 3.6 | 43.9 | 0.0558 | 2.8 | 44.0 | 0.0791 | 3.4 | 42.5 | 0.0151 | 3.4 | 46.5 | 0.2788 |
| T | 96.6 | 50.2 | 96.4 | 50.2 | 97.2 | 50.2 | 96.6 | 50.3 | 96.6 | 50.1 | |||||||
| 26 | rs10735810 | A | 38.6 | 48.7 | 0.02112 | 37.2 | 48.5 | 0.0815 | 37.2 | 48.6 | 0.0818 | 38.2 | 48.5 | 0.0510 | 39.0 | 49.0 | 0.1963 |
| G | 61.4 | 50.8 | 62.8 | 50.9 | 62.8 | 50.9 | 61.8 | 50.9 | 61.0 | 50.7 | |||||||
| 28 | rs2238136 | A | 25.6% | 48.8 | 0.1142 | 25.4 | 48.6 | 0.2111 | 22.2 | 48.7 | 0.2577 | 25.8 | 49.9 | 0.8923 | 25.4 | 47.7 | 0.0304 |
| G | 74.4 | 50.4 | 74.6 | 50.5% | 77.8 | 50.4 | 74.2 | 50.0 | 74.6 | 50.8 | |||||||
| 30 | rs4760648 | A | 41.4 | 51.2 | 0.01831 | 40.5 | 51.6 | 0.0474 | 40.1 | 51.2 | 0.1133 | 41.3 | 50.5 | 0.4714 | 41.5 | 52.0 | 0.0074 |
| G | 58.6 | 49.1 | 59.5 | 48.9 | 59.9 | 49.2 | 58.7 | 49.6 | 58.5 | 48.6 | |||||||
Abbreviations: Freq., overall allele frequency in all family members; SNP, single nucleotide polymorphism; T %, transmission rate.
Sequenom (SQN) genotyping platform
A second data release of the VDR SNP data (July 2008) provided a number corrected genotyping calls. The combined dataset showed nominal results for the three SNPs rs6580639, rs10735810 (a synonymous FokI), and rs4760648. The SNP rs6580639 showed an overall P-value of 0.0269, with significant distortion in males (P = 0.0054). SNP rs10735810 (FokI) exhibited an overall P = 0.0170, whereas rs2238136 (P = 0.0244) had an effect only in females and rs4760648 had significant distortion overall (P = 0.0304) and in females (P = 0.0138).
Discussion
The T1D candidate gene, VDR, has been examined in several populations both by case/control cohorts and by family transmission analysis. Conflicting results have been reported in the literature; a meta-analysis concluded that there was no association with T1D when taking all studies into account. The current results from the collection of ASP families assembled by the T1DGC are consistent with no evidence of association of VDR polymorphism with T1D, after taking into account subgroup analyses and adjusted P-value threshold of P < 0.001 to account for the 38 SNPs tested.
Recently, a meta-regression analysis was performed that correlated the geographical location of the T1D VDR association studies with the lowest UV-exposure (satellite data from Januaries). This analysis reported evidence for associations with SNPs at the restriction sites, FokI, BsmI, and TaqI if UV radiation increased. The earlier reported association of T1D with the VDR TaqI polymorphism was reduced in winter months.20 Although this meta-regression analysis was performed only on case–control studies, it raises the issue of analyzing the genotype in the context of the environment, the level of circulating vitamin D. This is particularly important as several epidemiological studies point to an association of UV irradiation and the protection from T1D.7 The currently presented data do not take this into account. Thus, future VDR gene associations with T1D and with other immune-mediated diseases could attempt to take into account seasonal variations in vitamin D metabolism.
Materials and methods
Subjects
The T1DGC consortium is a worldwide network of T1D research groups whose collaboration has achieved one of the largest collection of T1D ASP families in history.21,22 In total, there were 1654 families and 6707 individuals of whom 3399 were affected.
Genotyping
Genotyping was performed T1DGC ASP families using two platforms. One platform was the Illumina GoldenGate assay system (ILMN) and the other was the Sequenom iPlex (SQN). All genotyping and calls were performed by the facility at the Broad Institute of Harvard/MIT. Thirty-two tagging SNPs were selected and genotyped for the VDR gene. HLA DR-DQ genotyping in these same samples, used as stratification variable, was performed as described.2
Analytic methods
Transmission differences between parents to cases with T1D were analyzed by using UNPHASED software (version 2.403). This software is available from the website: http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/.23,24
Acknowledgements
This research uses resources provided by the Type I Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. The work was also supported by grants to JAT from the Wellcome Trust, Juvenile Diabetes Research Foundation and the National Institute for Health Research Biomedical Research Centre award to Cambridge. Genotyping was performed at the Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Etten E, Verlinden L, Giulietti A, Ramos-Lopez E, Branisteanu DD, Ferreira GB, et al. The vitamin D receptor gene FokI polymorphism: functional impact on the immune system. Eur J Immunol. 2007;37:395–405. doi: 10.1002/eji.200636043. [DOI] [PubMed] [Google Scholar]
- 4.Giulietti A, Gysemans C, Stoffels K, van Etten E, Decallonne B, Overbergh L, et al. Vitamin D deficiency in early life accelerates Type 1 diabetes in non-obese diabetic mice. Diabetologia. 2004;47:451–462. doi: 10.1007/s00125-004-1329-3. [DOI] [PubMed] [Google Scholar]
- 5.Mathieu C, Waer M, Laureys J, Rutgeerts O, Bouillon R. Prevention of autoimmune diabetes in NOD mice by 1,25 dihydroxyvitamin D3. Diabetologia. 1994;37:552–558. doi: 10.1007/BF00403372. [DOI] [PubMed] [Google Scholar]
- 6.Hypponen E, Laara E, Reunanen A, Jarvelin MR, Virtanen SM. Intake of vitamin D and risk of type 1 diabetes: a birth-cohort study. Lancet. 2001;358:1500–1503. doi: 10.1016/S0140-6736(01)06580-1. [DOI] [PubMed] [Google Scholar]
- 7.Mohr SB, Garland CF, Gorham ED, Garland FC. The association between ultraviolet B irradiance, vitamin D status and incidence rates of type 1 diabetes in 51 regions worldwide. Diabetologia. 2008;51:1391–1398. doi: 10.1007/s00125-008-1061-5. [DOI] [PubMed] [Google Scholar]
- 8.Greer RM, Rogers MA, Bowling FG, Buntain HM, Harris M, Leong GM, et al. Australian children and adolescents with type 1 diabetes have low vitamin D levels. Med J Aust. 2007;187:59–60. doi: 10.5694/j.1326-5377.2007.tb01130.x. [DOI] [PubMed] [Google Scholar]
- 9.Brekke HK, Ludvigsson J. Vitamin D supplementation and diabetes-related autoimmunity in the ABIS study. Pediatr Diabetes. 2007;8:11–14. doi: 10.1111/j.1399-5448.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 10.Holick MF. Vitamin D: importance in the prevention of cancers, type 1 diabetes, heart disease, and osteoporosis. Am J Clin Nutr. 2004;79:362–371. doi: 10.1093/ajcn/79.3.362. [DOI] [PubMed] [Google Scholar]
- 11.Gysemans C, van Etten E, Overbergh L, Giulietti A, Eelen G, Waer M, et al. Unaltered diabetes presentation in NOD mice lacking the vitamin D receptor. Diabetes. 2008;57:269–275. doi: 10.2337/db07-1095. [DOI] [PubMed] [Google Scholar]
- 12.Mathieu C, van Etten E, Gysemans C, Decallonne B, Kato S, Laureys J, et al. In vitro and in vivo analysis of the immune system of vitamin D receptor knockout mice. J Bone Miner Res. 2001;16:2057–2065. doi: 10.1359/jbmr.2001.16.11.2057. [DOI] [PubMed] [Google Scholar]
- 13.Pani MA, Knapp M, Donner H, Braun J, Baur MP, Usadel KH, et al. Vitamin D receptor allele combinations influence genetic susceptibility to IDDM in Germans. Diabetes. 2000;49:504–507. doi: 10.2337/diabetes.49.3.504. [DOI] [PubMed] [Google Scholar]
- 14.Nejentsev S, Cooper JD, Godfrey L, Howson JM, Rance H, Nutland S, et al. Analysis of the vitamin D receptor gene sequence variants in type 1 diabetes. Diabetes. 2004;53:2709–2712. doi: 10.2337/diabetes.53.10.2709. [DOI] [PubMed] [Google Scholar]
- 15.Lemos MC, Fagulha A, Coutinho E, Gomes L, Bastos M, Barros L, et al. Lack of association of vitamin D receptor gene polymorphisms with susceptibility to type 1 diabetes mellitus in the Portuguese population. Hum Immunol. 2008;69:134–138. doi: 10.1016/j.humimm.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Mimbacas A, Trujillo J, Gascue C, Javiel G, Cardoso H. Prevalence of vitamin D receptor gene polymorphism in a Uruguayan population and its relation to type 1 diabetes mellitus. Genet Mol Res. 2007;6:534–542. [PubMed] [Google Scholar]
- 17.Guo SW, Magnuson VL, Schiller JJ, Wang X, Wu Y, Ghosh S. Meta-analysis of vitamin D receptor polymorphisms and type 1 diabetes: a HuGE review of genetic association studies. Am J Epidemiol. 2006;164:711–724. doi: 10.1093/aje/kwj278. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen M, d’Alesio A, Pascussi JM, Kumar R, Griffin MD, Dong X, et al. Vitamin D-resistant rickets and type 1 diabetes in a child with compound heterozygous mutations of the vitamin D receptor (L263R and R391S): dissociated responses of the CYP-24 and rel-B promoters to 1,25-dihydroxyvitamin D3. J Bone Miner Res. 2006;21:886–894. doi: 10.1359/jbmr.060307. [DOI] [PubMed] [Google Scholar]
- 19.Baker AR, McDonnell DP, Hughes M, Crisp TM, Mangelsdorf DJ, Haussler MR, et al. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci USA. 1988;85:3294–3298. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponsonby AL, Pezic A, Ellis J, Morley R, Cameron F, Carlin J, et al. Variation in associations between allelic variants of the vitamin D receptor gene and onset of type 1 diabetes mellitus by ambient winter ultraviolet radiation levels: a meta-regression analysis. Am J Epidemiol. 2008;168:358–365. doi: 10.1093/aje/kwn142. [DOI] [PubMed] [Google Scholar]
- 21.Concannon P, Erlich HA, Julier C, Morahan G, Nerup J, Pociot F, et al. Type 1 diabetes: evidence for susceptibility loci from four genome-wide linkage scans in 1,435 multiplex families. Diabetes. 2005;54:2995–3001. doi: 10.2337/diabetes.54.10.2995. [DOI] [PubMed] [Google Scholar]
- 22.Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 23.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]