Abstract
Proteins of the G-protein coupled receptor (GPCR) family present numerous attractive targets for rational drug design, but also a formidable challenge for identification and conformational modeling of their 3D structure. A recently performed assessment of blind predictions of adenosine A2a receptor (AA2AR) structure in complex with ZM241385 (ZMA) antagonist provided a first example of unbiased evaluation of the current modeling algorithms on a GPCR target with ~30% sequence identity to the closest structural template. Several of the 29 groups participating in this assessment exercise (Michino et al., doi:10.1038/nrd2877) successfully predicted the overall position of the ligand ZMA in the AA2AR ligand binding pocket, however models from only three groups captured more than 40% of the ligand-receptor contacts.
Here we describe two of these top performing approaches, in which all-atom models of the AA2AR were generated by homology modeling followed by ligand guided backbone ensemble receptor optimization (LiBERO). The resulting AA2AR-ZMA models, along with the best models from other groups are assessed here for their virtual ligand screening (VLS) performance on a large set of GPCR ligands.
We show that ligand guided optimization was critical for improvement of both ligand-receptor contacts and VLS performance as compared to the initial raw homology models. The best blindly predicted models performed on par with the crystal structure of AA2AR in selecting known antagonists from decoys, as well as from antagonists for other adenosine subtypes and AA2AR agonists. These results suggest that despite certain inaccuracies, the optimized homology models can be useful in the drug discovery process.
Keywords: Adenosine receptor, GPCR structure, G-protein, antagonist, subtype selectivity, homology model, flexible docking, virtual screening, ligand guided optimization, normal mode analysis
Introduction
The current drug discovery process greatly benefits from analysis of 3D structures of the target receptors and their interactions with ligands. Application of the structure based approach to GPCR targets could be especially rewarding 1, given functional and clinical importance of these receptors2–4. About 800 seven-transmembrane (7TM) proteins of the GPCR family are involved in signaling and regulation in CNS, cardiovascular, immune and other major systems in our bodies2. Moreover, GPCRs are targets for almost half of the existing drugs, and the range of novel targets and investigational drugs in this field is rapidly expanding2,3. Unfortunately, 3D modeling of GPCRs was long hampered by the lack of relevant structural data, with rhodopsin (Rho) being the only GPCR with its crystal structure solved5. The situation is rapidly changing thanks to the recently determined high resolution structures of β-adrenergic (β2AR6,7, β1AR8) and adenosine A2a receptors (AA2AR)9, which enable accurate 3D modeling of whole families of clinically relevant GPCRs. In addition, these structures offer excellent templates for virtual screening of antagonists, and with some modifications, agonists of these GPCR targets10–12. Significant hurdles in GPCR crystallization still remain13,14, however, which make “on demand” crystallization of a given receptor subtype with a given drug scaffold much less accessible than for other large classes of drug targets, e.g. proteases or kinases15.
This inevitable sparseness of GPCR crystal structures highlights the role of 3D homology modeling as the major source of structural models for rational drug design. In the first line of modeling applications, minor differences between a crystal structure of a GPCR target and its closely related subtypes can be predicted quickly and accurately, providing a powerful tool to study ligand subtype specificity and helping the design of more selective and safer drugs. However, structural templates for such “close-range” homology modeling (>50% sequence identity, ID) are currently available only for a handful of receptors, while there is a high demand for much more challenging applications involving “mid-range” (~30–50% ID) and “long-range” (<30% ID) GPCR homology modeling.
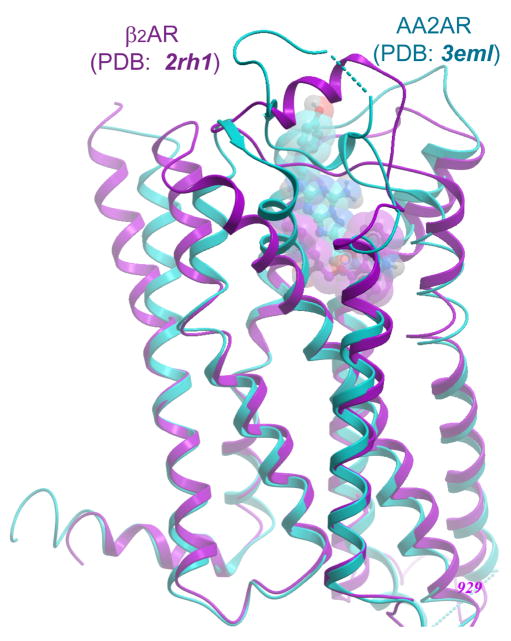
In fact, the common 7TM structure and the presence of universally conserved residues in each of the TM helices make it possible to build rough models of the 7TM helical bundle for even the most diverse GPCRs with as little as ~20% sequence identity (ID) to the structural template. Automated tools for fast modeling of GPCRs have been developed based on Rho homology16 and “ab initio” modeling17,18. Though accuracy of these models in terms of Cα RMSD in the 7TM helical bundle (RMSDCα7TM) was estimated to be within 3 Å, this seemingly low RMSD value can be a rather deceptive measure of model quality. For example, the overall structures of Rho, β2AR and AA2AR6,9 differ by only ~2 Å RMSDCα7TM, but a simple comparison shows that positions of some Cα atoms, especially in extracellular tips of some TM helices can deviate as much as 6 Å (see Figure 1). These deviations result from various shifts, kinks, and local distortions in the secondary structures of TM helices. Even more radical variations can be found in the extracellular loops of GPCRs (EL2 and EL3), which have very different amino acid sequences and lengths, and form very distinct secondary structure patterns. Also, N-terminal domains of various lengths that are present in many GPCRs can add to the diversity. All of these elements can be involved in binding of natural and therapeutic ligands, which is usually mediated by extracellular parts of GPCRs.
Figure 1.
Structures of β2AR/carazolol (PDB code 2rh1) and AA2AR/ZMA (PDB code 3eml) complexes are shown (magenta and cyan respectively), with superimposed Cα atoms of seven transmembrane helices (7TM).
Because of the remarkable diversity in the extracellular ligand binding regions of GPCRs, one may wonder whether current tools can yield models useful for virtual screening and other drug design applications based on mid-range and long-range homology predictions. A recently performed assessment of GPCR modeling19, based on blind predictions of AA2AR-ZMA complex structure9 was very helpful in providing a first unbiased look at this problem. In this exercise, organized similarly to CASP and CAPRI experiments20,21, twenty nine groups of GPCR modeling experts submitted their blind predictions (up to 10 models per group, 206 total models submitted) before the publication of the crystal structure. The assessment study subsequently compared each model to the crystal structure (PDB code 3EML) in terms of receptor and ligand RMSD values and of the number of correctly predicted contacts between ligand and receptor19.
According to Michino et al19, the three top-scoring groups in the assessment were able to predict correctly more than 40% of the 75 receptor-ligand contacts (40 contacts for Katritch/Abagyan, 34 for Costanzi, 33 for Lam/Abagyan best models), which is a dramatic improvement over previous Rho-based modeling attempts22–25. All these three groups used homology modeling with β2AR and β1AR structural templates (~30% sequence identity) to generate initial models; none of the top 10 predictions were obtained by using only Rho structural template or ab initio modeling.
Two of the top three groups in the assessment (Katritch/Abagyan and Lam/Abagyan) employed a so called Ligand guided Backbone Ensemble Receptor Optimization (LiBERO) approach, where structure-activity information (SAR) for a representative series of ZMA analogues23 was used to predict the binding site and optimize the receptor conformation. In general, the ligand guided approaches are based on (i) generation of multiple conformations of receptor and (ii) ranking conformations according to their performance in VLS enrichment for known ligands in a random decoy set26,27. This concept has proved to be efficient in previous applications to GPCR modeling, including modeling of dopamine D3, adrenergic β1, cannabanoid CB2 and Neurokinin I receptors28–31, as well as design of new chemical scaffolds for Melanin-Concentrating Hormone Receptor 1 (MCH-R1)32. Recently, the approach was also applied to prediction of agonist induced changes in β2AR binding pockets10,11. In the application to AA2AR modeling19 described in detail in this study, we have extended the ligand guided method to generate significant variations of the protein backbone in multiple receptor conformations by using either Monte Carlo sampling or elastic network normal mode analysis (EN·NMA)33 techniques.
While Michino et al19 analysis is focused on geometry of AA2AR models, submitted in the course of the assessment exercise, here we analyze these models in terms of their performance in a large scale virtual ligand screening (VLS) benchmark, which is directly related to their potential usefulness for drug discovery applications. The top models from our group were found highly efficient in the VLS based on a comprehensive GLIDA dataset of 14000 GPCR ligands containing 345 AA2AR-specific antagonists 34. These results also show a good correlation between improved VLS performance and the number of correctly predicted ligand-receptor contacts, suggesting that ligand guided approach is capable of “adding value” to the initial homology models19,35. On the other hand, certain differences between the β2AR template and adenosine AA2AR receptor were not predicted by any of the groups participating in the modeling assessment, suggesting that more advanced modeling methods and/or additional experimentally derived spatial restraints would be beneficial for more accurate modeling of GPCRs.
Methods
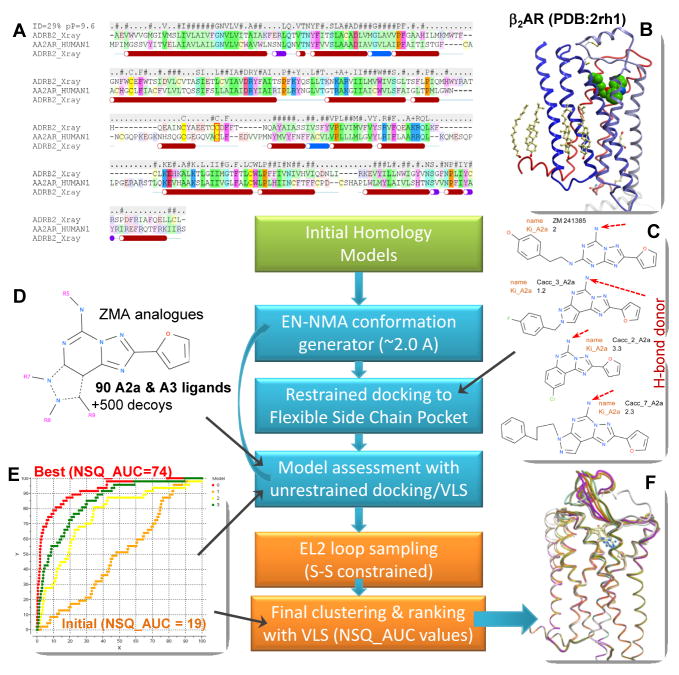
The combined homology modeling and Ligand guided Backbone Ensemble Receptor Optimization algorithm (LiBERO), employed by Katritch/Abagyan group includes the following steps, illustrated in Figure 2.
Figure 2.
A flowchart of the modeling algorithm. Initial homology modeling (green block) uses AA2AR/β2AR alignment (A) and β2AR structural template (B). The ligand-guided optimization procedure (cyan blocks) generates multiple conformations of the protein backbone with EN·NMA algorithm, which is followed by docking selected ligands (C) into the models with flexible side chains. Resulting models of receptor are evaluated by fast (rigid) docking of a set of AA2AR ligands and decoys (D), and the models with the best NSQ_AUC values (E) are selected. Final modeling steps (orange blocks) include loop modeling and ranking of the final AA2AR-ZMA models (F).
Initial homology model generation
Initial 3D models of the AA2AR were obtained with a standard homology modeling function BuildModel36 using ZEGA alignment algorithm37 implemented in an ICM molecular modeling package (ICM version 3.6.-1b, Molsoft LLC). High resolution structure of the β2AR6,7 with removed T4L fusion domain (Figure 2B) was used as a template with about 30% identical residues (ID). (Note that bovine rhodopsin has only ~19% ID with AA2AR). The automatic ZEGA alignment was manually adjusted to eliminate minor gaps in TM1, TM5 and TM7 domains, the final alignment is shown in Figure 2A. Also, an adjustment was made to ensure alignment between the last cysteins in EL2 loops of AA2AR and β2AR (Cys166 and Cys191 respectively, shown as red box); the unaligned portion of the extracellular loop 2 (EL2), residues G142-A165, was not included in the initial AA2AR model. After 3D placement of the AA2AR polypeptide chain according to the alignment and the 2rh1 PDB coordinates, a limited energy-based optimization of side chain and loop conformations was performed.
Note on residue numbering
Amino acid residues in the models are numbered according to human AA2AR sequence (accession # P29274). Superscript numbers for TM residues (as in Asn2536.55) indicate their corresponding TM helix and their relative position in this helix as described in ref.38. Briefly, the positions are calculated relative to the most conserved reference residue in that helix, which is assigned number 50; numbering decreases toward the N terminus and increases toward the C terminus. The superscript numbering is not used in loop regions.
Generation of the pocket conformational ensemble
Multiple conformations of the model were generated using all heavy-atom Elastic Network Normal Modes analysis39, as described previously33. With this approach, the interaction energy between two atoms is described by a Hookean potential where the initial distances are taken to be at the energy minimum, and the spring constant assumes an inverse exponential relationship with the distance. The force constant matrix of the system is described by the Hessian (3N × 3N matrix, being N the number of atoms), obtained as the partial second derivatives of the potential with respect to the coordinates. Diagonalization of the Hessian yields 3N - 6 eigenvectors ranked according to their corresponding eigenvalues. The eigenvectors provide the collective directive of motions of atoms, and the eigenvalues give the energy cost of deforming the system along the eigenvectors.
We included in the EN·NMA calculation all the amino acids defining the 7 transmembrane helices, including 2246 heavy atoms from 288 residues. More than 100 conformations (from the important NMA subspace) were generated at each step of the ligand guided optimization procedure, and the chemical distortions in each model were corrected by using 25 steps of Cartesian minimization.
Flexible ligand receptor docking
For each of the conformations generated by EN·NMA, ZMA compound and its three high affinity analogues (compounds #2, #3, #7 from ref.23, Figure 2C) were docked into the all atom representation of the complete AA2AR model with flexible side chains.
The set of AA2AR antagonists in ref.23 represents a comprehensive SAR for pyrazolo-triazolo-pyrimidine (PTP) scaffold that yields highly specific ligands for both A1 and A3 adenosine receptor subtypes. The analysis of the ligand affinity data in ref.23 points to the importance of the H-bond donor in position N5 for the high affinity binding of (PTP) analogues, though an acceptor partner for N5 cannot be unambiguously assigned from the experimental data. A potential contact of the N5 donor with one of the five possible acceptors in the side chains, His2506.52, Asn2536.55, His2787.43, Tyr2717.36 or Thr883.36, all located in the pocket found by ICM PocketFinder algorithm 40, was therefore used as an alternative harmonic distance restraint to guide initial placement of the ligand. Energy optimization was performed for the all atom models with flexible ligand and flexible receptor side chains in the 8 Å proximity of the binding pocket, using ICM biased-probability Monte Carlo algorithm41. The resulting ligand-receptor models with acceptable conformational energy were clustered according to the conformations of the binding pocket residues, yielding more than 400 non-redundant models.
Small scale VLS assessment
These models of the AA2AR binding pocket were assessed in virtual ligand screening (VLS) benchmark11 for their ability to discriminate 64 AA2AR subtype specific antagonists from 26 AA3R antagonist and 500 decoys randomly selected from the ChemDiv drug-like compound database (www.chemdiv.com). The dataset of AA2AR and AA3R antagonists derived by removing excessive redundancy from ligands described in ref.23 and ref.25, represents diverse compounds built on both adenine-like and PTP scaffolds (see Supplementary Materials).
The ligand and decoy compounds were docked into the AA2AR models, represented as pre-calculated potential grids, and then sorted according to their ICM binding scores. No distance restraints or any other experimentally derived information was used in the ligand docking procedure at the model assessment stage.
We used several different metrics to assess VLS performance, including Area Under ROC Curve (AUC) and enrichment factor at 1% dataset cutoff, EF(1%). However, these commonly used metrics have their own deficiencies, widely discussed in literature (see e.g.42). Recently, we devised a new Normalized Square root AUC (NSQ_AUC) metric which combines overall selectivity of AUC with “early enrichment” of EF(1%) measures. We used NSQ_AUC to select the best models at each optimizations step and to rank the final models (see definition below).
A model with the best NSQ_AUC values, chosen out of 200 models at the first iteration of the ligand guided optimization procedure was used as a starting model for a second iteration. In the second iteration, the three best out of 200 models were selected according to their NSQ_AUC values. These three models with the corresponding poses of ZMA compound were used to generate a full model of the AA2AR complete with loops.
Note that while the optimization and model selection/ranking procedure relies on experimentally derived SAR data for a series of adenosine receptor antagonists, our approach does not use mutagenic data in modeling or decision making (see Discussion).
NSQ_AUC definition
Similar to AUC, the value of NSQ_AUC is based on a calculation of the area under the ROC curve42. The difference is that the area (AUC*) is calculated for the ROC curve plotted with X coordinate calculated as the square root of “False Positive %”, X=Sqrt(FP). The Normalized Square root AUC (NSQ_AUC) is then calculated as:
Thus, the value of NSQ_AUC is more sensitive to initial enrichment than the commonly used linear AUC. The NSQ_AUC measure returns the value of 100 for any perfect separation of signal from noise and values close to 0 for a random subset of noise.
Full model generation and EL2 modeling
The backbone conformations generated with EN·NMA methodology can deviate significantly (~2 Å) from the original homology model both in the ligand binding region and in the cytoplasmic part of the 7TM bundle. In order to avoid unjustified deviations in the cytoplasmic half of the receptor, we used the following procedure. The homology model was superimposed onto a selected EN·NMA model, and heavy atoms of all residues within 8 Å distance from the ligand in the homology model were tethered by harmonic distance restraints to the corresponding heavy atoms of the EN·NMA model. At the same time, the cytoplasmic half of the homology model was tethered to a rigid copy of itself. The homology model was subsequently optimized with respect to both conformational energy and distance restraints. This fast procedure results in a “hybrid” model that reproduces both the ligand-optimized binding pocket residue conformations of the EN·NMA model and the original template-based conformation of the cytoplasmic half of AA2AR with an RMSD deviation lower than 0.2 Å.
Modeling of the non-conserved part of the extracellular loop EL2 (residues G142 to A165) was performed with the ICM loop modeling algorithm based on global optimization of conformational energy with disulfide bonding restraints. Five possible disulfide bonding configurations were tested for the 6 cysteins in the loop and 7TM domain, only one of them (C71–C159, C74–146, C77–C166) resulting in an energetically feasible conformation of the loop backbone. The two 3D conformations of the EL2 with the best predicted conformational energy were further refined in the context of the three ligand binding models generated in the previous step.
Model ranking and submission
The optimized binding pocket and EL2 loop combinations comprised the first six submitted models of AA2AR-ZMA complex, and were ranked according to their overall conformational energy. The remaining four models (#7 to #10) included possible alternative conformations of the ligand with the hydrogen bond donor N5 in contact with His2506.52 side chain or Ala813.29 main chain acceptors, as well as an alternative (β2AR-like) conformation of the EL2 loop.
Large scale VLS evaluation of the AA2AR models
We performed a rigorous large-scale VLS performance evaluation for the top models submitted to the GPCR assessment19. For this purpose we used GLIDA database34, which comprises 14000 diverse GPCR ligands compounds, including 348 AA2AR antagonists. The database also has compounds marked as selective AA3R antagonists (82 compounds) and AA1R antagonists (194 compounds), as well as 58 AA2AR agonists, which were used in selectivity assessment of our top model.
All atom protein models for docking were prepared from heavy atom coordinates (available at http://jcimpt.scripps.edu/gpcr_dock.html) using an ICM “convert” procedure that builds and energy-optimizes polar hydrogens; for AA2AR crystal structure (PDB code 3EML), the side chains of Asn, Glu and His residues were also optimized.
Calculations of correct ligand-receptor contacts and RMSD for all AA2AR-ZMA models were performed as described in ref.19, but using a somewhat different definition of 7TM helices (7TM). The “TM I-VII Cα RMSD” definition in Table 1 of ref.19 comprised all residues of AA2AR between Ser6 and Ile292, assigned as “helical” in the 3EML PDB entry, including some short helices (e.g. Ser35-Gln38) in the extracellular and intracellular loop regions. A more conventional definition used here includes only seven membrane-spanning helices themselves (residues 6–34, 40–67, 73–107, 117–142, 173–205, 227–259, 267–292), yielding generally smaller values of RMSDCα7TM than in ref.19.
Results
Improvement of the raw homology model through optimization steps
Multiple models of AA2AR-ZMA complex, more than 400 total, were generated at different stages of the homology modeling and ligand-guided optimization (LiBERO) procedure, starting from an initial raw homology model built with the β2AR structural template and progressing to final models submitted for GPCR assessment19. Evaluation and selection of the most promising models at each stage were based on results of docking and small scale virtual ligand screening (VLS) for a set of ZMA analogues with adenine-like and PTP scaffolds.
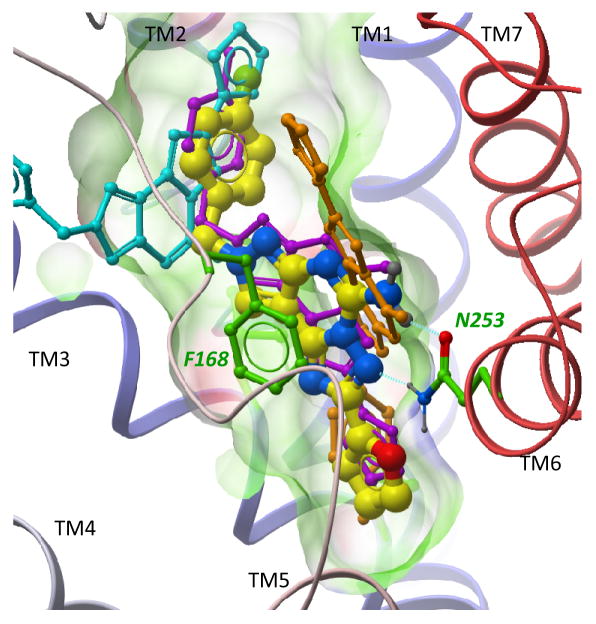
Figure 3 shows four examples of the best-scoring poses for one of the PTP compounds (#3 from ref.23) docked into AA2AR models generated at different steps of the procedure. In the initial raw models the ligand docks with the phenoxy group buried deep in the pocket or even in a different part of the pocket (orange and cyan sticks respectively). After the first iteration of ligand-guided optimization, the predicted ligand pose (magenta sticks) starts to resemble the top ranked pose with hydrogen bonding to Asn253 and aromatic stacking with Phe168. Finally, the best performing second-iteration models, which were submitted to the assessment (mod2upu, mod1cf1 and mod5ano in ref19 supplementary material) consistently bind compound #3 and most other ZMA analogues in similar binding poses, with the furanyl ring buried in the pocket (thick sticks with yellow carbons). These poses are also characterized by aromatic stacking of the ligand aromatic moiety with Phe1685.29 side chain and polar interactions between exocyclic amine group and Asn2536.55 side chain (see Supplementary Materials for 3D structures of the mod2upu receptor model with other AA2AR ligands of the optimization set.)
Figure 3.
Examples of best scoring poses for compound #3 from ref.23, docked into AA2AR models at different optimization stages: (1) raw homology modeling (orange) (2) side chains refinement (cyan) (3) first iteration of ligand-guided refinement (magenta) (4) second iteration of ligand-guided refinement (thick balls and sticks with yellow carbons). The model of AA2AR (as in mod2upu) is shown by ribbon colored from N- to C-terminal (from blue to red), ligand binding pocket of the model shown by transparent skin.
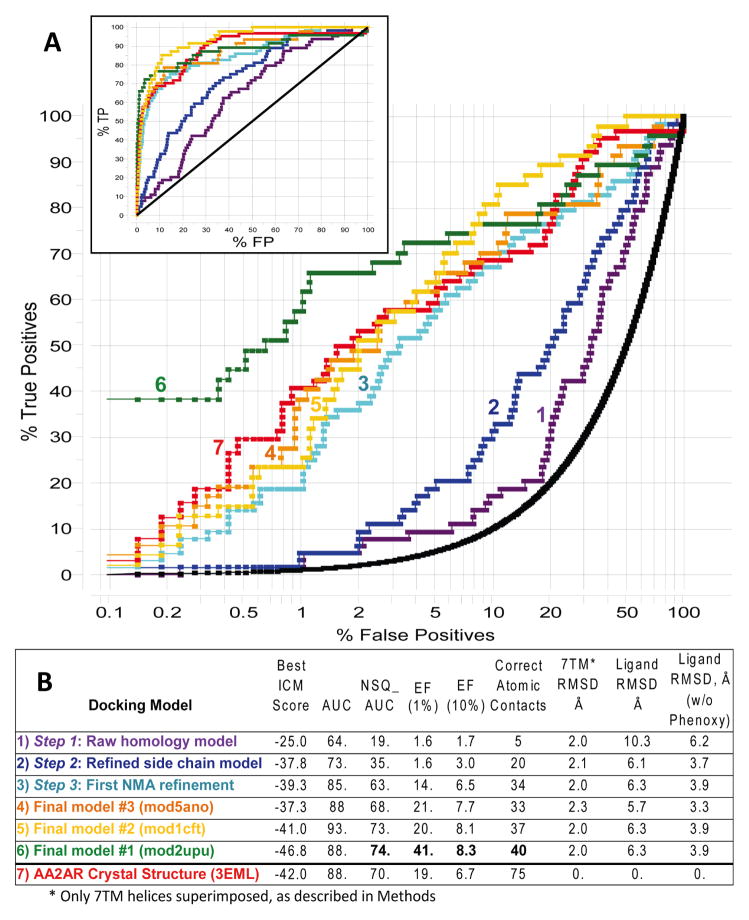
Figure 4 sums up results of a small scale VLS for the best-performing models at each optimization step. Note that the raw homology model (Step 1) allowed only suboptimal docking (the best ICM Docking Score = −25) and very low selectivity of AA2AR antagonists (area under AUC = 64, NSQ_AUC=19). Side chain refinement with flexible ligands (Step 2) somewhat improved docking scores, AUC and NSQ_AUC values, but the initial enrichment factor (EF_1%) remained very low. Dramatic improvement of VLS performance was achieved only in Step 3 through generation of multiple conformations of the receptor model using EN·NMA methodology. The best performing model (AUC=85, NSQ_AUC=63) was used as a starting template for a second iteration of EN·NMA procedure, and three models with the best NSQ_AUC values were selected for EL2 loop reconstruction. The three selected final models demonstrated very good VLS selectivity for AA2AR antagonists, which later turned out to be comparable to the VLS performance of the AA2AR crystal structure (3EML)9. Note that our best-ranking model even outperformed the 3EML model, especially in terms of initial enrichment. This can be partially explained by an explicit bias of our models towards the limited compound set used in the model optimization; more stringent assessment of the models is given in the sections below with the comprehensive GLIDA ligand dataset34.
Figure 4.
Results of small-scale VLS evaluation for our AA2AR models at different stages of the optimization procedure, as well as for our three best models submitted to assessment. A. ROC curves for each of the models, shown in logarithmic scale to emphasize initial enrichment at 1% of the dataset (Insert shows a standard view of the ROC curves). B. The table listing key characteristics of the corresponding ROC curves and the overall quality of the model structures for the models as compared to the crystal structure (PDB code 3eml). The values of NSQ_AUC, enrichment factors and number of contacts are shown in bold for our rank#1 model.
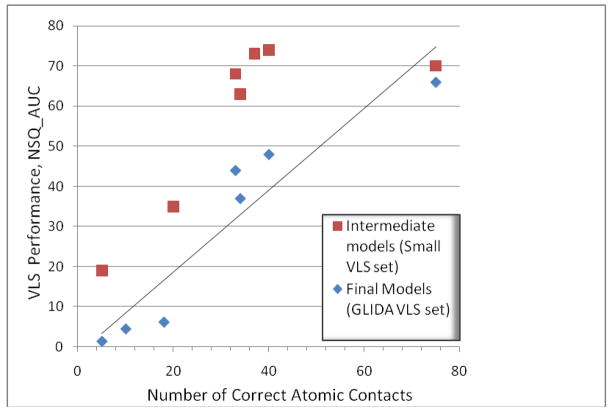
Interestingly, comparison of our intermediate and final models with the 3eml crystal structure shows that VLS performance (measured by NSQ_AUC value) improved in parallel with quality of the model (measured as a number of correctly predicted atomic contacts). In contrast, values of protein and ligand RMSDs did not seem to correlate with VLS performance and did not significantly improve through the refinement steps.
Blind prediction models compared to crystal structure
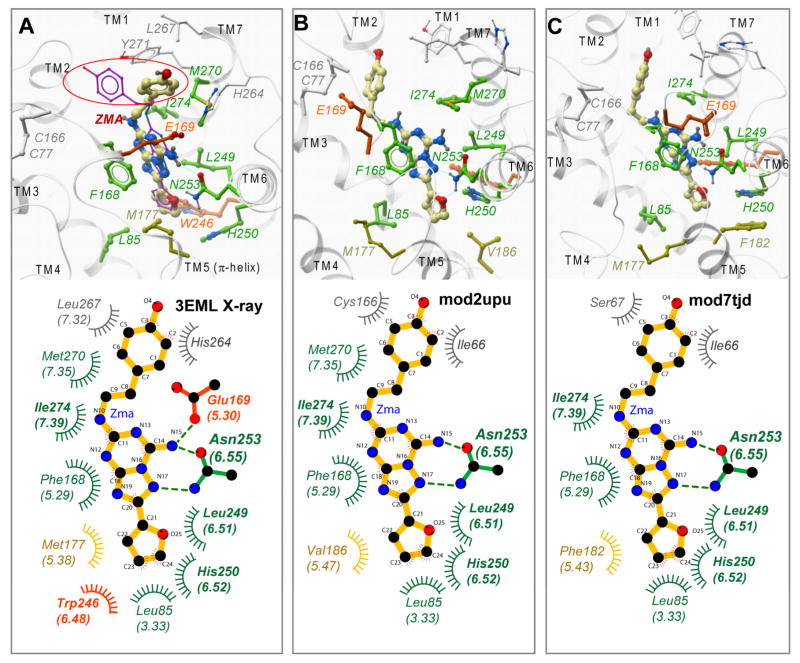
The top ranking models predicted by Katritch/Abagyan and Lam/Abagyan groups19 are shown in Figure 5 (B and C respectively) in comparison to the crystal structure of AA2AR-ZMA complex (A). Both models correctly predict the overall positioning of the ligand, which is reflected in 53% (B) and 44% (C) of correctly predicted atomic contacts between ligand and receptor.
Figure 5.
Comparison of the AA2AR-ZMA crystal structure9 (A) with the top ranked models from Katritch/Abagyan (B) and Lam/Abagyan (C) groups. Top panels show 3D snapshots and bottom panels show 2D plots of ligand interacting side chains of the receptor. The ligand is shown with yellow carbons, the protein backbone is shown by grey ribbons. Ligand-receptor hydrogens bonds are shown by cyan dashed lines. An alternative (magenta) conformation in the top panel A represents ZMA ligand docked by ICM into the crystal structure of AA2AR9, phenoxy moiety of ZMA circled red. The protein side chains in both 2D and 3D presentations are colored according to the ZMA-residue contact predictions: green residues, contacts correctly predicted by mod2upu model; orange residues, contacts not predicted; yellow residues, hydrophobic contacts replaced by another side chain; grey residues, contacts of the phenoxy ring that do not have major contribution into ligand binding. Note that contact residues suggested by previous mutation analysis are in bold font.
All of the correct contacts in both models are between the ligand ”core” (without phenoxy ring) and TM3, TM6, TM7, and EL2 side chains listed in Figure 5D. The key polar interaction is the hydrogen bonding network of the Asn2536.55 side chain in TM6 with the exocyclic amine and the triazole nitrogen in the ligand core (with the donor-acceptor distances 3.0 and 2.8 Å respectively). The furanyl oxygen of the ligand is also located in close proximity (2.9 Å) to Asn2536.55 amide nitrogen, though its role as H-bond acceptor is likely to be less pronounced 43,44, and its contribution to binding is not well defined in available ligand SAR data (e.g. 25). Another critical interaction is the aromatic stacking between the F1685.29 side chain and the ligand bicyclic ring, which in our model has the contact area of ~32 Å2, as compared to ~30 Å2 in the crystal structure9. Other correctly predicted ligand contacts are hydrophobic in nature and include side chains of Leu853.33, Leu2496.51, His2506.52, Met2707.35 and Ile2747.39 that define the shape of the binding pocket.
Comparison of Figures 5A and 5B shows a striking difference in position of the flexible phenoxy ring of the ligand in the predicted model and in the crystal structure, which results in very high full ligand RMSD (~ 6Å) for our best model. Note however, that such a discrepancy owes largely to the high conformation flexibility of the ligand’s phenoxy moiety in the AA2AR-ZMA complex. Thus, in the AA2AR-ZMA crystal structure6 (3EML), the phenoxy group of the ligand has an exceptionally high B-factor (>100 A2) as compared with the core of the ligand (~50) (see Supplementary Materials, Figure SM1). This moiety of the ligand is highly solvent accessible and has only a few contacts with the receptor in a relatively wide opening in the extracellular part of the binding pocket between the loop regions EL2 and EL3. Moreover, docking of ZMA and its PTP analogues into the crystal structure based models suggests some alternative positions of the phenoxy ring in this opening. (Alternative positions of the ZMA phenoxy ring were also reported for the docked ZMA ligand in ref.19). The conformational variability of this moiety is also supported by ligand SAR data that suggest high tolerability of PTP and similar scaffolds to a range of very diverse (small and bulky, hydrophobic and hydrophilic) substitutions for the phenoxy ring. In this light we believe that deviations in the ZMA phenoxy group position are not critical for assessment of the modeling accuracy or performance.
At the same time, we should point to two other important deviations of our predicted models from AA2AR-ZMA crystal structure, which can impact VLS performance.
The first significant error in the models was the lack of a polar interaction between Glu1695.30 side chain (shown in orange sticks in Figure 5) with the ligand exocyclic amino group. In the crystal structure of AA2AR9 the Glu1695.30 residue is a part of an unusual small 1-turn helical structure in the EL2 loop, which is also stabilized by interaction with His2646.66 of the EL3 loop. The Glu1695.30-ZMA interaction contributes to binding energy and seems to be a major selectivity factor that distinguishes A2a from A3 subtype (which has V1695.30 instead). Interestingly, this hard-to-predict structural feature was not captured in any of the models submitted to the assessment19, despite availability of mutagenesis data inferring this side chain in binding of some AA2AR ligands45. Judicial use of subtype selectivity and mutation data could possibly lead to more accurate predictions in this region of the model, though reliance on mutagenesis data is not always beneficial (see Discussion).
The second problematic area included several residues in TM5 helix, most notably Met1775.38 (colored yellow in Figure 5), which in the crystal structure is in contact with ZMA furan moiety deep in the binding pocket. Incorrect position of this side chain made for an enlarged opening in the model binding pocket, resulting in the ligand shifted “down” from its correct position. Note that the lack of ligand contact with Met1775.38 was an issue for all models in the modeling assessment. The reason for this consistent error, again, lies in an unusual secondary structure in AA2AR, where the whole extracellular portion of TM5 (~3 turns) is comprised of π-helix, not a canonical α-helix, as in β1AR and β2AR. Because π-helix has different helical repeat than α-helix (i+5 instead of i+4), the Met1775.38 and other residues in this helix are “rotated” away by more than 60 degrees. Such non-canonical secondary structure features would be very hard to predict computationally, though there are hints at a possibility of some structural deviations in this region such as a weak local alignment between β2AR and AA2AR sequences and a crowding by 5 aromatic residues in 3 helical turns on the same face in the hypothetical canonical α-helix.
Assessment of AA2AR models with large scale GLIDA benchmark
An extended set of 14000 GPCR ligands from GLIDA database, containing 348 AA2AR antagonists, was used to assess VLS performance of our best AA2AR model and compare it to models from other groups and the crystal structure.
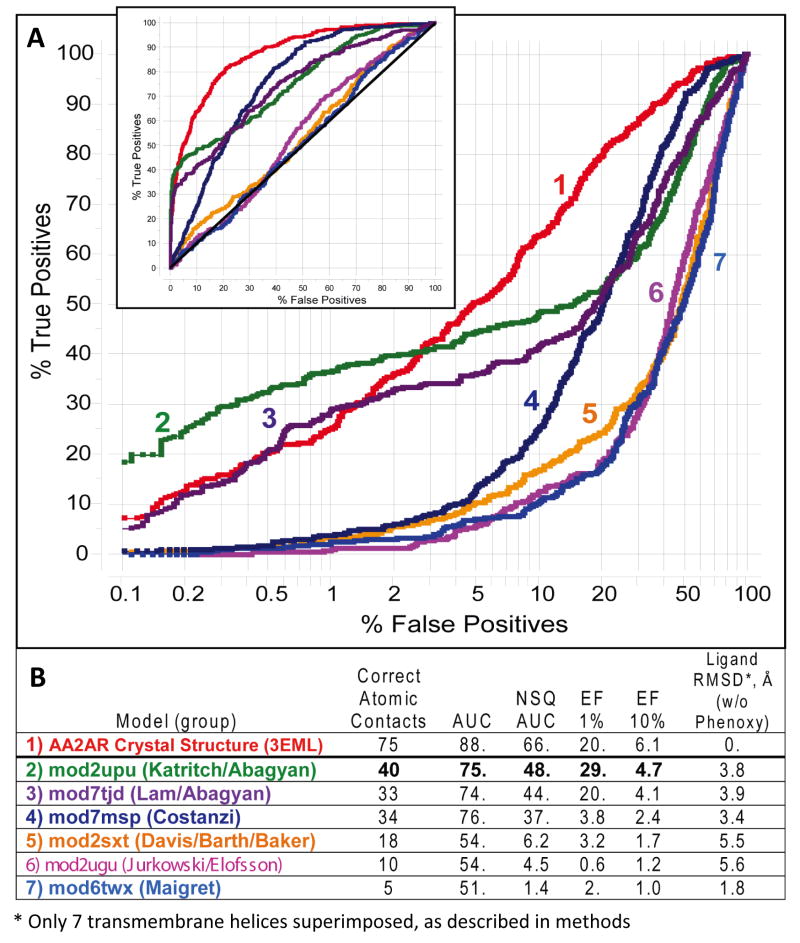
Figure 6 presents the results of this large scale assessment, suggesting VLS performance of our models is on par with the crystal structure. Note that the best model from Katritch/Abagyan group (mod2upu) is even more effective in the initial enrichment (up to 2% of the database cutoff) of AA2AR antagonists than the 3EML model, though the overall performance of the 3EML model is better. A top model from Lam/Abagyan group, built with a different β1AR structural template, also had a very good initial enrichment and overall performed similarly to our top model. The Lam/Abagyan models were also optimized using a similar ligand guided concept, though the method employed a somewhat different ligand set, different way to generate alternative conformations and different model selection criteria (see Discussion). Interestingly, the VLS performance of the Costanzi model (mod7msp) was on par with our models in terms of AUC value, but had a distinct shape of the ROC curve characterized by low initial enrichment, EF(1%) < 4. As one can see in Figure 6, top models from other groups did not show any substantial enrichment over random baseline. Similar to results for our intermediate models in Figure 4B, the results in Figure 6B suggest a good correlation between VLS performance and model quality in terms of correct ligand/receptor contacts; both sets of data are plotted in Figure 7 (see Discussion).
Figure 6.
Results of large-scale VLS evaluation with GLIDA database ligands for the top six models described in Table I of ref.19. A. ROC curves for each of the models, shown in logarithmic scale to emphasize initial enrichment at 1% of the dataset (Insert shows a regular view of the ROC curves). B. The table listing key characteristics of the corresponding ROC curves, number of atomic contact and ligand RMSD for the models, as compared to the crystal structure (PDB code 3eml). The values of NSQ_AUC, enrichment factors and number of contacts are shown in bold for our rank#1 model.
Figure 7.
Correlation between the VLS performance and the number of correct atomic contacts for the AA2AR models. Brown squares show results for our intermediate models evaluated with the small ligand set as listed in Figure 4. Blue diamonds show results for the top 6 models from ref19, evaluated with the large GLIDA dataset, as listed in Figure 6. Regression line is shown only for GLIDA dataset results.
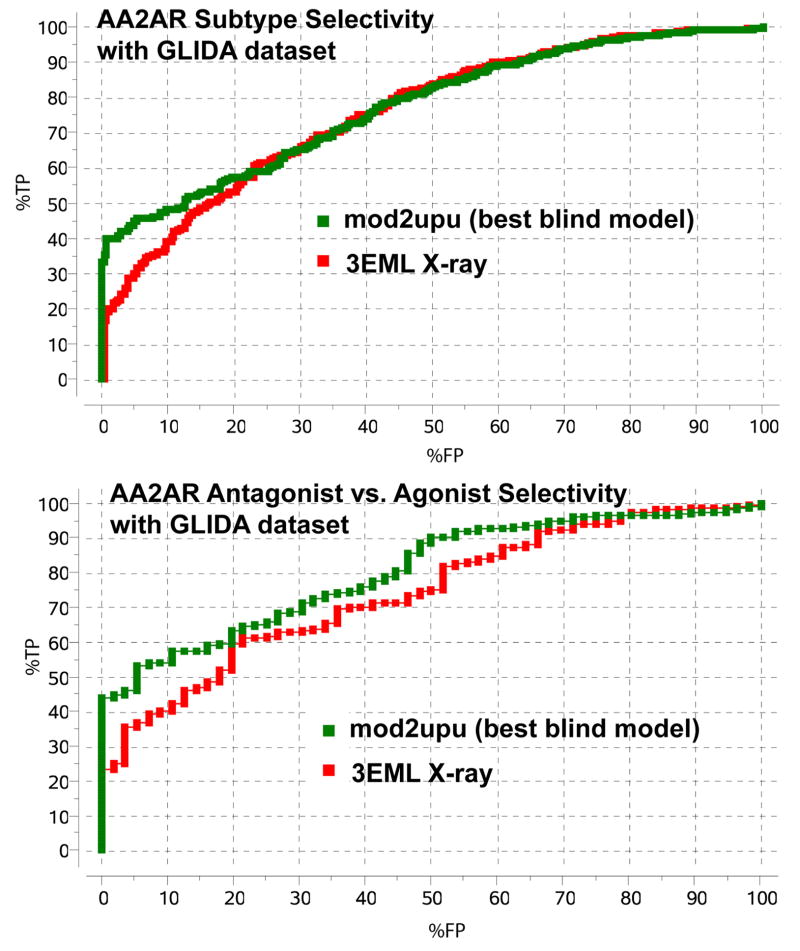
The results of large-scale GLIDA assessment also point to the ability of our best model to discriminate between AA2AR antagonists and antagonists selective to other adenosine receptors, especially the AA3R subtype (Figure 8A). This was somewhat unexpected, because our model did not correctly predict conformation of Glu1695.30 side chain interacting with the exocyclic amine, which turned out to be the major factor in AA2AR vs. AA3R selectivity. Inspection of the model (mod3upu) however, suggests that instead of Glu1695.30, another side chain Met2707.35 “caps” the exocyclic amine (see Figure 5B). The Met2707.35 “capping” effectively precludes our AA2AR model from binding of A3-selective ligands, most of which have a bulky substituent in place of one of the exocyclic amine hydrogens. The general applicability of the model to selection AA2AR-specific ligands may be limited because of different properties of Glu and Met side chains and differences in local interaction geometry.
Figure 8.
Selectivity profile for our model ranked #1 (mod2upu). (A) Subtype selectivity, AA2AR vs. adenosine receptor subtypes AA1R and AA3R. (B) Antagonist vs. Agonist selectivity.
The mod2upu model was also found effective in selecting AA2AR antagonists vs. AA2AR agonists (Figure 8B). Note that several of the AA2AR agonists from GLIDA dataset consistently docked into our models and into the crystal structure of the AA2AR, with adenine group assuming similar positions and interactions as the heteroaromatic group of ZMA in the AA2AR-ZMA complex. However the binding scores of agonists, and specifically polar interactions of their ribose moieties were suboptimal even when docked to the crystal structure of the AA2AR, suggesting possible conformational changes in the receptor upon agonist binding (Katritch et al, manuscript in preparation).
Discussion
Model evaluation with RMSD, ligand-receptor contacts and VLS performance
In most benchmarks, quality of 3D protein modeling and ligand docking is commonly assessed by comparing the predicted models with a “true” crystal structure in terms of protein and ligand RMSD and correct ligand-receptor contacts46. However, in real drug discovery applications of homology models, the crystal structure of the target (the “answer”) does not exist. Therefore, the quality of the model can only be evaluated based on the model’s capability to reproduce available experimental information, such as mutagenesis and ligand binding affinity data. Ability of the model to efficiently select known high affinity binders (ligands) vs. non-binders (decoys) can be especially useful as an internal measure of quality, since it is the most direct predictor of the model performance in selecting new candidate inhibitors in VLS. This measure has already been widely employed for selection of optimal receptor conformations for VLS screening10–12,28–32,47.
The results of this GPCR assessment19 suggest that even with the available β1AR and β2AR structural templates, accurate modeling of AA2AR remains highly challenging. While three of the top groups correctly predicted more than 40% AA2AR and ZMA atomic contacts and overall orientation of the ligand, even the best AA2AR models missed several features of the AA2AR important for ligand binding, such as positions of Glu1695.30 and Met1775.38 side chains. Nevertheless, even such “imperfect” models can be very useful in drug discovery process, as the assessment of their VLS performance suggests in the current study. Thus, the results of the comprehensive GLIDA dataset34 screening in Figure 6 (14000 GPCR ligands) show that the values of AUC and NSQ_AUC for our best models are comparable to those obtained with a crystal structure. Moreover, our rank#1 model (mod2upu) even outperformed the crystal structure in terms of initial enrichment EF(1%) for known ligands in VLS.
Of course, such a good performance of our models partially owes to the fact that they were optimized towards recognition of the most popular class of AA2AR selective compounds (adenine and PTP analogues of ZMA). At the same time our models successfully reproduce a vast diversity of high affinity AA2AR binders within these two scaffold classes (Tanimoto distance as high as 0.55) and even within several other scaffolds not employed in model optimization. An example of docking of a xanthine analogue from GLIDA database, which represents another prominent AA2AR antagonist scaffold48 is shown in Figure 9. The predicted binding motif of the methyl-xanthine scaffold resembles ZMA core interactions, characterized by aromatic stacking with Phe1685.29 phenol and polar interaction with Asn2536.55 side chain. The predicted ICM binding scores for xanthine analogues, however were significantly reduced as compared to adenine or PTP-like compounds, both in the blind models and in the crystal structure. While xanthines lack exocyclic amine group and the corresponding polar interactions, which partially explain their lower binding scores, the models may also be missing some other interactions that confer high affinity AA2AR binding of xanthines. Note that due to high flexibility and propensity for induced fit in GPCRs, a single static model of the protein is actually not expected to accommodate all ligands, therefore the use of models biased (or specialized) towards certain scaffold classes is justified in VLS 49.
Figure 9.
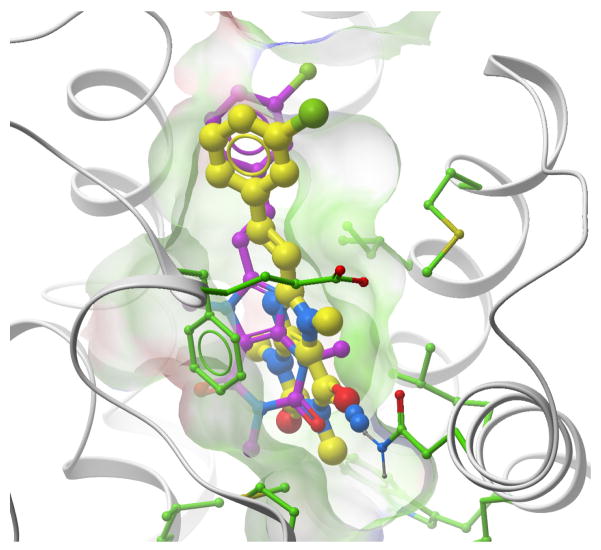
A xanthine analogue (GLIDA ID#L011922) docked into the AA2AR. The crystal structure (3eml) of the receptor is shown by grey ribbon sticks with green carbons. The ligand conformation docked into the crystal structure has yellow colored carbon atoms, the conformation docked into the blindly predicted model (mod2upu) has magenta colored carbons.
Another observation from our GLIDA screening assessment was the ability of our best model to select AA2AR selective ligands vs. those selective towards other adenosine subtypes A1 and A3, (no sufficient data for A2b subtype), as well as to select AA2AR antagonists vs. AA2AR agonists (Figure 8). Utility of these selectivity predictions in ligand optimization and rational drug design applications, however may be limited due to remaining inaccuracies in protein geometry, which are probably inevitable for such long-distance homology models.
Interestingly, our results in Figure 7 suggest a good correlation between VLS performance and number of correct contacts between ligand and receptor atoms. This correlation holds well not only for our models at various stages of optimization, but also for other top six models in AA2AR assessment. At the same time, no significant correlation of ligand RMSD with either VLS performance or number of contacts was observed, e.g. the model with the best ligand RMSD (mod6twx), had only 5 correct contacts and no VLS enrichment at all. Importance of contact-based metrics has long been demonstrated in evaluation of protein homology models46,50 and our current results suggest that analysis of ligand-receptor contacts can be widely used in quality assessments of molecular docking.
Importance of ligand guided optimization
As results in Figures 4 and 8 suggest, ligand-guided optimization was critical for improving both ligand-receptor contacts and VLS performance of our models, as compared to starting raw homology model of AA2AR. In general, the ligand guided approaches26,27,32 use a basic evolutionary concept: at each iteration, (i) a wide diversity of the receptor conformations is generated, which is followed by (ii) selection of the best conformations by their VLS performance. Both Katritch/Abagyan and Lam/Abagyan groups used this ligand guided optimization concept, but with different numbers of iterations, different ligand sets, and most importantly with very different ways to generate structural diversity in AA2AR.
The Katritch/Abagyan group used β2AR structural template (PDB code 2rh1) for initial homology modeling and employed a normal mode analysis (EN·NMA) algorithm33 to generate large variations in the model backbone and side chain conformations. The EN·NMA algorithm is especially beneficial in this case since it is very fast, allows conformational sampling with large movements of backbone (~2Å and more), and it also can modulate secondary structure (e.g. bend TM helices) without breaking it. In the case of AA2AR predictions, two consecutive iterations of the algorithm (100 NMA-generated conformations each) allowed dramatic improvements of VLS performance, though the number of iterations and conformations could be increased further with additional computational resources.
The Lam/Abagyan group19 in contrast, used turkey β1AR template (PDB code: 2VT4) and employed a Monte Carlo based conformational sampling using a hybrid full atom/grid model of the receptor ligand complex. The automated algorithm used 12 independent runs with up to 30 sampling/evaluation iterations in each run, resulting in about 300 models used in final ranking and evaluation. The evaluation ligand sets in both algorithms included 40 or more diverse AA2AR antagonists based on adenine-like and PTP scaffolds; Katritch/Abagyan also added to the mixture PTP ligands with A3 subtype specificity.
Despite such variations in details, both of the ligand guided algorithms achieved respectable VLS performance for AA2AR models, which also tightly correlate with the model quality in terms of number of correct ligand/receptor contact. Thus, starting from raw homology models with a few correct contacts, both algorithms were able to iteratively improve the number of contacts.
The ability of the ligand-guided algorithms to find correct receptor conformations, of course, is limited to those cases when local discrepancy between the starting raw model and the “true” structure is relatively minor. Thus, the algorithm was capable of finding correct ligand contacts for most of the side chains that were positioned in the ligand binding site in the initial β2AR based homology model. In contrast, the algorithm failed to identify conformation for Met1775.38 contact residue, which is a part of unusual π-helix in AA2AR. Curiously, some hard-to-predict contacts can be “compensated” by other contacts, as long as the “fake” contact improves VLS performance. Thus, in our best performing model (mod2upu) the lack of the correct contact between ligand exocyclic amine (N5) and Glu1695.30, is “compensated” by N5 contact with Met2707.35 side chain; this conformation turns out to be as efficient as the true crystal structure for reproducing A2a vs. A3 subtype selectivity in PTP scaffolds (see Figure 8). These observations point out the limitations of the current computational methods, including ligand-guided optimization. At the same time our results also suggest that ligand-optimized models even with a few local conformational mistakes can still be useful in virtual screening applications.
Technically, the several other algorithms presented in the GPCR assessment19 can be described as “ligand-guided” as long as they used ability of the AA2AR models to bind ZMA ligand to select models. What sets our approach apart is the use of binding data for multiple and highly diverse ZMA analogues as a criterion for model selection and ranking, and doing it iteratively. This approach captures much more information than a single ligand binding and provides much more stringent criteria for selection of “best” models at each optimization stage.
Use of mutagenesis data in AA2AR-ZMA structure optimization
While our algorithms rely heavily on the available ligand binding data, we did not use mutagenic information at any stage of model building, optimization and ranking. Though extensive data on point mutations for the adenosine receptor family are available in the literature (e.g. reviewed in ref 51), our previous experience with GPCRs and other highly flexible and allosteric receptors suggests that interpretation of mutation effects may sometimes be treacherous. For example, for certain point mutations in β2AR, e.g. S204A mutations, a drop in ligand affinity and/or activity can be explained by indirect effects including changes in conformational preferences of the protein itself, rather than direct ligand contacts 10,11.
In the AA2AR case, site directed mutagenesis has previously identified at least 10 residues, in which mutations result in a complete loss or significant drop in affinity of representative antagonists (e.g. summarized in ref22 and51). However, when 3EML structure became available, only 4 of these residues (Glu1695.30, His2506.52, Asn2536.55, Ile2747.39) were found to be in direct contact with ZMA antagonist. Two residues were found in the binding pocket but not in contact with ZMA (Leu843.32, His2787.43) and the other four residues (Glu151(EL2), Phe1825.43, Phe2576.59, Ser2817.46) were located outside of the binding pocket. These latter side chains are very unlikely to interact directly with any other high affinity ligand, and effect of the corresponding mutations could be explained by their indirect impact on the shape of the ligand pocket. The mutation data are also incomplete due to time/cost limitations of the experiments, for example only 4 of 15 contact residues in the AA2AR binding pocket were identified correctly from 46 mutation experiments listed in ref22; among the important contacts missed in experiments were Phe1685.29, Met1775.38 and Leu2496.51.
Of course the use of reliable mutation information can still be beneficial for modeling accuracy in some cases. Thus, the use of a distance restraint between ligand amine and Glu1695.30 carboxyl45, for example, does improve ligand RMSD and overall number of contacts in our model (results not shown). In practical applications however, this type of restraints should be used very cautiously, and the results thoroughly cross-verified by other computational methods and experiments.
Accuracy of TM helical bundle predictions
Our models of AA2AR, like most of the models participating in GPCR assessment were capable of predicting conformation of Cα backbone of AA2AR 7TM helices with RMSDCα7TM about 2.0 Å. This result is similar to RMSDCα7TM between the crystal structures bRho, AA2AR and β2AR, which is also about 2.0 Å. (Note difference in calculations of RMSDCα7TM here and in ref19 as described in Methods section).
Certain local deviations in backbone structure between β2AR template and AA2AR however, are much higher, and they are driven by significant differences in the secondary structure of these GPCRs. Those differences are unlikely to be corrected by any existing optimization algorithms, unless reliable experimentally derived restraints can be used to drive these corrections. We already mentioned some of the new, unexpected features in AA2AR such as folding of the extracellular part of TM5 in π-helical instead of α-helical configuration. Other notable deviations between the homology modeling template (β2AR) and the AA2AR structures include (i) difference in TM1 tilting, resulting in ~ 5 Å distance between TM1 helix “tops”, (ii) different orientation of TM2 extracellular portion above proline-induced “bulge” yielding ~5 Å distance between tops of TM2 helices (iii) very peculiar proline-independent deviation of the top two turns of theTM3 helix, stabilized by a disulfide bond to EL2 loop (Cys74/Cys146).
Visual inspection of the 3EML and 2RH1 structures in Figure 1 suggests that the extracellular part of the helical bundle in 3EML is significantly “skewed” as compared to 2RH1 with RMSDCα7TM in this half of the TM bundle estimated at ~2.4 Å. Interestingly, the deviations in the cytoplasmic half are less dramatic and the RMSDCα7TM is only about 0.8 Å. This observation suggests more structural variability in the extracellular, ligand binding part of GPCRs, and more conserved cytoplasmic half, which share common interactions with a limited set of G-proteins.
Challenges in Loop predictions
Great diversity in the loop sequences and structures between GPCRs and their intrinsic conformational flexibility present even higher level challenges for accurate prediction of loop conformations and ligand interactions. Indeed, the results of the AA2AR modeling assessment show that though some of the modeling approaches allowed correct assignment of all three disulfide bonds in EL2 loop, even for the best predictions the overall Cα RMSD in the loop was higher than 7 Å (note that Costanzi model omits most of the EL2 residues)19.
Nevertheless, models generated with our approach and another few top models were able to correctly predict important elements of EL2 loop, critical for ligand binding. The stretch of the loop between Cys166 of EL2 and Met174 of TM5 proved to be the key ligand recognition site, primarily because of Phe1685.29 phenol aromatic stacking with the rings of adenine like and PTP scaffolds. Note, that correct prediction of Phe1685.29 orientation and interactions owes mostly to conservation of this structural element in the β2AR template that was used for homology modeling. Though prediction of such structural features with ab initio loop modeling methods is unlikely, at least without reliable experimentally derived restraints, further accumulation of GPCR crystal structures may yield a growing library of key structural elements in both extracellular and intracellular loop areas. These libraries might be used as initial templates for modeling loops.
Conclusions
Accurate prediction of 3D structure and ligand interactions remains a highly challenging endeavor for a majority of GPCR subfamilies that do not have a crystal structure resolved. Nevertheless, as the example of the AA2AR blind modeling assessment suggests, a combination of homology modeling and advanced optimization tools can be used successfully to predict ligand binding interactions and generate models potentially useful for VLS and compound prioritization. Although the current prediction exercise was limited to AA2AR, it can provide some important hints for GPCR modeling in general:
Homology modeling seems to be more accurate in prediction of overall GPCR structure and ligand binding pocket than ab initio modeling; its accuracy has already improved by switching from rhodopsin to β2AR structural template, and will improve further with the increasing number of available GPCR templates.
Ligand-guided optimization has significant benefits, yielding models with the highest number of correct ligand-receptor contacts and the best performance in large-scale VLS benchmarks. The ligand guided optimization helps to shape up the binding pocket environment, and while it still cannot correct certain types of large local differences between the template and the target protein backbones, it can often “compensate” for missing contacts.
In case of cross-family homology modeling of GPCRs, structure optimization has to consider backbone movements in the TM bundle. This can include not only rigid-body shifts/rotations of helices, but also local bending and even local changes in secondary structure (like in TM5). Using modeling methods that allow switches in secondary structure (e.g. from α- to π-helix) may be helpful in some cases.
Accurate ab initio modeling of long and flexible loops like EL2 is beyond capabilities of the current modeling resources. However, conserved parts, especially those involved in ligand binding can be identified and predicted accurately.
Judicious use of ligand binding activity, mutation analysis and biophysical data can dramatically improve quality of the models and their predictive power. However, caution should be exercised in interpreting mutagenic data as evidence of direct residue contacts, as they often reflect indirect effects. Charge-reversal or hydrogen bond donor-acceptor reversal mutagenesis data are much more useful and reliable.
Accurate modeling of GPCRs still requires an individualized approach to each receptor subfamily based on its level of homology to 3D templates, specific structural features and any additional experimental information that can be used in initial model generation. At the same time, the ligand guided approaches, including our LiBERO algorithm, may provide a reliable tool for automated receptor optimization and selection of the best models suitable for VLS applications.
Supplementary Material
Supplementary material includes (i) Figure SM1 (ii) Table SM1 (iii) A zip file with 64 blindly predicted complexes of our top ranked AA2AR model (mod2upu) with A2a ligands from Table SM1.
Supplementary Material
Acknowledgments
This study was partially funded by NIH grants R01-GM071872 and R01-GM074832 to RA. We would like to thank Charles Brooks, Scott Dixon, John Moult and Raymond Stevens for organizing the “Critical Assessment of GPCR Structure Modeling and Docking 2008”, and Karie Wright for help with manuscript preparation.
Abbreviations
- AA2AR
adenosine A2a receptor
- β2AR
β2-adrenergic receptor
- GPCR
G protein-coupled receptor
- RMSD
root mean square deviation
- TM
transmembrane
- EL2
extracellular loop 2
- VLS
Virtual Ligand Screening
- AUC
Area Under Curve
- NSQ_AUC
Normalized Square root AUC
- EN·NMA
Elastic Network Normal Mode Analysis
References
- 1.Reynolds K, Abagyan R, Katritch V. 3d structure and modeling of GPCRs: implications for drug discovery. In: Gilchrist A, editor. Shifting Paradigms in G Protein Coupled Receptors. Hoboken, NJ: Wiley & Sons, Inc; 2009. in press. [Google Scholar]
- 2.Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1(4):405–421. doi: 10.2174/1573406054368675. [DOI] [PubMed] [Google Scholar]
- 3.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7(4):339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 4.Jacoby E, Bouhelal R, Gerspacher M, Seuwen K. The 7 TM G-protein-coupled receptor target family. ChemMedChem. 2006;1(8):761–782. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 8.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322(5905):1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katritch V, Reynolds KA, Cherezov V, Hanson MA, Roth CB, Yeager M, Abagyan R. Analysis of full and partial agonists binding to beta(2)-adrenergic receptor suggests a role of transmembrane helix V in agonist-specific conformational changes. J Mol Recognit. 2009 doi: 10.1002/jmr.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds KA, Katritch V, Abagyan R. Identifying conformational changes of the beta(2) adrenoceptor that enable accurate prediction of ligand/receptor interactions and screening for GPCR modulators. J Comput Aided Mol Des. 2009;23(5):273–288. doi: 10.1007/s10822-008-9257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Graaf C, Rognan D. Selective structure-based virtual screening for full and partial agonists of the beta2 adrenergic receptor. J Med Chem. 2008;51(16):4978–4985. doi: 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
- 13.Kobilka B, Schertler GF. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol Sci. 2008 doi: 10.1016/j.tips.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Hanson MA, Stevens RC. Discovery of new GPCR biology: one receptor structure at a time. Structure. 2009;17(1):8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedorov O, Sundstrom M, Marsden B, Knapp S. Insights for the development of specific kinase inhibitors by targeted structural genomics. Drug Discov Today. 2007;12(9–10):365–372. doi: 10.1016/j.drudis.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Bissantz C, Logean A, Rognan D. High-throughput modeling of human G-protein coupled receptors: amino acid sequence alignment, three-dimensional model building, and receptor library screening. J Chem Inf Comput Sci. 2004;44(3):1162–1176. doi: 10.1021/ci034181a. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2(2):e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Goddard WA., 3rd Prediction of structure of G-protein coupled receptors and of bound ligands, with applications for drug design. Pac Symp Biocomput. 2008:344–353. [PubMed] [Google Scholar]
- 19.Michino M, Abola E, Brooks CL, III, Dixon JS, Moult J, Stevens RC. Critical assessment of GPCR structure modeling and docking 2008. Nature Reviews: Drug Discovery. 2009 doi: 10.1038/nrd2877. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lensink MF, Mendez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd Edition. Proteins. 2007;69(4):704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 21.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SK, Gao ZG, Van Rompaey P, Gross AS, Chen A, Van Calenbergh S, Jacobson KA. Modeling the adenosine receptors: comparison of the binding domains of A2A agonists and antagonists. J Med Chem. 2003;46(23):4847–4859. doi: 10.1021/jm0300431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cacciari B, Bolcato C, Spalluto G, Klotz KN, Bacilieri M, Deflorian F, Moro S. Pyrazolo-triazolo-pyrimidines as adenosine receptor antagonists: A complete structure-activity profile. Purinergic Signal. 2007;3(3):183–193. doi: 10.1007/s11302-006-9027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Graaf C, Foata N, Engkvist O, Rognan D. Molecular modeling of the second extracellular loop of G-protein coupled receptors and its implication on structure-based virtual screening. Proteins. 2008;71(2):599–620. doi: 10.1002/prot.21724. [DOI] [PubMed] [Google Scholar]
- 25.Michielan L, Bacilieri M, Schiesaro A, Bolcato C, Pastorin G, Spalluto G, Cacciari B, Klotz KN, Kaseda C, Moro S. Linear and nonlinear 3D-QSAR approaches in tandem with ligand-based homology modeling as a computational strategy to depict the pyrazolo-triazolo-pyrimidine antagonists binding site of the human adenosine A2A receptor. J Chem Inf Model. 2008;48(2):350–363. doi: 10.1021/ci700300w. [DOI] [PubMed] [Google Scholar]
- 26.Schapira M, Abagyan R, Totrov M. Nuclear hormone receptor targeted virtual screening. J Med Chem. 2003;46(14):3045–3059. doi: 10.1021/jm0300173. [DOI] [PubMed] [Google Scholar]
- 27.Bisson WH, Cheltsov AV, Bruey-Sedano N, Lin B, Chen J, Goldberger N, May LT, Christopoulos A, Dalton JT, Sexton PM, Zhang XK, Abagyan R. Discovery of antiandrogen activity of nonsteroidal scaffolds of marketed drugs. Proc Natl Acad Sci U S A. 2007;104(29):11927–11932. doi: 10.1073/pnas.0609752104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JZ, Wang J, Xie XQ. GPCR structure-based virtual screening approach for CB2 antagonist search. J Chem Inf Model. 2007;47(4):1626–1637. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- 29.Evers A, Klebe G. Successful virtual screening for a submicromolar antagonist of the neurokinin-1 receptor based on a ligand-supported homology model. J Med Chem. 2004;47(22):5381–5392. doi: 10.1021/jm0311487. [DOI] [PubMed] [Google Scholar]
- 30.Evers A, Klabunde T. Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the alpha1A adrenergic receptor. J Med Chem. 2005;48(4):1088–1097. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 31.Varady J, Wu X, Fang X, Min J, Hu Z, Levant B, Wang S. Molecular modeling of the three-dimensional structure of dopamine 3 (D3) subtype receptor: discovery of novel and potent D3 ligands through a hybrid pharmacophore- and structure-based database searching approach. J Med Chem. 2003;46(21):4377–4392. doi: 10.1021/jm030085p. [DOI] [PubMed] [Google Scholar]
- 32.Cavasotto CN, Orry AJ, Murgolo NJ, Czarniecki MF, Kocsi SA, Hawes BE, O’Neill KA, Hine H, Burton MS, Voigt JH, Abagyan RA, Bayne ML, Monsma FJ., Jr Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J Med Chem. 2008;51(3):581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 33.Rueda M, Bottegoni G, Abagyan R. Consistent Improvement of Cross-Docking Results Using Binding Site Ensembles Generated with Elastic Network Normal Modes. J Chem Inf Model. 2009 doi: 10.1021/ci8003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR--ligand database for chemical genomics drug discovery--database and tools update. Nucleic Acids Res. 2008;36(Database issue):D907–912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read RJ, Chavali G. Assessment of CASP7 predictions in the high accuracy template-based modeling category. Proteins. 2007;69(Suppl 8):27–37. doi: 10.1002/prot.21662. [DOI] [PubMed] [Google Scholar]
- 36.Cardozo T, Totrov M, Abagyan R. Homology modeling by the ICM method. Proteins. 1995;23(3):403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- 37.Abagyan RA, Batalov S. Do aligned sequences share the same fold? J Mol Biol. 1997;273(1):355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 38.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 39.Tirion MM. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys Rev Lett. 1996;77(9):1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 40.An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4(6):752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;(Suppl 1):215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 42.Truchon JF, Bayly CI. Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J Chem Inf Model. 2007;47(2):488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 43.Böhm H-J, Brode S, Hesse U, Klebe G. Oxygen and nitrogen in competitive situations: which is the hydrogen-bond acceptor? G Chem Eur J. 1996;2:1509–1513. [Google Scholar]
- 44.Kubinyi H. Drug research: myths, hype and reality. Nat Rev Drug Discov. 2003;2(8):665–668. doi: 10.1038/nrd1156. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Jiang Q, Glashofer M, Yehle S, Wess J, Jacobson KA. Glutamate residues in the second extracellular loop of the human A2a adenosine receptor are required for ligand recognition. Mol Pharmacol. 1996;49(4):683–691. [PMC free article] [PubMed] [Google Scholar]
- 46.DeWeese-Scott C, Moult J. Molecular modeling of protein function regions. Proteins. 2004;55(4):942–961. doi: 10.1002/prot.10519. [DOI] [PubMed] [Google Scholar]
- 47.Cavasotto CN, Orry AJ. Ligand docking and structure-based virtual screening in drug discovery. Curr Top Med Chem. 2007;7(10):1006–1014. doi: 10.2174/156802607780906753. [DOI] [PubMed] [Google Scholar]
- 48.Jacobson KA, Gao ZG. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 2006;5(3):247–264. doi: 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J Med Chem. 2009;52(2):397–406. doi: 10.1021/jm8009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abagyan RA, Totrov MM. Contact area difference (CAD): a robust measure to evaluate accuracy of protein models. J Mol Biol. 1997;268(3):678–685. doi: 10.1006/jmbi.1997.0994. [DOI] [PubMed] [Google Scholar]
- 51.Cristalli G, Lambertucci C, Marucci G, Volpini R, Dal Ben D. A2A adenosine receptor and its modulators: overview on a druggable GPCR and on structure-activity relationship analysis and binding requirements of agonists and antagonists. Curr Pharm Des. 2008;14(15):1525–1552. doi: 10.2174/138161208784480081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.