Abstract
With antigenically novel epidemic and pandemic influenza strains persistently on the horizon it is of fundamental importance that we understand whether heterosubtypic antibodies gained from exposures to circulating human influenzas exist and can protect against emerging novel strains. Our studies of IVIG obtained from an infection-naive population (Australian) enabled us to reveal heterosubtypic influenza antibodies that cross react with H5N1. We now expand those findings for an Australian donor population to include IVIG formulations from a variety of northern hemisphere populations. Examination of IVIGs from European and South East-Asian (Malaysian) blood donor populations further reveal heterosubtypic antibodies to H5N1 in humans from different global regions. Importantly these protect against highly pathogenic avian H5N1 infection in vitro, albeit at low titres of inhibition. Although there were qualitative and quantitative differences in binding and protection between globally different formulations, the heterosubtypic antibody activities for the respective IVIGs were in general quite similar. Of particular note because of the relative geographic proximity to the epicentre of H5N1 and the majority of human infections, was the similarity in the antibody binding responses between IVIGs from the Malayan peninsula, Europe and Australia. These findings highlight the value of employing IVIGs for the study of herd immunity, and particularly heterosubtypic antibody responses to viral antigens such as those conserved between circulating human influenzas and emerging influenza strains such as H5N1. They also open a window into a somewhat ill defined arena of antibody immunity, namely heterosubtypic immunity.
Keywords: Heterosubtypic, antibody, IVIG, influenza, H5N1, H1N1, H3N2
INTRODUCTION
Intravenous immunoglobulins (IVIGs) are widely used as highly-efficacious therapies to treat a broad range of conditions (Simon and Spath, 2003). Because IVIGs are highly-purified IgG products prepared from the plasmas of tens of thousands of blood donors they effectively reflect the herd immunity of their respective adult blood donor populations, and a sampling of the spectrum of antibodies to common infectious agents, such as influenza viruses. We earlier investigated whether Australian blood donors – uniquely geographically separated from the rest of the world on an island continent and naive to H5N1 exposure – may contain heterosubtypic antibodies, which cross-react with H5N1. Indeed, we provided evidence for heterosubtypic antibodies that bound to many of the proteins of H5N1, including both surface as well as internal proteins (Lynch et al, 2008).
There are considerable uncertainties that surround the current expansion of circulating H1N1, H5N1 and H9N2 strains and their potential to evolve and re-assort to feed highly-pathogenic strains capable of human-to-human pandemic transmission. Consequently, the issue of whether there is a level of heterosubtypic antibody protection in the community at large to protect against newly emergent influenzas is of considerable importance. In this study we assessed whether intravenous immunoglobulins (IVIGs) from a range of different regions across the globe contain heterosubtypic antibodies reactive to avian H5N1 influenza. Utilizing IVIGs as a gauge of the herd IgG immunity of adult populations we determined whether there are heterosubtypic antibodies to H5N1 in IVIGs prepared from European and South East Asian (Malaysian) blood donors as we have found with Australian-derived IVIG.
This study was designed to; i) assess the herd antibody immunity to a novel influenza, such as H5N1, ii) compare the herd immunities between geographically separate populations, and following positive affirmation, iii) profile the proteins of avian H5N1 influenza that IVIG cross-reactive antibodies bind to, and of most importance, iv) determine whether heterosubtypic IVIG antibodies from different regions are capable of protecting against in vitro H5N1 infection.
MATERIALS AND METHODS
IVIGs
The IVIGs studied were from a variety of different preparations, obtained from suppliers listed in Table 1. These highly-purified IgG preparations (≥ 96% pure) were isolated from plasma pools of up to 60,000 normal blood donors from the indicated geographical communities. IVIGs dilutions were prepared with TBS or PBS as indicated in the text.
Table 1:
Intravenous immunoglobulin (IVIG) preparations
| RegFormulation name | Prepared by | Donor plasma |
|---|---|---|
| Intragam P-Aust | CSL | Australian |
| Intragam P-Malay | CSL | Malaysian |
| Sandoglobulin-Eur | ZLBBioplasma | European |
| Octagam-Eur | Octapharma | European |
| Carrimune-US | CSLBehring | United States |
| Gamunex-US | Talecris | United States |
| Octagam-US | Octapharma | United States |
Influenza viruses
A list of influenza viruses is provided in Table 2. The human viruses for this study were representative vaccine strains of previously circulating viruses, and were included in northern and southern hemisphere influenza vaccines between 1992 and 2000. These had been expanded in chicken eggs, and included two purified H1N1 and two H3N2 influenza isolates from humans obtained from the WHO laboratory, Parkville, Victoria, Australia. The avian influenza viruses examined included three H5N1 isolates, and for comparison H7N7 and H9N2 isolates. The avian viruses were grown in the allantoic sac of embryonated chicken eggs and purified in sucrose gradients. Briefly, allantoic fluid was collected from infected eggs, pooled and clarified by low speed centrifugation (10,000xg for 10min). Virus was pelleted in an ultracentrifuge (80,000xg for 60min), resuspended in 5 ml TNE buffer (0.01 M Tris, 0.001M EDTA, 0.1M NaCl, pH 7.2) and purified by ultracentrifugation through a 20-50% discontinuous sucrose density gradient (80,000xg for 3hrs). The visible band of virus was extracted, and virus pelleted in an ultracentrifuge (80,000xg for 60min) and re-suspended in TNE buffer to 1% of the original volume of allantoic fluid. The A/Texas/36/1991 strain was represented in northern hemiphere vaccines (NHV) from 1992 to 1995, and for southern hemisphere vaccines (SHV) for 1997: the A/Beijing/32/1992 NHV 1993 to 1994; and the A/Sydney/5/1997 NHV 1997 to 2000 and SHV for 1998 to 2000.
Table 2:
Influenza strains
| Label | Strain | Subtype | GeneBank# | Country |
|---|---|---|---|---|
| α | A/Texas/36/1991 | H1N1 | ABF21312 | United States |
| β | A/Johannesburg/82/1996 | H1N1 | CY033610 | South Africa |
| γ | A/Beijing/32/1992 | H3N2 | GQ46319 | China |
| δ | A/Sydney/5/1997 | H3N2 | AB271703 | Australia |
| ζ | A/Chicken/Vietnam/8/'04 | H5N1-1 | AY724788 | Vietnam |
| θ | A/Chicken/Victoria/1985 | H7N7 | CY025069 | Australia |
| ι | A/Duck/Malacca/2003 | H9N2 | DQ139321 | Malaysia |
The growth and expansion of the HPAI avian viruses was performed at the CSIRO Australian Animal Health Laboratories (AAHL), Geelong, Victoria. To permit removal from the AAHL secure area and to allow the virus to be handled safely for antibody binding and protein analyses the viruses were inactivated by exposure to 5 megarads of radiation. The purity of the viruses was accessed by their SDS-PAGE profiles (see below and Lynch et al, 2008).
Infection studies
All experiments utilizing live HPAI viruses were performed in the high-security laboratories at AAHL. Virus neutralization assays were done in MDCK (Madin-Darby Canine Kidney – ATCC CCL-34) cells or Vero (African green monkey kidney – ATCC CCL-81) using a modification of a standard protocol (Cottey et al, 2001). For this, 100 tissue culture infectious doses of each virus were added to two-fold dilutions of each serum or IVIG preparation in microtitre plate wells and incubated for 60min at 37°C. Cells were added and the plates incubated at 37°C for 3 days. Viral activation by trypsin was unnecessary as all the viruses used in this study have a multi-basic cleavage site in their hemagglutinin protein. The neutralizing titre of serum or IVIG was the highest dilution giving complete neutralization of the virus.
Two-dimensional (2D) gel electrophoresis
Proteins were separated using 11cm precast immobilized pH gradients (IPG) across pH ranges 4-7 (Biorad, Life Sciences, Hercules, Ca, USA) and 6-11 (GE Healthcare, Uppsala, Sweden). 300μg of protein was loaded for each sample. Isoelectric focusing of proteins was conducted according to published techniques, in the pH range 4-7 (Nouwens et al, 2000) and proteins were focused for a total of 35kVh. Proteins spanning pH 6-11 were cup loaded and focused as previously described (Nouwens et al, 2000) for a total of 38kvh. The IPG strips were reduced, alkylated, and the detergent was exchanged, prior to second dimension separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Nouwens et al, 2000). SDS-PAGE was carried out in 11cm 8-16% (w/v) polyacrylamide/Tris-HCl gels (Bio-Rad) using a Criterion Dodeca Cell (Bio-Rad). Gels were fixed in 10% (v/v) methanol, 7% (v/v) acetic acid for 1hr, and then stained with Sypro Ruby (Molecular Probes, OR, USA) overnight. Gels were de-stained for 2hr in 10% (v/v) methanol and 7% (v/v) acetic acid and scanned using a ProXPRESS Proteomic Imaging System (PerkinElmer, MA, USA).
One-dimensional SDS-PAGE and immunoblotting
Viruses were subjected to SDS-solubilization and electrophoretic separation in criterion (Biorad, Life Sciences, Hercules, CA, USA) 4-16% (w/v) gradient gels (SDS-PAGE) under non-reducing conditions (Sloane et al, 2005). The relative molecular weights of influenza proteins were calculated from in-gel migration of pre-stained molecular weight standards (Sea Blue Plus2; Invitrogen Life Sciences). As the mobility of some pre-stained proteins can vary between batches as well as gel type, to account for this each batch used was compared in-house against a panel of unstained proteins over the molecular weight range, and where necessary the assigned molecular weight were recalibrated for our tests. As indicated the SDS-PAGE gel separated viral proteins were visualised by Coomassie Blue R (Sigma-Aldrich) or silver staining (Biorad).
For immunoblotting the SDS-PAGE separated viral proteins were electrotransfered onto 0.2μm Protranreg nitrocellulose (Schleicher and Schuell, Whatman, PE- Life & Analytical Sciences, Boston, MA), followed by blocking with 5% (w/v) skimmed milk powder in TBS. The antibody-binding step was performed using 1/200-1/500 dilutions of IVIG (50mg/ml), incubated for 1hr at room temperature (RT). After washing with TBS, blots were incubated at RT for 1hr with a 1/40,000 dilution of a horseradish peroxidase (HRP) goat anti-whole human antibody (Sigma-Aldrich chemicals, Mo). Following the removal of unbound secondary antibody by washing the immunoreactive bands were identified by chemi-luminescence detection (HRP detection kit, Amersham Biosciences UK Ltd. Buckinghamshire, England) (Sloane et al, 2005).
Antibody Binding Assays (MAP)
To assess antibodies binding to a variety of common viruses, including human influenza viruses, globally different IVIG products were subjected to bead-based multianalyte profiling (MAP) at Rules-Based Medicine (www.rulesbasedmedicine.com) (Austin, Texas, USA), as described in detail by Bertenshaw and colleagues (Bertenshaw et al, 2008). The Influenza antigens used for the MAP analyses were from sonicates of purified Influenza A H3N2 strains A/Texas/1/1977 and A/Shandong/9/1993; and Influenza B, B/Hong Kong/5/1972.
Viral microarray
For binding of IVIG antibodies to influenza antigens, whole influenza antigen was robotically spotted in a protein array, and detected by influenza-specific antibodies in IVIG. Serial dilutions of influenza strains A/Sydney/5/1997 (H3N2) and A/Johannesburg/82/1996 (H1N1), Tetanus toxoid, and BSA, were spotted (10nl/spot) onto nitrocellulose coated glass slides (Schleicher & Schuell, Dassel, Germany) using a Piezorray robotic arrayer (Perkin Elmer). Slides were cured overnight at 4°C then blocked with 5% (w/v) skimmed milk in PBS. One slide without milk blocking was stained with Ponceau Red to visualise total protein. Reactivity to the spotted antigen was determined by incubating blocked slides with IVIG (1mg/ml in PBS/0.05%, w/v, Tween-20) for 2hr, followed by 2hr with anti-hIgG-HRP (1:2000 in PBS/Tween-20), and visualized with a peroxidase substrate kit (Becton Dickinson) according to the manufacturer's instructions.
RESULTS
Antibodies in IVIGs to human and avian H5N1 influenzas
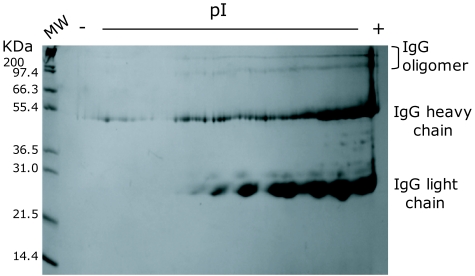
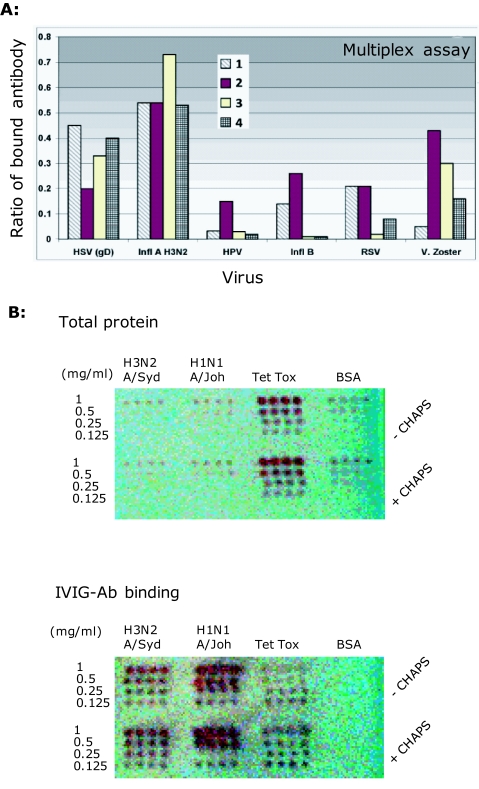
IVIGs are highly-purified IgG preparations prepared from large pools of human plasma and the immunoglobulin subtypes in IVIG formulations (i.e., IgG1, IgG2, etc) reflect those of normal plasma. The highly-purified nature of IVIGs is shown in Figure 1 of a protein stained 2D-gel electrophoresis map, and revealed heavy and light chains of these natural antibody preparations and their broad pI properties. In antibody binding studies using multiplex (Rules Based Medicine) and microarray binding assays (Figure 2A and B), there were quantitative and qualitative differences in anti-influenza antibody binding to the antigens of H3N2 influenza A/Shandong/9/93, both between different IVIG preparations (data not shown), and within batches of a single IVIG preparation. Along with quantitative differences in antibody binding of immobilized, H1N1 and H3N2 strains of human influenza (Figure 2B). The relative binding of human antibodies in IVIGs was compared by the multianalyte profiling of antibodies to antigens of a variety of other viruses that commonly infect humans, namely herpes simplex virus (HSV), human papilloma virus (HPV), Influenza B (Infl B), respiratory syncytial virus (RSV) and Varicella Zoster (V. Zoster). As might be predicted the antibody responses against the highly antigenic herpes simplex and influenza A viruses that commonly infect humans predominate over quantitative responses to the other viruses examined.
Figure 1.
2D gel electrophoretic profile of a representative IVIG preparation, with IPG isoelectric fractionation over 4-7 pI range. The separation of protein molecular weight standard markers in the acrylamide separation phase is shown on the left hand side of the 2D-separated immunoglobulin fractions. Note, the respective IVIG formulations used in this study (Table 1) have been de-identified, but each assigned a label A-G. The formulation used for 2D analysis was product D.
Figure 2.
Anti-human influenza binding antibodies in IVIGs: Assays for detection of Ab binding to conformational/ discontinuous influenza epitopes. A. Multiplex assay for anti-influenza IVIG Ab binding showing a comparison of the antibody binding activities of 4 different batches of a single IVIG formulation. IVIG antibody binding to Influenza A: H3N2 A/Shandong/9/1993 and Influenza B: B/Hong Kong/5/1972 antigens displayed on microbeads and performed at the Rules Based Medicine biomarker testing laboratory using multianalyte bead Fluorescence (www.rulesbasedmedicine.com). Binding to herpes simplex virus (HSV), human pappilloma virus (HPV), Influenza B (Infl B) respiratory syncytial virus (RSV) and varicella zoster (V. Zoster) are also included for comparison. The Y-axis values show the amount of IVIG antibody that binds to the respective antigens coated on beads and expressed as the mean ratio of total antibody binding to the respective viral antigens compared to highly-validated RBM internal-reference control standards. B. Microarray slides spotted with viral dilutions (1-0.125mg/ml) of H3N2 (A/Sydney/5/1997), H1N1 (A/Johannesburg/82/1996), tetanus toxoid (Tet Tox) or BSA. IVIG-antibody (D-3) bind robotically arrayed whole inactivated influenza and tetanus toxoid. The BSA is negative. Spots are 10nl and 0.8mm apart. [Note: antigens were bound in duplicate arrays with (top array panel of each slide) and without (bottom array panel on each slide) 0.005% CHAPS. CHAPS was tested as a means of improving liquid flow rates in the Piezorray, but as shown, had no influence on protein binding to the array substrate.]
Comparison of IVIG antibody binding to H5N1 and human influenzas
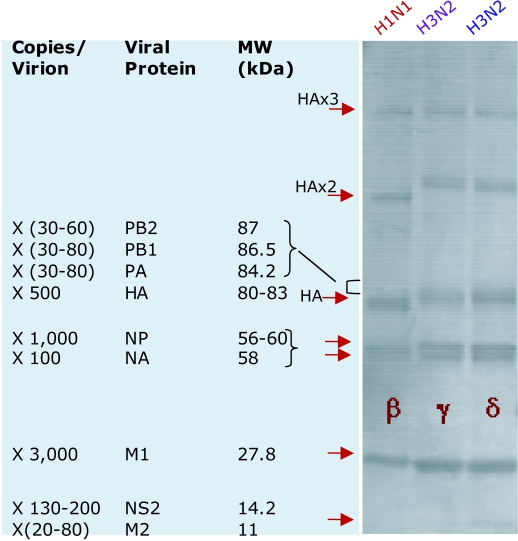
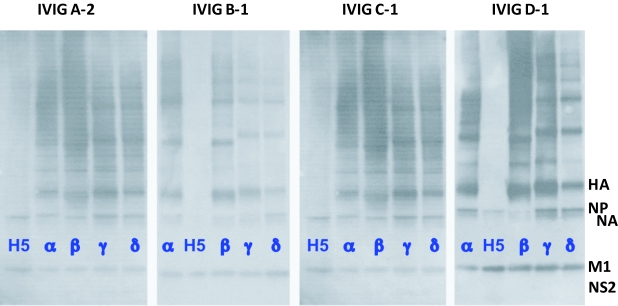
For comparison with our studies of H5N1 influenza proteins separated by SDS-PAGE, shown in Figure 3 are the protein bandings of the most prominent proteins of 3 human influenza strains. Included are their relative molecular weights and understood per virion abundance (Privalsky and Penhoet, 1978; Lamb and Krug, 2001; Lynch et al, 2008). Between the protein profiles of the H1N1 and H3N2 strains, the stand out feature is the faster migrating HA (and oligomer) bands of H1N1 compared with H3N2 virus, reflecting glycosylation differences (namely, H3 > H1) (Lynch et al, 2008). For our studies we have equalized viral loading onto SDS-PAGE gels on the basis of protein equivalence, using the most abundant and detectable protein of influenza, M1, as an internal standard (Lynch et al, 2008). By immunoblotting using IVIGs from geographically quite separate populations from Europe (i.e., Sandoglobulin, Octagam), Australia (i.e., Intragam-P) and Malaysia (i.e., Intragam-Pm), that were isolated prior to December 2005 (i.e., in the period June 2004 to November 2005) we demonstrated that as for our earlier studies of human antibodies in Australian IVIG, that similar binding activities for avian H5N1 influenza proteins are widely found in all of the different IVIG preparations (Figure 4). IVIG antibodies from the various products bound equivalently to the highly conserved M1 protein across H5N1, H1N1 and H3N2 influenza virusus. From these immunoblotting comparisons we find equivalent IVIG antibody binding to the abundant and conserved internal M1 and NP proteins between H5N1, H1N1 and H3N2 isolates. With minimal binding to the far more heterogeneous HA and NA envelope proteins of H5N1 compared to the human isolates, for which the latter show marked binding activities to the surface and immunodominant HA proteins (including oligomeric forms) of each of the human strains (Figure 4). When matched side-by-side differences in binding profiles for the human influenza isolates between the IVIG preparations undoubtedly reflects differences in the environmental strain-specific exposure and vaccination between the respective donor communities. As reported earlier (Lynch et al, 2008) and here, low levels of antibodies that bind to the HA of H5N1 are revealed by long exposure times. The specificity and heterosubtypic properties of this H5N1 antibody binding activity in IVIG and normal human serum was demonstrated by various means, including selective depletion by adsorption to human H3N2 and H1N1 influenza proteins in the absence of quantitative non-specific depletion of the total IVIG pool of IgGs (Lynch et al, 2008; Stelzer-Braid et al, 2008; and unpublished data).
Figure 3.
SDS-PAGE gel separations of influenza proteins showing coomassie-stained protein signatures of purified preparations of the human H1N1 A/Johannesbeg/82/1996 (β), and H3N2 strains A/Beijing/ 32/1992 (γ) and A/Sydney/5/97 (δ). The viruses used have been assigned a greek letters (Table 2). Relative molecular weights (Privalsky and Penhoet, 1978; Lynch et al, 2008) and approximate copies per virion (Lamb and Krug, 2001) for each of the influenza proteins are listed in the left panel.
Figure 4.
IVIG mmunoblotting profiles of human H1N1, H3N2 and avian H5N1 influenza proteins that bind human antibodies. Purified influenza samples were applied to 5-15% (w/v) SDS-PAGE gels and immunoblotted with the European (Sando-globulin-Eur, Octagam-Eur), Australian (Intragam P-Aust) or Malaysian (Intragam P-Malay) IVIGs. The Greek alphabets denote IVIG products, while the numbers A-2, B-1, C-1 and D-1 represent the particular batch of the product. The H5N1 virus (i.e., purified A/Chicken/Vietnam /8/2004 and identified as H5), is compared with the human H1N1 strains A/Texas/36/1991 (α) and A/Johannesbeg /82/1996 (β), and the H3N2 strains A/Beijing/32/1992 (γ) and A/Sydney/5/97 (δ).
Heterosubtypic human antibodies to avian influenza
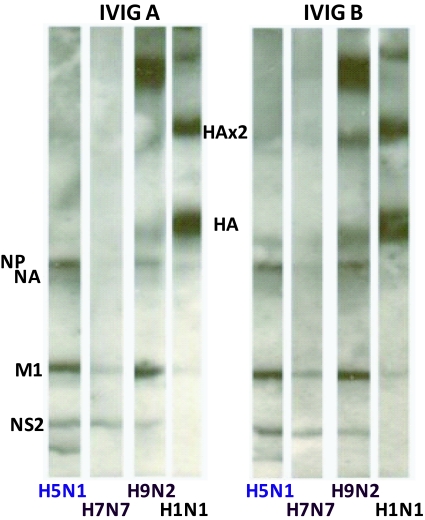
Optimization of chemiluminescence exposure times for avian H5N1 immunoblots has enabled more finite detection of human antibodies in IVIGs to many of the proteins of H5N1 (Lynch et al, 2008). This is further demonstrated in Figure 5, where several different avian isolates were examined by SDS-PAGE immunoblotting and compared directly with human H1N1 influenza at a 1/10 lower protein loading. As demonstrated in our earlier report, when optimized for signal detection human antibodies in globally different IVIG preparations revealed antibodies that bind with many of the H5N1 protein, none-the-least to H5 HA (Lynch et al, 2008). The faster migrating H5 HA on SDS-PAGE is indicative of lower total carbohydrate compositions than for the HA of H1 or H3 of human influenza viruses. These observations were made from consistent findings for the respective avian strains but we acknowledge a limited sampling and need for a more expansive examination of additional avian isolates. Consistent in our studies were apparent differences in the quantitative binding of antibodies in IVIGs for avian HAs with H9> H5> minimal if any H7 binding; and qualitative binding showing prominent apparent binding to the high molecular weight polymeric forms of H9 HA compared to H5 HA. More in-depth examination of avian strains is now needed to access and substantiate indicated differences in human IVIG antibody binding profiles between strains.
Figure 5.
Identification of Heterotypicheterosubtypic IVIG antibody binding to the proteins of avian viruses by immunoblotting. IVIG antibody binding H5N1 protein signatures have been observed for Intragam P-Aust, Sandoglobulin-Eur, and Octagam-Eur IVIGs, or Intragam P-Malay. Two different IVIG formulations reactive to the proteins of H5N1 (A/Chicken/ Vietnam/8/2004), H7N7 (A/Chicken/Victoria/1985) and H9N2 (A/Duck/Mellacca /2003) avian strains are shown, and for comparison a human H1N1 (A/Johannesburg/82/1996) is shown at a 1/10 protein loading compared to the avian viruses.
Human antibodies in IVIG protect against H5N1 infection in cell culture
As indicated from the comparative immunoblotting profiling of IVIGs antibodies above the low levels of heterosubtypic human antibodies that bind to H5N1 HA is of the order of 100-1000 times less than antibody binding to human influenza (H3N2, H1N1) HAs. Strain-specific human serum antibodies routinely give high neutralization titres in, in vitro, infection assays of human influenza infection. In contrast, any human antibody disruption of infection of a completely foreign influenza strain, such as H5N1, would likely be minimal, particularly considering the level of binding to H5N1 observed above. Despite this, when northern hemisphere IVIGs were tested in cell culture for H5N1 neutralization, we observed bona fide protection, albeit with modest titres with 100% inhibition (Table 3). These activities were similar to the neutralization levels observed within the same experiments in assays of southern hemisphere, Australian-derived IVIG, as reported earlier (Lynch et al, 2008) and duplicated her for direct comparison. Each of the IVIG preparations inhibited infections of 3 strains of H5N1 (i.e., 2 clade representative strains – one from human and two from avian infections), when tested in both Vero and MDCK cell culture infections. As each of the IVIG preparations inhibit the infections of 3 strains of H5N1 but not the infection of H7N7 virus, strain-specific heterosubtypic protection is implicated. The individual and combined roles of anti-HA and NA antibodies (as well as the more abundant antibodies to NP, M1, NS2) in providing this protection remain to be defined.
Table 3:
IVIG neutralization of H5N1 infection of Vero and MDCK cells. IVIG products from Europe (Octagam-Europe) (2 batches), Australia (Intragam P-Australia) and Malaysia (Intragam P-Malay) were examined in triplicate. These respective IVIG preparations are listed in this table using de-identified labels, A to D. [*Denotes duplicate experiments]
| IVIG | HAPI influenza A virus strain – Vero cell infection | |||
|---|---|---|---|---|
| Chick/viet/8/'04 H5N1 (Vietnam) | Chick/wates/1/'05 H5N1 (Indonesia) | Human/viet/1203/'04 H5N1 (Vietnam) | Chick/victoria/'85 H7N7 (Australia) | |
| A | 8-16* | 32 | 32* | <2* |
| B | <2 & 2* | 8 | 2-8* | <2* |
| C | 2-4* | 8 | 8-16* | <2* |
| D | 8* | 64 | 8-32* | <2* |
| MDCK cell infections | ||||
| A | 8 | 1024 | 128 | ND |
| B | 4 | 1024 | 32 | ND |
| C | 16 | 512 | 32 | ND |
| D | 16 | 512 | 256 | ND |
DISCUSSION
H5N1 continues to be a major global health concern, with 60% mortality among humans and 100% among some bird species. Understanding the immune mechanisms that underpin the immune protection in human survivors is therefore of fundamental importance, in order to firstly understand natural protection against any future expansion of H5N1 or other emerging new influenzas (e.g., the 2009 H1N1), and secondly to harness any heterosubtypic immune protection through the design of new vaccines and antivirals. Undoubtedly, innate and heterosubtypic cellular immune mechanisms protect against infection or disease of novel influenza viruses, but these are unlikely to explain the observed 40% level of ‘natural’ protection against H5N1 human infections. We believe our findings of heterosubtypic anti-H5N1 reactive antibodies in different unexposed populations offer an insight into a tenable process of adaptive antibody immunity to protect against novel influenzas such as H5N1. Following on from our earlier reports (Lynch et al, 2008; Stelzer-Braid et al, 2008), where we identified anti-H5N1 directed human antibodies in sera and IVIG preparations from Australian blood donors we now show that IVIGs from different geographic populations from the northern hemisphere, and notably from European blood donations collected prior to the emergence of avian H5N1 influenza in Europe, similarly share this property. This suggests that some pre-existing heterosubtypic ‘herd’ antibody immunity to avian H5N1 influenza may be a universal characteristic of populations across the globe. To demarcate a heterosubtypic response from the well-studied ‘seasonal’ adaptive response that current vaccines stimulate we refer to this particular type of humoral immunity as a ‘seasoned’ adaptive response.
While the value of strain-specific antibodies to protect against seasonal influenzas is well appreciated and understood, any suggestion of broad acting antibodies to protect against hitherto unseen novel influenza viruses (e.g., from birds or pigs) has till now been overlooked. From our observations we propose that pre-existing heterosubtypic antibodies in the community may offer a molecular mechanism to explain protection against H5N1 and past pandemic strains, and together with innate and heterosubtypic cellular immune protection could provide a significant barrier of protection for novel viruses. Our findings here and in previous reports give direct evidence that heterosubtypic ‘seasoned’ antibody responses to novel influenza viruses do exist, but what we do not yet know are the sites and mechanisms involved, and most importantly, in the wider context the significance of this response for natural protection against emerging infections and whether it could be harnessed for new styled anti-virals.
This proposition gives rise to a number of predictions, which themselves can be examined and challenged, and if found to be of value, appropriately validated. These relate to the influenza protein epitopes that heterosubtypic antibodies target, and how an adaptive heterosubtypic humoral response may be established and progressed. We predict the development of antibodies to sites that are highly-conserved across influenza viruses, such that, when a new and antigenically novel influenza virus arises antibodies to exposed conserved sites, irrespective of the strain, could bind and disrupt viral infection and growth. In support of this concept the extent of binding of heterosubtypic antibodies of human serum and IVIGs to influenza target proteins directly correlates with the extent of the viral protein cross-strain sequence conservation. In making this comparison we benefit from the bioinformatic assessments of Bui et al (2007) and Heiny et al (2008) of conserved sites across a wide variety of influenza strains. Particularly significant is the study by Heiny and colleagues involving examination of 36,343 influenza A sequences of linear stretches of 9 amino acids or more that were conserved across influenza subtypes (Heiny et al, 2007). On those criteria, they identified a wide range of conserved sequences between the different proteins. Notably, they found that 50% of the protein sequence of polymerase protein PB2 is widely conserved followed by 36% conservation of PB1, 27% of M1, 25% of NP, 16% of PA, and only 2% of HA. On this basis, none of the proteins NA, PB1-F2, M2, NS1 or NS2 had fully conserved regions across this large sequence data base (Heiny et al, 2007). Within the sensitivity constraints of our studies and restricted to the most abundant proteins (see Figure 3) we broadly found that antibody binding was preferential for the H5N1 proteins with the highest sequence conservations with human influenzas (Figure 5). One notable example was that of the most abundant influenza protein, the matrix protein M1 of H5N1 with 27% conserved stretches of sequence, which we found to have equivalent IVIG antibody binding as for the M1 protein of human H1N1 and H3N2 influenza strains (also see Lynch et al, 2008). These considerations do not however take into account the contributions of other heterosubtypic antibody responses to conserved, linear regions of less than 9 amino acids, or to conserved discontinuous epitopes. Unequivocal validation of each of the antibody-epitope interactions is now required to define the respective antibody binding kinetics and saturation, and by peptide and protease fragmentation studies coupled with mass spectrometry to definitively resolve the target-epitopes and match against conserved sites predicted from bioinformatics data-base screening and molecular modelling.
In this study we used IVIGs as a tool to broadly study human antibodies for H5N1. However, IVIGs as a potential therapeutic option in the event of the sudden emergence of a HPI pandemic could also be considered. IVIGs are well-tolerated therapies for primary and secondary immune-deficiencies and autoimmune disorders (Simon and Spath, 2003). However, while IVIGs and hyper-immune immunoglobulins are highly valued and studied therapies for a variety of infectious agents, including CMV and hepatitis viruses (Gupta et al, 1996), there is a notable paucity of information in the literature on the anti-viral properties of IVIGs for common viral agents, such as for influenzas. If it was possible to identify a cohort of individuals with solid pre-existing seasoned immunity for hyper-immune heterosubtypic anti-influenza antibodies for passive immune therapy, this could add an additional therapeutic asset to complement small molecule anti-virals and vaccines. If hyper-immune anti-influenza IVIG were possible, in addition to harvesting pandemic convalescent influenza antibodies they would offer an already licensed production-capable fallback therapy option during a HPI pandemic. The passive transfer of antibodies for the protection of infection-naïve individuals at risk from pandemic influenza was successfully deployed in 1918 and more recently in the successful treatment of an individual infected with H5N1, and to this day convalescent ‘Spanish Flu’ survivors have retained protective antibody responses to the 1918 H1N1 influenza (Luke et al, 2006). The antibody repertoires of convalescent individuals would be particularly beneficial, as they would possess a complement of both seasoned and seasonal antibodies to protect against epidemic and pandemic influenzas. Furthermore, as there is growing evidence implicating the prominent role of secondary infections in the mortality of pandemic influenzas, IVIGs, as broad anti-infective agents, may also be beneficial in a pandemic as a backup for antibiotics, to keep secondary infections in check (e.g., bacterial pneumonia, CMV, tuberculosis) (Klugman et al, 2009a; Klugman et al, 2009b) and to suppress overt host cytokine-storm responses particularly in younger individuals, who are prime targets for pandemic influenza mortality.
As the quest in the design of universal vaccines and inhibitors for influenza gains momentum we may well learn many essential lessons from how heterosubtypic antibody immunity has developed in nature to protect against new infectious challenges.
ACKNOWLEDGEMENTS
Competitive Grant Support for the study of IVIG and avian H5N1 antibody immunity was gained from the Australian Red Cross Blood Service Grant for GWL and from the University of Sydney for GWL and JSS. We wish to acknowledge the Ramaciotti Centre for Gene Function Analysis (The University of New South Wales, Randwick, NSW Australia) for access and use of the Piezorrayer.
LIST OF ABBREVIATIONS
- Ab
Antibody
- CHAPS
3-[(3-Cholamindopropyl)dimethylamminio-1-propanesulfate
- HPI
High-pathogenic influenza
- HPV
Human papilloma virus
- HRP
Horseradish peroxidase
- HSV
Herpes simplex virus
- IgG
Immunoglobulin G
- IVIG
Intravenous immunoglobulins
- KDa
Kilodalton
- MAP
Multianalyte profiling
- MDCK
Madin-Darby canine kidney
- MW
Molecular weight
- PBS
Phosphate-buffered saline
- RSV
Respiratory syncytial virus
- RSV
Respiratory syncytial virus
- TBS
Tris-buffered saline
- TNE
Tris/NaCl/EDTA buffer
COMPETING INTERESTS
None declared.
REFERENCES
- Lynch GW, Selleck PW, Axell AM, et al. Cross-Reactive anti-Avian H5N1 Influenza Neutralizing Antibodies in a Normal ‘Exposure-Naive’ Australian Blood Donor Population. The Open Immunol J. 2008;1:13–19. [Google Scholar]
- Simon HU, Spath PJ. IVIG-mechanisms of action. Allergy. 2003;58:543–552. doi: 10.1034/j.1398-9995.2003.00239.x. [DOI] [PubMed] [Google Scholar]
- Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci USA. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiny AT, Miotto O, Srinivasan KN, et al. Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets. PLoS One. 2007;2:e1190. doi: 10.1371/journal.pone.0001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottey R, Rowe CA, Bender BS. Current Protocols in Immunology: Influenza Virus. New York, USA: John Wiley and Sons Inc; 2001. Chapter 19, Unit 19-11. [DOI] [PubMed] [Google Scholar]
- Nouwens AS, Cordwell SJ, Larsen MR, et al. Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis. 2000;21:3797–3809. doi: 10.1002/1522-2683(200011)21:17<3797::AID-ELPS3797>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Sloane AJ, Raso V, Dimitrov DS, et al. Marked structural and functional heterogeneity in CXCR4: separation of HIV-1 and SDF-1alpha responses. Immunol Cell Biol. 2005;83:129–143. doi: 10.1111/j.1440-1711.2004.01304.x. [DOI] [PubMed] [Google Scholar]
- Bertenshaw GP, Yip P, Seshaiah P, et al. Multianalyte profiling of serum antigens and autoimmune and infectious disease molecules to identify biomarkers dysregulated in epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2872–2881. doi: 10.1158/1055-9965.EPI-08-0464. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Krug RM. Orthomyxoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fundamental Virology. 4th edition. Philadelphia, USA: Lippincot Williams & Wilkins; 2001. pp. 725–770. [Google Scholar]
- Privalsky ML, Penhoet EE. Influenza virus proteins: identity, synthesis, and modification analyzed by two-dimensional gel electrophoresis. Proc Natl Acad Sci USA. 1978;75:3625–3629. doi: 10.1073/pnas.75.8.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer-Braid S, Wong B, Robertson P, et al. A commercial ELISA detects high levels of human H5 antibody but cross-reacts with influenza A antibodies. J Clin Virol. 2008;43:241–243. doi: 10.1016/j.jcv.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Gupta CK, Leszczynski J, Gupta RK, Siber GR. IgG subclass antibodies to human cytomegalovirus (CMV) in normal human plasma samples and immune globulins and their neutralizing activities. Biologicals. 1996;24:117–124. doi: 10.1006/biol.1996.0015. [DOI] [PubMed] [Google Scholar]
- Luke TC, Kilbane EM, Jackson JL, Hoffman SL. Meta-analysis: Convalescent blood products for Spanish influenza pneumonia: A future H5N1 treatment? Ann Intern Med. 2006;145:631–632. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- Klugman KP, Astley CM, Lipsitch M. Time from illness onset to death, 1918 influenza and pneumococcal pneumonia. Emerg Infect Dis. 2009;15:346–347. doi: 10.3201/eid1502.081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugman KP, Chien YW, Madhi SA. Pneumococcal pneumonia and influenza: A deadly combination. Vaccine. 2009;27(Suppl 3):C9–C14. doi: 10.1016/j.vaccine.2009.06.007. [DOI] [PubMed] [Google Scholar]