Abstract
How DNA helical tension is constrained along the linear chromosomes of eukaryotic cells is poorly understood. In this study, we induced the accumulation of DNA (+) helical tension in Saccharomyces cerevisiae cells and examined how DNA transcription was affected along yeast chromosomes. The results revealed that, whereas the overwinding of DNA produced a general impairment of transcription initiation, genes situated at <100 kb from the chromosomal ends gradually escaped from the transcription stall. This novel positional effect seemed to be a simple function of the gene distance to the telomere: It occurred evenly in all 32 chromosome extremities and was independent of the atypical structure and transcription activity of subtelomeric chromatin. These results suggest that DNA helical tension dissipates at chromosomal ends and, therefore, provides a functional indication that yeast chromosome extremities are topologically open. The gradual escape from the transcription stall along the chromosomal flanks also indicates that friction restrictions to DNA twist diffusion, rather than tight topological boundaries, might suffice to confine DNA helical tension along eukaryotic chromatin.
Keywords: DNA supercoiling, DNA twist, telomere, topoisomerase, transcription
Introduction
The helical tension of DNA has deep implications in most genome transactions. This condition of altered twist facilitates or hinders the melting of the duplex as well as its interactions with structural and regulatory factors (Wang et al, 1983; Vologodskii and Cozzarelli, 1994). DNA helical tension also promotes the formation of supercoils that contribute to the juxtaposition of distant DNA sites and to the global folding of DNA (Huang et al, 2001). In bacteria, chromosomes are circular and DNA seems to be organized into independent topological domains (Delius and Worcel, 1974; Sinden and Pettijohn, 1981; Postow et al, 2004), in which different levels of helical tension can be modulated. In eukaryotic cells, however, chromosomes are linear and DNA is folded into more complex chromatin fibres. Therefore, the issue of whether DNA is organized into closed topological domains in which helical tension is constrained is less clear and remains controversial (Eissenberg et al, 1985; Esposito and Sinden, 1988; Freeman and Garrard, 1992). Addressing this issue will require better tools to examine the helical state of chromosomal DNA, as well as a better understanding on the multiplicity of factors that determine the generation, transmission, and dissipation of DNA twisting forces in vivo. Thus far, DNA tracking processes (such as transcription and replication) (Liu and Wang, 1987; Brill and Sternglanz, 1988; Giaever and Wang, 1988) and the activity of different topoisomerases (Salceda et al, 2006) are the main factors known to be involved in the generation and relaxation of DNA helical tension in eukaryotic cells.
DNA transcription enforces axial rotation of the duplex relative to the large RNA polymerase complex. This rotation is quickly hindered by nearby interactions that anchor DNA to other structures or, simply, by the large rotational drag of DNA folded in chromatin. Consequently, positive (+) DNA helical tension increases in front of an advancing polymerase and negative (−) DNA helical tension arises behind it (Liu and Wang, 1987). Analogously, (+) DNA helical tension also builds up in front of DNA replication forks (Schvartzman and Stasiak, 2004). DNA topoisomerases relax this helical stress by producing temporary single- or double-strand DNA breaks (Champoux, 2001; Wang, 2002). In eukaryotic cells, topoisomerase I (encoded by TOP1) cleaves one strand of the duplex allowing the DNA to rotate in either direction around the uncleaved strand; and topoisomerase II (encoded by TOP2) removes supercoil crossings by transporting one segment of duplex DNA through a transient double-strand break in another (Champoux, 2001; Wang, 2002).
In the yeast Saccharomyces cerevisiae, as both topoisomerase I and II can relax (+) and (−) helical tension, the presence of either one of the two enzymes suffices for transcription to proceed (Kim and Wang, 1989). Inactivation of both topoisomerases in Δtop1 top2ts double mutants alters rRNA and polyA+ RNA synthesis (Brill et al, 1987; Yamagishi and Nomura, 1988), although transcription is not broadly reduced, as concurrent (+) and (−) supercoiled domains can eventually cancel each other out (Stupina and Wang, 2004). However, a large reduction of global RNA synthesis occurs when the inactivation of topoisomerases I and II is combined with the ectopic expression of the TopA gene that encodes Escherichia coli topoisomerase I (Gartenberg and Wang, 1992). This bacterial enzyme operates only on underwound DNA and yields, therefore, an asymmetric relaxation of (+) and (−) supercoils (Giaever and Wang, 1988). As a result, (+) helical tension accumulates along intracellular DNA up to specific linking number differences of about 4% (Salceda et al, 2006). This threshold value is likely to reflect the point at which the increasing overwinding of the duplex precludes DNA transcription, either at the initiation or elongation steps (Salceda et al, 2006).
In this study, we induced the accumulation of (+) DNA helical stress in yeast and used microarray analysis to examine how transcriptome alterations spread throughout the yeast chromosomes. The results exposed a striking positional effect in all 32 yeast chromosomal arms: whereas most genes reduced their transcript levels by several fold, genes situated at <100 kb from the chromosomal ends gradually escaped from the transcription stall.
These results are consistent with a dissipation of DNA helical tension at the chromosomal ends and provide, thus, a functional indication that yeast chromosome extremities are topologically open. The gradual escape from the transcription stall observed along all chromosomal flanks also denotes that tight topological boundaries are scarce in yeast chromatin.
Results
Functional response of the yeast transcriptome to the induction of DNA helical stress
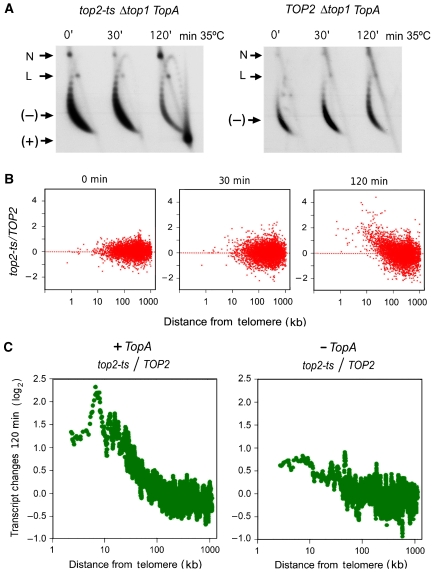
We induced (+) helical stress of DNA in S. cerevisiae Δtop1 top2ts mutant cells that constitutively expressed the E. coli TopA gene by thermal inactivation of the topoisomerase II temperature-sensitive allele. We monitored the accumulation of helical tension at different time points of thermal shift (0, 30, and 120 min at 35°C) by analysing the supercoiling of DNA in yeast circular minichromosomes (Figure 1A, left). After 30 min, DNA topology was not significantly altered. However, after 120 min of topoisomerase II inactivation, most minichromosome molecules seemed highly positively supercoiled. We conducted the same thermal shift with the control strain, Δtop1 TOP2 that also expressed the E. coli TopA gene, and no trace of positively supercoiled molecules was observed (Figure 1A, right).
Figure 1.
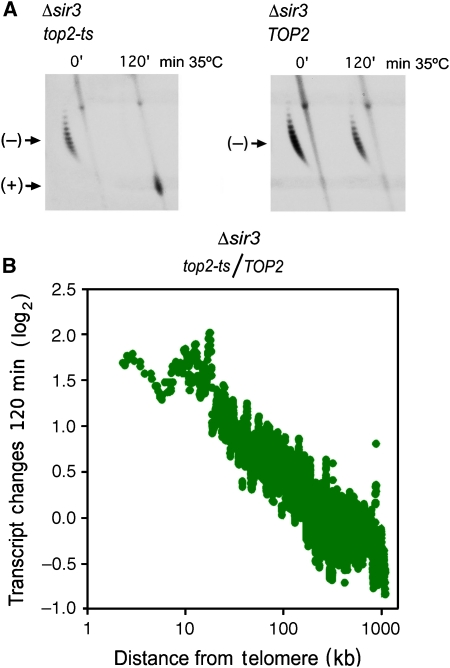
DNA positive helical stress alters the yeast transcriptome according to the chromosomal position of the genes. (A) Two-dimensional agarose gel electrophoresis of DNA extracted from Δtop1 top2ts TopA (left) and Δtop1 TOP2 TopA (right) yeast strains after 0, 30, and 120 min of heat inactivation (35°C) of topoisomerase II. The gel-blot shows the conformers of the 2μ circular minichromosome. (−), negatively supercoiled DNA circles; (+), positively supercoiled DNA circles; N, nicked DNA circles; L, linear DNA. (B) The variation of transcript levels (log2 ratio top2ts/TOP2) after 0, 30, and 120 min of topoisomerase II inactivation is plotted for all analysed genes against their distance (bp) from the telomere. (C) Comparison of the variation of transcript levels (log2 ratio top2ts/TOP2) after 120 min of topoisomerase II inactivation with (left) and without (right) the expression of the E. coli TopA gene. Both graphs show averaged values for genes situated to a similar distance from the telomere in all the S. cerevisiae chromosomal arms (each point averages 20 genes). Data were adjusted to obtain an average ratio of 1 for all genes analysed.
We extracted total RNA from the above top2ts and TOP2 cells after 0, 30, and 120 min of topoisomerase II inactivation and used microarrays to compare transcript levels between both strains. The microarray data exposed apparently modest changes in the transcriptome of top2ts cells as compared with that of TOP2 cells (Supplementary Table I). At the 30 min time point, about 5% of genes increased the relative abundance of their transcripts by a factor of two or more, and about 5.2% decreased it by a similar factor. At the 120 min time point, the fractions of relatively increased and decreased transcripts changed to 8.8 and 6.4%, respectively. Analysis of the functional categories of the affected genes revealed that, at the 30 min time point, the main traits were a severe reduction in the transcription of ribosomal genes and an increase in that of carbohydrate metabolism genes. This response is typical for many forms of cellular stress (Gasch et al, 2000; Rojas et al, 2008). However, when (+) supercoiling of DNA was evident at the 120 min time point (Figure 1A), the functional characteristics of altered genes changed and Ty-related transcripts became the predominant over-represented category (Supplementary Table I).
Physical clustering of altered genes on accumulation of DNA helical stress
To examine how transcriptome alterations between the above top2ts and TOP2 strains spread throughout the yeast chromosomes after the accumulation of DNA (+) helical stress, we plotted the relative transcript variations (after 0, 30 and 120 min of topoisomerase II inactivation) versus the respective gene distance from the telomere (Figure 1B). At the 0 and 30 min time points, the overall transcript variations did not expose any deviation with respect to gene position. However, a striking asymmetry occurred at the 120 min time point. When DNA helical stress was evident, the relative abundance of transcripts of most genes closer than 100 kb from the telomere increased gradually towards the chromosomal end. At this time point, about half of the over-represented transcripts (>45% of total, P=1.4 × 10−95, χ2 test) were from genes located at <50 kb from the telomere, a compartment that confines about 15% of yeast genes. Conversely, decreased transcripts in the same section were strongly under-represented (<5% of total, P=4 × 10−9, χ2 test).
Although the differential response of the chromosome flanking genes seemed associated to the accumulation of DNA (+) helical stress at the 120 min time point, we had to discard that this positional effect could be simply consequent to the prolonged lack of topoisomerase II activity. We conducted, therefore, a control experiment by comparing the transcriptome of cells with wild type and thermo-sensitive topoisomerase II, but no TopA expression and thus no (+) DNA supercoiling. After 120 min of topoisomerase II inactivation, relative abundance of transcripts from genes at the chromosomal flanks were not significantly increased (Figure 1C, right), in sharp contrast to that observed on accumulation of DNA (+) helical stress (Figure 1C, left).
Chromosomal flanks escape from the transcription stall produced by DNA helical stress
As a global reduction of RNA synthesis was expected on accumulation of DNA (+) helical stress (Gartenberg and Wang, 1992), we used qRT–PCR to determine the absolute value of transcript levels in our strains, as well as to validate the differential response of the chromosome flanks uncovered by the microarray data. Notice that, because microarray data are routinely normalized against average values, changes affecting a majority of genes would seem as the opposite effect in the minority of non-affected ones. In contrast, qRT–PCR data correspond to straight changes in Ct values, with no standard reference, except for the amount of cDNA loaded in each reaction.
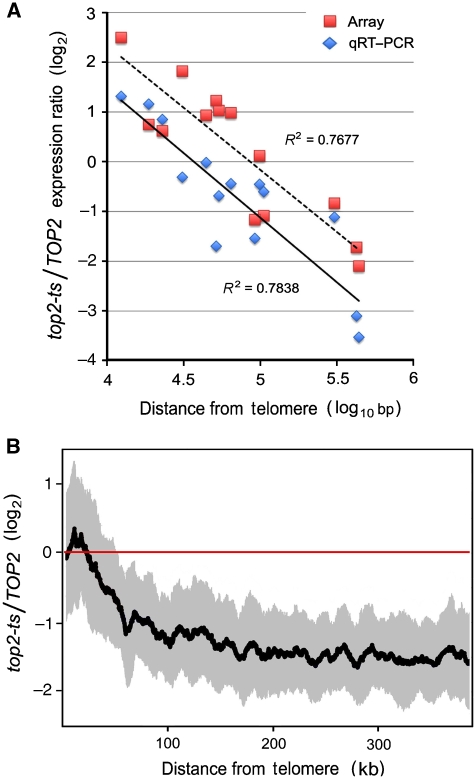
To conduct the qRT–PCR analysis, we selected a subset of genes from different chromosomes and distances from the telomeres (Supplementary Table 2). The PCR data exposed a clear gradient of transcript ratios between top2ts and TOP2 strains as genes vary their distance to the chromosomal ends, with a similar slope to that observed in the microarray dataset (Figure 2A). However, the transcript levels obtained from qRT–PCR were approximately one log2 lower than the corresponding values obtained from microarrays. The qRT–PCR results indicated, therefore, that DNA (+) helical stress reduces by two- to six-fold transcription of most yeast genes (over 80% of the genome). Then, the relative increase of transcripts from genes located at chromosomal flanks exposed by the microarray data was indeed reflecting a lesser reduction or nearly no effect of the induced DNA helical stress, as genes get closer to the chromosomal ends (Figure 2B).
Figure 2.
Chromosomal flanks escape from the global reduction of transcript levels. (A) Correlation between microarray and qRT–PCR data. Microarray data of selected genes are given as ratios (log2) between top2ts and TOP2 strains after 120 min of topoisomerase II inactivation (red dots). The respective qRT–PCR values correspond to differences in Ct values at time 0 and 120 min after topoisomerase II inactivation in the top2ts strain (blue dots). Note that Ct values are linearly correlated to the log2 of the concentration of mRNA for each specific gene. Regression lines for both sets of data are shown. (B) The microarray data ([top2-ts/TOP2]120 min/[top2-ts)/TOP2]0 min) is represented by fitting the fold reduction values of the above selected genes according to qRT–PCR data. The graph plots (log2) a sliding mean and standard deviation (black and grey, respectively) of 20 consecutive genes against the gene distance (bp) from the chromosomal end.
All chromosomal flanks show the same positional response to DNA helical stress
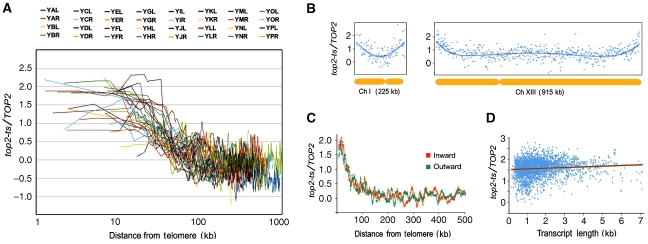
Next, we inspected whether the differential response of chromosome flanking genes to DNA helical stress was consistent in all yeast chromosomes. We plotted the relative transcript variations versus the gene distances from the telomere for the individual 32 chromosomal arms (Figure 3A). In all cases, relative transcript levels gradually increased, as genes got closer to the chromosomal end. Moreover, regardless of the respective chromosomal length, all flanks followed similar slopes starting around 100 kb from the telomere. By this circumstance, in short chromosomes (such as Chr. I, 225 kb) the positional effect covered nearly the entire chromosome (Figure 3B). In longer chromosomes (such as Chr. XIII, 915kb), only the flanks exposed the differential response, without similar deviations of the transcript levels at more internal or core regions (Figure 3B). We also observed that the positional effect equally affected genes transcribed in the direction towards and away from the chromosomal end (Figure 3C), and that the different response of chromosome flanking genes to DNA helical stress was independent of their respective transcript length (Figure 3D).
Figure 3.
Positional response to DNA helical stress occurs similarly in all chromosomal flanks and independently of chromosome length, transcript length, and transcript direction. (A) The variation of transcript levels (log2) on the accumulation of DNA (+) helical stress is plotted in each of the 32 yeast chromosomal arms against the corresponding gene distance (bp) from the telomere. Coloured splines average values for nine consecutive genes. Distances from telomeres correspond to the central gene. (B) The relative variation of individual transcript levels (log2) is plotted along the physical length of yeast chromosomes I (225 kb) and XIII (915 kb). Polynomial tendency lanes are shown. (C) The relative variation of inwards and outwards transcripts plotted against their gene distance (kb) from the corresponding telomere. Values are calculated as in Figure 2B. (D) The relative variations of transcript level (log2) of the genes located at <100 kb from the telomere is plotted against their respective transcript length (kb).
Differential response of chromosomal flanks to DNA helical stress is not related to a reduced transcription activity of subtelomeric genes
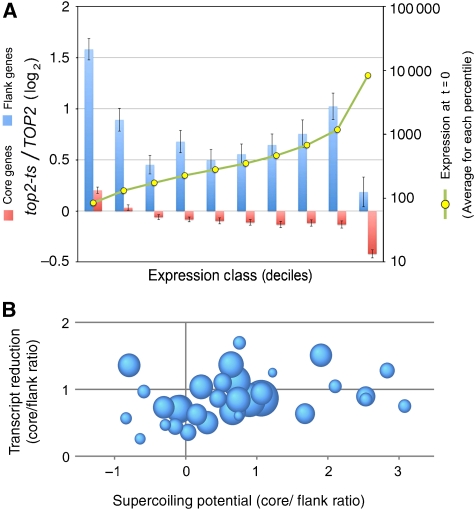
It is well documented that genes close to the telomere (up to 20 kb from chromosomal ends) tend to be transcriptionally inefficient (Gottschling et al, 1990; Vegas-Palas et al, 1997; for review, see Ottaviani et al, 2008). This silencing effect varies substantially from telomere to telomere and affects individual genes distinctively (Pryde and Louis, 1999; Mondoux and Zakian, 2007). Therefore, although the escape from the transcription stall was evenly observed in all chromosomal flanks on accumulation of helical stress, we discarded that this outcome could be simply a consequence of the low transcript abundance of some subtelomeric genes. To test whether the extent of transcript reduction was biased because of the initial mRNA abundance, we classified all yeast genes by their starting mRNA abundance in 10 equal groups (decile classes). We then compared, in each group, the effect of (+) helical tension between genes located at <50 kb from the telomere (flank genes) and the rest (core genes). The result of this analysis visibly indicated that flank genes behave always differently from core genes, irrespective of their initial mRNA abundance (Figure 4A).
Figure 4.
The positional response to DNA helical stress is not consequent to different levels of transcription activity between chromosomal core and flanking genes. (A) Comparison between the effect of DNA (+) helical stress in chromosome flanking genes (at <50 kb form the telomere) and chromosome core genes (the rest), which are classified by their transcript levels before accumulation of helical stress. The classes are deciles (10 equal groups of about 480 genes) of genes with similar mRNA abundance at time 0 (yellow dots indicate averaged fluorescence units). The resulting graph illustrates that flanking genes (blue) behave always different from core genes (pink), irrespectively of their initial transcript levels. (B) Comparison of the capacity to generate transcription-driven supercoils between chromosomal core and flanking regions. Each of the 32 spheres corresponds to an individual chromosomal arm, being the sphere area proportional to the arm length. The horizontal axis plots the core/flank ratio (log2) of supercoiling potential for each individual arm (see text for details). Note how some flanks can generate more transcription-driven supercoils than their respective core regions. The vertical axis plots the core/flank ratio of transcript level change (log2) for each individual arm after the accumulation of DNA helical stress.
We also considered that, because gene activity and gene density in subtelomeric regions could be lower than the chromosomal average, these regions could have less potential to generate DNA helical tension, leading to little change in their transcription output. To examine this possibility, we compared the capacity to generate transcription-driven supercoils per unit length between the flank (up to 50 kb from the telomere) and core regions (the rest) of each chromosomal arm. As the potential to generate DNA helical tension depends on the amount of transcribed DNA (bp) per time unit, we estimated this parameter as the transcript length times gene transcription rate (García-Martínez et al, 2004). This analysis revealed that 9 of the 32 chromosomal flank regions had a capacity to generate transcription-driven supercoils higher than their respective core regions (Figure 4B). We then compared the flank/core ratios of supercoiling potential with the corresponding flank/core ratios of transcript reduction observed after accumulation of (+) helical stress. The resulting plot clearly illustrated that both parameters are independent (Figure 4B). All the above analyses led us to conclude, hence, that the differential response of chromosomal flanking genes to the accumulation of (+) helical tension is not consequent to the atypical transcription activity of subtelomeric regions. Thus far, the effect simply seems to be a function of the gene distance to the telomere, which applies equally to all chromosomal arms and spreads up to 100 kb inwards.
Differential response of chromosomal flanks to DNA helical stress occurs independently of the subtelomeric chromatin structure
It is well established that telomeric silencing, as regarded above, is determined by the interaction of SIR complexes along subtelomeric chromatin (for review, see Rusche et al, 2003). A heterochromatin-like structure originates at the telomere and spreads inwards a few kb, with the distance of spread determined by the concentration of available Sir3 protein (Renauld et al, 1993; Maillet et al, 1996; Fourel et al, 1999) and the activity of other cofactors (Pryde and Louis, 1999). We then considered that, if subtelomeric chromatin were spreading far inwards in all chromosomal arms, this structural change could cause the differential response of chromosomal flanks to the accumulation of DNA (+) helical tension. For instance, subtelomeric chromatin could preclude the activity of E. coli topoisomerase I during transcription and thus (+) helical tension could not built up; or subtelomeric chromatin could constrain (+) helical tension in a way that allowed transcription to proceed close to normal rates. Although these scenarios do not seem likely, we formally discarded them by repeating the above transcriptome analyses in Δsir3 backgrounds.
We replaced the SIR3 gene by the selectable marker NAT1 in both Δtop1 TOP2 and Δtop1 top2ts strains. After 120 min of heat inactivation of topoisomerase II, DNA (+) helical tension accumulated in Δtop1 top2ts Δsir3 cells expressing the E. coli TopA gene (Figure 5A). In the different replicates of the experiment, the amount of plasmid becoming highly supercoiled in the Δsir3 cells at the 120 min time point was comparable with that of SIR3 cells. We evaluated transcriptome alterations associated to DNA helical stress from the transcript ratios of the sir3 strains as above (top2ts versus TOP2). The microarray data clearly denoted that disruption of subtelomeric chromatin does not alter the differential behaviour of genes located at chromosomal flanks (Figure 5B). The gradual decrease of transcript levels as genes become more distant from the chromosomal ends was alike in both SIR3 and Δsir3 backgrounds.
Figure 5.
The differential response of chromosomal flanks to DNA helical stress also occurs in Δsir3 backgrounds. (A) DNA extracted from Δtop1 TOP2 sir3 and Δtop1 top2ts Δsir3 yeast strains after 0 and 120 min of thermal inactivation (35°C) of topoisomerase II. Both strains constitutively expressed a plasmid borne E. coli TopA gene. The gel-blot shows the conformers of the yeast 2μ circular minichromosome. (−), negatively supercoiled DNA circles; (+), positively supercoiled DNA. (B) The variation of transcript levels (log2) induced by (+) helical tension (120 min of topoisomerase II inactivation) in sir3 strains. The graph is constructed as those in Figure 1C.
Discussion
Accumulation of (+) helical tension of intracellular DNA produces a striking pattern of changes in the yeast transcriptome. Transcript levels of different genes are not determined by their function or regulatory pathways, but rather by the gene distance to the chromosomal end. Whereas the bulk of genes reduce their transcript levels by two- to six-fold, genes located at <100 kb from the telomere escape gradually from the transcription shutdown. The overall reduction of transcript levels essentially corroborates the observations of Gartenberg and Wang (1992), who reported that transcription is greatly diminished in highly positively supercoiled yeast circular minichromosomes. Gartenberg and Wang concluded that overwinding of DNA after a critical threshold of (+) helical tension should stall transcription at the initiation or elongation steps. On that respect, our data reveal that the reduction extent of individual transcripts is not dependent on transcript length (Figure 3D), thus suggesting a general impairment of transcription initiation rather than elongation.
The intriguing question here is why all chromosomal flanks escape from the global transcription stall. The simplest explanation for this neat positional effect is that DNA helical stress cannot build up in the chromosome flanking regions because DNA is torsionally unconstrained at the chromosomal ends. Still, our results could have alternative explanations related to other structural and functional traits that are known to characterize subtelomeric regions. These traits vary substantially from telomere to telomere and are responsible for transcriptional silencing of genes located at <10–20 kb from chromosomal ends (Gottschling et al, 1990; Renauld et al, 1993; Pryde and Louis, 1999). We found, however, that the positional response discovered here is independent of the reduced transcription activity of subtelomeric genes or to the distinctive structure of subtelomeric chromatin. The observed effect seems to be a simple physical function of the gene distance to the telomere. In contrast to telomere silencing effects, this functionality occurs just after accumulation of DNA (+) helical tension, applies evenly to all 32 chromosomal extremities, and spreads up to 100 kb inwards. We conclude, therefore, that the dissipation of DNA helical stress at the chromosomal ends is the most likely cause for the differential response of chromosomal flanking genes reported here (Figure 6A).
Figure 6.
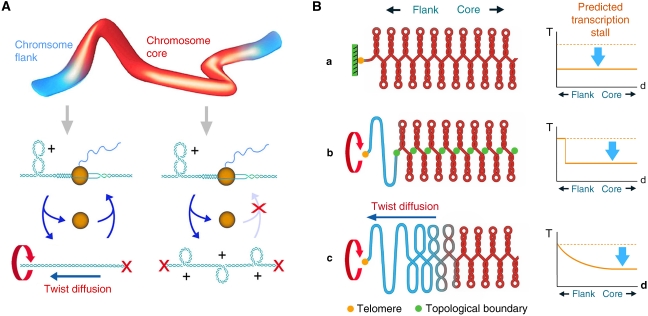
Summary model. (A) Generation and diffusion of DNA helical stress. In the absence of cellular topoisomerase I and II activities and the presence of E. coli topoisomerase I, (+) helical tension is generated all along the yeast chromosomes because of unbalanced relaxation of the DNA supercoils produced during DNA transcription. In internal regions of the chromosome (core), DNA (+) helical tension cannot diffuse and accumulates until over-twisting of the duplex precludes transcription re-initiation. However, (+) helical tension can dissipate at the chromosomal ends, so allowing transcription to re-initiate at nearby regions of the chromosome (flanks). (B) DNA topological constrains along yeast chromosomes. (a) If DNA (+) helical stress could not dissipate at chromosome ends, a general stall of transcription would be expected throughout the entire chromosome. Our results discard this model. (b) If DNA (+) helical stress could dissipate at chromosome ends, but chromosomal DNA were organized as a succession of tight topological domains, a sharp transcription stall would be observed between the relaxed terminal domains and the rest of the chromosome. Our results do not support this model, unless distal topological boundaries were located beyond 100 kb from the telomere in all chromosomal arms. (c) If DNA (+) helical stress can dissipate at chromosome ends, but DNA twist diffusion is mainly restricted by the large rotational drag of chromatin, a gradual escape from the transcription stall would be expected in all chromosomal flanks, alike the observed in our results. As less DNA torque is needed in the chromosomal flanks to overcome the rotational drag of chromatin, the probability of transcription initiation is gradually restored towards the chromosomal ends.
Our inference that DNA is torsionally unconstrained at the chromosomal ends has relevant implications on the nature of the anchoring of telomeres to subnuclear structures. In principle, the rotation of chromosomal DNA ends might seem hampered by the complex folding of telomeric DNA (Rhodes et al, 2002; Neidle and Parkinson, 2003) and the tethering of telomeres to the nuclear envelope (Heun et al, 2001; Taddei et al, 2004). Our findings denote, however, that these structural traits are compatible with the dissipation of DNA helical stress. To that regard, we should realize that helical tension can dissipate at chromosomal ends not only by the spinning of discontinued DNA strands around the duplex axis, but also by the rotation of the entire macromolecular ensemble in which the telomeric DNA strands are embedded. We should also notice that tethering of telomeres to subnuclear structures does not necessarily preclude the axial rotation of nearby DNA or chromatin. For instance, DNA could rotate inside the anchoring complex or, simply, the anchoring complex itself could have freedom to rotate at its interface with the nuclear envelope. Finally, axial rotation of chromosomal ends would be fully impeded only if telomere anchorages were stiff and permanent. In vivo imaging studies had shown, however, that this is not the case. Although yeast telomeres seem confined to specific areas adjacent to the nuclear envelope, they are highly dynamic within a restricted volume (Hediger et al, 2002). These movements could reflect a fluid attachment (Rosa et al, 2006), which could provide the telomere freedom to rotate while remaining anchored to the nuclear envelope. Alternatively, telomere motions could reflect sporadic detachments, during which spinning bursts could relieve DNA helical stress.
If DNA helical stress can dissipate at chromosomal ends, an obvious question is why DNA is relaxed only at the chromosomal flanks and not along the entire chromosome. One possible answer is that helical tension is constrained within topologically closed DNA domains all along each linear chromosome, with the sole exception of their terminal compartments that are topologically open (Figure 6B). Such terminal compartments should be delimited by a topological boundary at some distance from the telomere. The transcription stall would then sharply disappear at such boundary, the location of which could vary from chromosome to chromosome. This architecture, however, does not fit in with our experimental data, which show a transcription stall gradually fading in all 32 chromosomal flanks.
A more plausible scenario to explain the regular response of flanking regions is that, with chromosome extremities being topologically open, the accumulation of DNA helical tension depends on restrictions to DNA twist diffusion that do not necessarily invoke tight domain boundaries (Figure 6B). In short linear DNA molecules (<2 kb), axial spinning of the duplex allows very quick twist diffusion. However, in chromatin fibres confining several kb of DNA, such spinning motion is severely hindered by the huge rotational drag inflicted by the many bends and interactions of DNA (Nelson, 1999; Stupina and Wang, 2004). Diffusion of helical stress thus relies on the rotation of the chromatin fibre; and such a regime will be feasible as long as the torque affected by DNA helical tension overcomes the viscous rotational drag of the revolving volume. We infer, therefore, that at internal regions of the chromosome, the levels of DNA helical tension needed to overpower the chromatin rotational drag must be above the threshold levels that stall transcription. So, even though some twist diffusion may occur, transcription can hardly re-initiate. However, at the chromosomal flanks, the revolving volume lessens and twist diffusion can take place with lower levels of torque. Consequently, helical tension builds up to lower values towards the telomere. The probability of transcription initiation should then improve beyond some point and reach quasi-normal levels close to the chromosomal end, as observed in our experimental data.
As the dissipation of helical stress by overpowering the chromatin rotational drag relies on a general property of the chromatin fibre, this effect should spread a similar distance in all the chromosomal arms, thus in good agreement with our results. A further inference on how DNA twist diffuses along yeast chromatin is difficult to attain from our data. We can anticipate, however, that twist diffusion is a slow process. If dissipation of helical stress near the chromosomal ends was fast enough to instantly counteract the (+) helical tension generated ahead of the transcribing RNA polymerase, we would observe also a different response between chromosome flanking genes transcribed towards and away from the telomere. Yet, the escape of these genes occurs equally regardless of their outwards or inwards direction of transcription (Figure 2C). Finally, we expect that the positional dependence associated to DNA torsional stress reported here might apply also to other DNA transactions. Future studies can examine, for instance, how DNA replication is altered in the proximity of yeast chromosomal ends.
In summary, the results of our study propose two novel traits regarding the architecture and the dynamic behaviour of yeast chromosomes. First, they suggest that DNA is not torsionally constrained at the chromosomal ends. This trait has relevant implications for the nature of the anchoring of telomeres to subnuclear structures; and also for the relevance of DNA helical tension to modulate genome transactions, which may be distinct at telomere-proximal and telomere-distal regions. Second, our results suggest that frictional restrictions to DNA twist diffusion might be an important determinant for constraining helical tension along eukaryotic chromatin. This trait invites to revise our simple view of the organization of chromosomal DNA as a steady succession of DNA domains separated by tight topological boundaries.
Materials and methods
Yeast strains and growth conditions
S. cerevisiae strains JCW27 (Δtop1 TOP2) and JCW28 (Δtop1 top2ts), carrying the null mutation Δtop1 or the thermo-sensitive mutation top2–4, are derivatives of FY251 (Roca et al, 1992). Disruption of the SIR3 gene was carried out by gene replacement with the dominant selectable marker NAT1. PCR analyses of the Δsir3:NAT1 loci confirmed the resulting strains JCW27-Δsir3 and JCW28-Δsir3. Plasmid JRW13, a derivative of YEp13, carries the E. coli TopA gene under constitutive pGPD yeast promoter. Yeast strains transformed with JRW13 were grown at 26°C to logarithmic phase in an appropriate synthetic drop out medium containing 2% glucose. At time 0 min, aliquots of the cultures were taken. The rest of the cultures were placed at 35°C and samples were taken after 30 and 120 min. Cells were pelleted and immediately stored at −80°C.
Two-dimensional agarose gel electrophoresis
Total DNA extracted from yeast cells was loaded into a 0.6% agarose gel containing TBE plus 0.6 μg/ml chloroquine. The electrophoresis was run for 20 h at 48V in the first dimension. The gel slab was then equilibrated with TBE plus 3 μg/ml chloroquine and electrophoresis in the orthogonal second dimension was run for 6 h at 66V. Gel-blot hybridization was carried out using 32P-labelled DNA probes.
RNA preparation
Yeast cells were washed twice with 5 ml MilliQ water by means of centrifugation (5000 g at 4°C). Total RNA was extracted with the RiboPure Yeast kit (Ambion, Austin, TX) and then treated with DNase I (F Hoffmann-La Roche, Basel Switzerland) to remove contaminating genomic DNA. The resulting total RNA was quantified by spectrophotometry in a NanoDrop ND-1000 (NanoDrop Technologies, Wilmintong DE) and its integrity checked by gel electrophoresis. Purified RNA aliquots were kept at −80°C.
DNA microarray hybridization and analysis
Microarrays were provided by the Genomics Unit of the Scientific Park of Madrid (Spain). They consist of 13 824 spots, each one corresponding to a synthetic oligonucleotide (70-mer, Yeast Genome Oligo Set, OPERON, Cologne, Germany) encompassing the complete set of 6306 ORFs coded by the S. cerevisiae genome. Each ORF was printed twice; 600 spots were used as negative controls, either void or printed with random oligonucleotides; a small subset of genes (ACT1, HSP104, NUP159, NUP82, RPL32, RPS6B, SWI1, TDH1, TDH2, TUB4, and UBI1) were printed between 6 and 12 times for testing reproducibility; 15 μg of total RNA were used for cDNA synthesis and labelling with Cy3-dUTP and Cy5-dUTP fluorescent nucleotides, after indirect labelling protocol (CyScribe post-labelling kit, GE-Healthcare, New York, NY). Labelling efficiency was evaluated by measuring Cy3 or Cy5 absorbance in Nanodrop Spectrophotometer. Microarray prehybridization was performed in 5 × SSC (SSC: 150 mM NaCl, 15 mM Na-citrate, pH 7.0), 0.1% SDS, 1%BSA at 42°C for 45 min (Fluka, Sigma-Aldrich, Buchs SG, Switzerland). Labelled cDNA was dried in a vacuum trap and used as probe after resuspension in 110 μl of hybridization solution (50% formamide, 5 × SSC, 0.1% SDS, 100 μg/ml salmon sperm from Invitrogen, Carlsbad, CA). Hybridization and washing were performed in a Lucidea Slide Pro System (GE Healthcare, Uppsala, Sweden). Arrays were scanned with a GenePix 4000B fluorescence scanner and analysed by Genepix Pro 6.0 software (Axon Instruments, MDS Analytical Technologies, Toronto, Canada). Data was filtered according to spot quality. Only those spots with intensities at least twice the background signal and with at least 75% of pixels with intensities above background plus two standard deviations were selected for further calculations. After these criteria, over 70% of spots in each array were usually found suitable for further analysis.
Data analysis
Microarray experiments were conducted by comparing pairs of top2ts versus reference TOP2 strains. The results for each gene were given as a ratio of pixel intensities (ratio of medians of the top2ts strain divided by the TOP2 strain). Ratios were normalized within the Genepix Pro 6.0 software. The experimental design provided for each condition (0, 30, 120 min) up to 6 determinations for each gene (three biological replicates and two replicated spots). Those genes for which a minimum of 12 (out of 18) data values passed the microarray quality standards were considered for statistical analyses (4639 genes). Data were calculated as binary logarithms (log2) of fluorescence ratios. Significant changes of expression values between the starting point and at 30 and 120 min after the temperature shift were determined by one-way ANOVA (P<10−3).
Quantitative real-time PCR analysis
An aliquot of RNA used in the microarray experiments was reserved for qRT–PCR follow-up studies. First strand cDNA was synthesized from 2 μg of total DNAseI-treated RNA in a 20 μl reaction volume using Omniscript RT Kit (Qiagen, Valencia, CA) following manufacture's instructions. qRT–PCR reactions were conducted in triplicate using the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and the ABI-PRISM 7000 Sequence Detection System (Applied Biosystems). Gene-specific primers were designed using Primer Express software (Applied Biosystems). Amplified fragments were confirmed by sequencing in a 3730 DNA Analyzer (Applied Biosystems) and comparison with the published genomic data at SGD. Real-time PCR conditions included an initial denaturation step at 95°C for 10 min, followed by 40 cycles of a two-step amplification protocol: denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. Given the singular pattern of transcriptional changes observed in our study, no reference gene could be used to compensate for inaccuracies in total RNA quantitation.
Supplementary Material
Supplementary Tables I and II
Review Process File
Acknowledgments
We thank Xavier Fernández and Marta Casado for technical assistance. This work was supported by the Plan Nacional I+D+I of Spain (Grants BIO2005-00840, BFU2008-00366, and AGL2000-0133-P4-03). RJ, BP, and JR did the experiments, data analysis, and data mining. BP and JR directed the study and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Brill SJ, Dinardo S, Voelkel-Meiman K, Sternglanz R (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326: 414–416 [DOI] [PubMed] [Google Scholar]
- Brill SJ, Sternglanz R (1988) Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell 54: 403–411 [DOI] [PubMed] [Google Scholar]
- Champoux JJ (2001) DNA topoisomerases: structure, function and mechanism. Annu Rev Biochem 70: 369–413 [DOI] [PubMed] [Google Scholar]
- Delius H, Worcel A (1974) Electron microscopic visualization of the folded chromosome of Escherichia coli. J Mol Biol 82: 107–109 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Cartwright IL, Thomas GH, Elgin SC (1985) Selected topics in chromatin structure. Annu Rev Genet 19: 485–536 [DOI] [PubMed] [Google Scholar]
- Esposito F, Sinden RR (1988) DNA supercoiling and eukaryotic gene expression. Oxf Surv Eukaryot Genes 5: 1–50 [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman LA, Garrard WT (1992) DNA supercoiling in chromatin structure and gene expression. Crit Rev Eukaryot Gene Expr 2: 165–209 [PubMed] [Google Scholar]
- García-Martínez J, Aranda A, Pérez-Ortín JE (2004) Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell 15: 303–313 [DOI] [PubMed] [Google Scholar]
- Gartenberg MR, Wang JC (1992) Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci USA 89: 11461–11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever GN, Wang JC (1988) Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell 55: 849–856 [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM (2002) Live imaging of telomeres, ku and sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM (2001) Chromosome dynamics in the yeast interphase. Science 294: 2181–2186 [DOI] [PubMed] [Google Scholar]
- Huang J, Schlick T, Vologodskii A (2001) Dynamics of site juxtaposition in supercoiled DNA. Proc Natl Acad Sci USA 98: 968–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC (1989) Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol 208: 257–267 [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA 84: 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Mondoux MA, Zakian VA (2007) Subtelomeric elements influence but do not determine silencing levels at Saccharomyces cerevisiae telomeres. Genetics 177: 2541–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S, Parkinson GN (2003) The structure of telomeric DNA. Curr Opin Struct Biol 3: 275–2783 [DOI] [PubMed] [Google Scholar]
- Nelson P (1999) Transport of torsional stress in DNA. Proc Natl Acad Sci USA 96: 14342–14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Gilson E, Magdinier F (2008) Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 90: 93–107 [DOI] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR (2004) Topological domain structure of the Escherichia coli chromosome. Genes Dev 18: 1766–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE (1993) Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Rhodes D, Fairall L, Simonsson T, Court R, Chapman L (2002) Telomere architecture. EMBO Rep 12: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, Gartenberg M, Oshima Y, Wang JC (1992) A hit-and-run system for targeted genetic manipulations in yeast. Nucleic Acids Res 20: 4671–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Casado M, Portugal J, Piña B (2008) Selective inhibition of yeast regulons by daunrubicin: a transcriptome-wide analysis. BMC Genomics 9: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Maddocks JH, Neumann FR, Gasser SM, Stasiak A (2006) Measuring limits of telomere movement on nuclear envelope. Biophys J 90: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Salceda J, Fernández X, Roca J (2006) Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J 25: 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JB, Stasiak A (2004) A topological view of the replicon. EMBO Rep 5: 256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE (1981) Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci USA 78: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Wang JC (2004) DNA axial rotation and the merge of oppositely supercoiled DNA domains in Escherichia coli: effects of DNA bends. Proc Natl Acad Sci USA 101: 8608–8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Gasser SM (2004) The function of nuclear architecture: a genetic approach. Annu Rev Genet 38: 305–345 [DOI] [PubMed] [Google Scholar]
- Vegas-Palas MA, Venditti A, Di Mauro E (1997) Telomeric transcriptional silencing in natural context. Nat Genet 15: 232–233 [DOI] [PubMed] [Google Scholar]
- Vologodskii AV, Cozzarelli NR (1994) Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct 23: 609–643 [DOI] [PubMed] [Google Scholar]
- Wang JC (2002) Cellular roles of DNA topoisomerase: molecular perspective. Nat Rev Mol Cell Biol 3: 430–449 [DOI] [PubMed] [Google Scholar]
- Wang JC, Peck LJ, Becherer K (1983) DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol 47: 85–91 [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Nomura M (1988) Deficiency in both type I and type II DNA topoisomerase activities differentially affect rRNA and ribosomal protein synthesis in Schizosaccharomyces pombe. Curr Genet 13: 305–314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables I and II
Review Process File