Abstract
A longstanding challenge is to understand at the atomic level how protein dynamics contribute to enzyme catalysis. X-ray crystallography can provide snapshots of conformational substates sampled during enzymatic reactions1, while NMR relaxation methods reveal the rates of interconversion between substates and the corresponding relative populations1,2. However, these current methods cannot simultaneously reveal the detailed atomic structures of the rare states and rationalize the finding that intrinsic motions in the free enzyme occur on a time scale similar to the catalytic turnover rate. Here we introduce dual strategies of ambient-temperature X-ray crystallographic data collection and automated electron-density sampling to structurally unravel interconverting substates of the human proline isomerase, cyclophilin A (CypA). A conservative mutation outside the active site was designed to stabilize features of the previously hidden minor conformation. This mutation not only inverts the equilibrium between the substates, but also causes large, parallel reductions in the conformational interconversion rates and the catalytic rate. These studies introduce crystallographic approaches to define functional minor protein conformations and, in combination with NMR analysis of the enzyme dynamics in solution, show how collective motions directly contribute to the catalytic power of an enzyme.
It has become widely accepted that not only the chemical steps of an enzymatic reaction, but also protein conformational rearrangements contribute to the rate acceleration of enzymes1,3-6. For human CypA, a peptidyl-prolyl cis/trans isomerase, NMR relaxation experiments revealed that millisecond motions during catalysis occur in a “dynamic network” that could be described as a two-state interconversion between enzyme conformations bound to the cis- and trans- Pro substrates. Further, the free enzyme samples the same two conformations on a similar time scale, but with the equilibrium shifted far toward one substate4. Although these “major” and “minor” conformations are sampled during turnover, studies aimed at modeling the catalytic mechanism of CypA7-10 have focused on the substrate peptide and have not explained enzyme conformational changes. Moreover, superposition of 48 CypA crystal structures, including complexes with peptides, inhibitors, and physiological substrates such as the HIV capsid protein11,12, shows no structural heterogeneity that could rationalize the NMR results 4 (Supplementary Fig. 1).
In search of the catalytically essential, minor conformer of free CypA detected in solution, we first extended the resolution of the crystal structure to 1.2 Å under cryogenic conditions. This high-resolution structure, however, yielded no evidence for an alternate conformation of the dynamic network. We then used a novel algorithm, Ringer13, to systematically sample the electron density around each dihedral angle to discover additional unmodeled side-chain conformers. In a test set of 402 structures determined at 1.5 Å resolution or higher, >15% of the residues with unbranched side-chains have unmodeled secondary electron-density peaks in the range of 0.3–1σ, below the normal noise threshold of 1σ. Strikingly, these peaks are significantly enriched at low–energy rotameric positions, supporting the interpretation that the peaks reflect true minor populations. While Ringer identified discrete side-chain heterogeneity for active-site residues Met61 and Arg55 in 1.2-Å CypA electron-density map (Supplementary Fig. 2), these alternate conformations were insufficient to explain the extension of the dynamic network away from the active site, particularly to Leu98 and Ser99, which showed the largest differences in NMR chemical shifts between the major and minor conformers4,14.
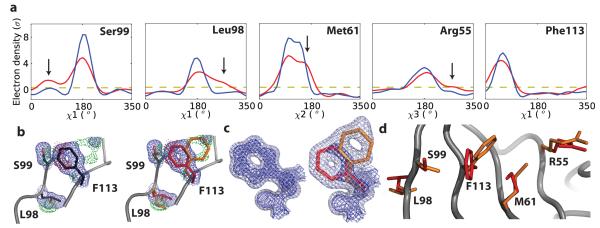
To address this discrepancy between the X-ray and NMR analyses, we collected 1.39-Å diffraction data at ambient temperature to explore the possibility that the low-temperature data collection might alter the conformational distribution in the crystal15,16. Ringer plots indeed uncovered 0.3-1 σ features for alternate rotamers of several residues including Leu98 and Ser99, in addition to the side-chain heterogeneity observed at cryogenic temperature (Fig. 1a). These results emphasize that crystal freezing can alter conformational distributions.
Figure 1. Room-temperature X-ray crystallography and Ringer analysis detect conformational substates in CypA.
a, Local maxima above the 0.3σ threshold (yellow line) in Ringer plots13 reveal alternate side-chain conformations in room-temperature (red line) but not cryogenic (blue line) electron density for Ser99, Leu98, Met61 and Arg55. b, Electron-density maps calculated using room-temperature X-ray data define the alternate conformers of Leu98, Ser99 and Phe113. 2Fo-Fc electron density (blue mesh; 1σ); positive (green) and negative (red) Fo-Fc difference density (3σ). c, 2Fo-Fc composite simulated-annealing omit electron density maps (1.0σ (dark blue) and 0.3σ (light blue)) show a unique conformation for Phe113 in the 1.2-Å-resolution cryogenic structure (blue) and distinct major (red) and minor (orange) conformers in the 1.39-Å-resolution room-temperature structure. Electron density around the main chain and the surrounding residues was omitted for clarity. d, Steric collisions across the network of major (red) and minor (orange) conformers of Arg55, Met61, Phe113 and Ser99 explain how side-chain motions link the active site to remote buried residues.
Modeling the minor conformation of Ser99 produced a clash of the side-chain hydroxyl group with the Phe113 ring. Inspection of the electron density surrounding Phe113 revealed an alternate conformation below the 1  threshold. Fo-Fc difference density maps calculated without bias from model rebuilding confirmed the alternate conformers of Leu98, Ser99 and Phe113 (Fig. 1b, c, Supplementary Fig. 3). Ringer did not identify the Phe113 minor rotamer due to a concomitant shift in the backbone that places Cγ within the envelope of the major conformer. This finding emphasizes that no algorithm can yet identify with complete fidelity all structural features without further scrutiny of the electron-density distribution. Refinement verified a network of alternate side-chain rotamers covering the entire active site and extending into the buried regions of the dynamic network previously detected by NMR (Fig. 1d).
threshold. Fo-Fc difference density maps calculated without bias from model rebuilding confirmed the alternate conformers of Leu98, Ser99 and Phe113 (Fig. 1b, c, Supplementary Fig. 3). Ringer did not identify the Phe113 minor rotamer due to a concomitant shift in the backbone that places Cγ within the envelope of the major conformer. This finding emphasizes that no algorithm can yet identify with complete fidelity all structural features without further scrutiny of the electron-density distribution. Refinement verified a network of alternate side-chain rotamers covering the entire active site and extending into the buried regions of the dynamic network previously detected by NMR (Fig. 1d).
The room-temperature diffraction data afford a possible structural rationale for the large NMR chemical-shift differences between the substates in free CypA. The alternate side-chain rotamers, particularly of Phe113, are predicted to alter the magnetic environment of the surrounding residues. Only a coupled switch of rotamers is consistent with the steric repulsion of major and minor rotamers, such as the Ser99/Phe113 clash in the dynamic network (Fig. 1b).
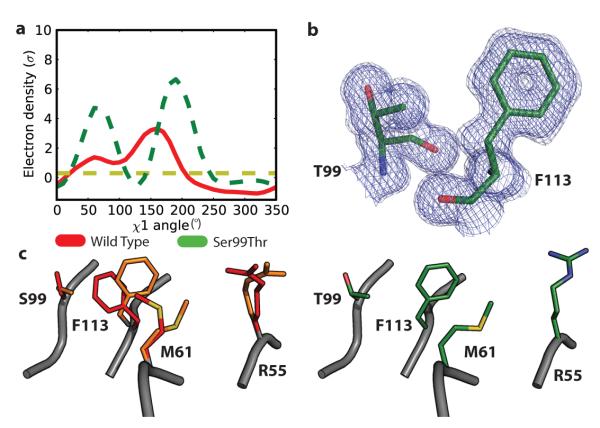
To critically test the idea that these two conformers interconvert during turnover, we designed a mutation distant from the active site to stabilize the minor CypA substate. Ser99, a buried residue in the dynamic network located >14 Å from the catalytic Arg55, was replaced by Thr to fill the space occupied by both Ser99 rotamers. This conservative change was designed to stabilize Phe113 in the “out” position by emulating the steric clash between the minor Ser99 rotamer and the Phe113 “in” position. Crystal structures of the Ser99Thr mutant, solved at 1.6-Å and 2.3-Å resolution, indeed showed Thr99 mimicking the alternate Ser99 conformations, and Phe113 was detected only in the exposed “out” rotamer (Fig. 2a, Supplementary Fig. 4). This change in rotamer populations was buttressed by 3-bond J-coupling solution NMR experiments showing that the dominant Phe113 χ1 angle changed from +60° in wild-type CypA to −60° in the Ser99Thr mutant (Supplementary Table 2). In the Ser99Thr variant structures, Thr 99, Phe113 and Met61 occupy the minor rotamers. The positions of Leu98 and Arg55 are consistent with either of the rotamers seen in wild-type CypA. This pattern corroborates the conclusions that Ser99, Phe113, Met61 and possibly Arg55 are conformationally coupled (Fig. 2c) and that the Ser99Thr mutation severely reduces the population of the major conformation seen in wild-type CypA.
Figure 2. The structure of the Ser99Thr mutant resembles the minor conformer of wild-type CypA.
a, χ1 Ringer plot (0.3σ threshold is shown as yellow line) of the Ser99Thr mutant (dashed green) and room-temperature, wild-type Ser99 CypA structure (red) show that Thr99 occupies both positions populated by the Ser99-OHγ group. The angular offset between the major peaks reflects a backbone shift. b, The 2Fo-Fc simulated-annealing omit electron density map of the Ser99Thr CypA mutant (1.0σ (dark blue) and 0.3σ (light blue)) shows apparently unique conformations for Thr99 and Phe113. The structure confirmed the prediction that rotation of Phe113 to the “out” position is coupled to rotation of the Ser99 hydroxyl to the minor rotamer. c, Phe113 and Met61 in Ser99Thr CypA (green, right) are detected exclusively in the position of the minor state of the wild-type enzyme (orange, left).
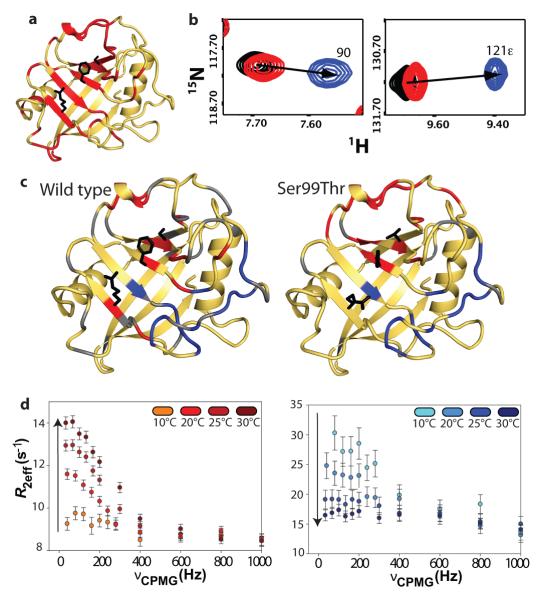
Connecting this interpretation of the crystal structures to the solution behaviour of the enzyme, NMR detected amide chemical-shift differences between wild-type and Ser99Thr CypA in most active-site and core residues of the dynamic network (Fig. 3a,b). Although it is tempting to speculate that the Ser99Thr mutation “traps” the minor state of wild-type CypA based on our crystallographic data (Fig. 2), NMR relaxation-dispersion analysis18 of the mutant enzyme revealed conformational exchange in both regions that showed collective motions in the wild-type enzyme (Fig. 3c). These regions include residues coupled to the active site (group I) and residues in the 65-85 loops (group II)4. In addition, for most of the group I residues, the peaks in Ser99Thr CypA shift relative to wild-type in the same direction as peaks in the previously characterized Lys82Ala variant, which displays a small increase in the population of the minor state4. The Ser99Thr mutation, however, causes much larger shifts (Fig. 3b, Supplementary Figs. 5,6), indicating that the populations are inverted. Together with our crystallographic and NMR J-coupling data, these results show that the Ser99Thr mutation shifts the structural equilibrium strongly towards a conformation that recapitulates key features of the previously undefined minor state of wild-type CypA.
Figure 3. The Ser99Thr mutation shifts the equilibrium toward the minor wild-type conformation and slows motions in the dynamic network in free CypA.
a, Significant 1H-15N chemical-shift differences between Ser99Thr and wild-type CypA (red) propagate through group I residues (Arg55, Phe113, and Ser99 shown as black sticks). b, Linear amide chemical shift changes (arrows) between wild-type (black), Lys82Ala (red) and Ser99Thr (blue) CypA reflect the inversion of the major/minor equilibrium due to the Ser99Thr mutation. c, Residues undergoing slow (red) or fast (blue) motions on the NMR time scale in Ser99Thr (right) coincide with previously identified group I (red) and group II (blue) residues in wild-type (left) CypA (amides in grey are prolines or overlapped peaks). d, Temperature dependence of CPMG 15N NMR relaxation data for group I (left) and group II (right) in Ser99Thr CypA reveal that the mutation impedes group I conformational dynamics (REX ~ k1 and REX increases with temperature). In contrast, group II residues are unaffected by the mutation and display the opposite temperature dependence characteristic of fast motions on the NMR time scale. Dispersion curves were normalized to R20 at 30 °C.
In addition to this dramatic population shift, the Ser99Thr mutation also specifically slows the motions of only group I residues by at least two orders of magnitude, pushing them into the slow NMR time regime. The slow dynamics of group I are easily identified by the characteristic increase in dispersion amplitude with increasing temperature, while the fast dynamics in the group II loop region, unaffected by the mutation, are characterized by the opposite trend (Fig. 3d, Supplementary Figs. 7,8). This result is reinforced by the static magnetic-field dependence (B0) of REX as determined by the α value19: at 25 °C the REX of group I residues is independent of B0 (<⟨>=0.16, Supplementary Table 3), characteristic of slow exchange, while for group II residues the field-dependence is quadratic (<⟨>=2.0), characteristic of fast exchange. Quantitative analysis of the group I residues yields a rate constant of conversion from the major to minor state (k1) for Ser99Thr CypA of 1.0+/−0.3 s−1 at 10 °C, in contrast to ~60 s−1 for wild-type CypA4. In the slow exchange regime of Ser99Thr, k1 is well-determined and the major peak represents the true chemical shift of the major state (Fig. 3b).
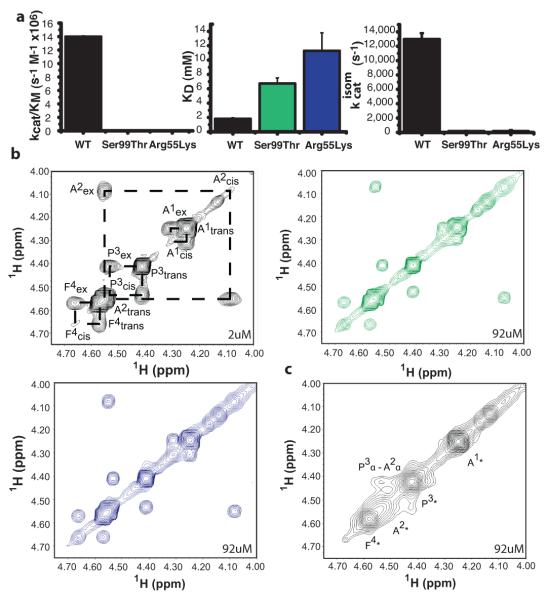
Given the slower conformational transitions in Ser99Thr CypA, what is the effect of this mutation on catalytic turnover? In a protease-coupled enzymatic assay20, the Ser99Thr mutant showed a 300-fold reduction in kcat/KM of the cis-to-trans isomerization of the peptide succinyl-AlaAlaProPhe-p-nitroanilide (AAPF) (Fig. 4a). This large reduction in catalytic efficiency resembles the effects of the Arg55Lys mutation, which removes the active-site residue thought to promote the chemical step of the reaction but does not perturb the enzyme dynamics4 or global structure (Supplementary Fig. 9). To separate the energetic contributions to the binding and isomerization steps, we measured the dissociation constants of the peptide substrate for the CypA variants using NMR titration experiments. Peptide binding (KD) was weakened only 3-6 fold by the Ser99Thr (6.7+/−0.8 mM) and Arg55Lys (11.3+/−2.5 mM) mutations (Fig. 4a, Supplementary Fig. 10) suggesting that the major effect of these mutations is on kcat and not on KM.
Figure 4. Impeded motions in the dynamic network severely reduce the catalytic power of a chemically competent enzyme.
a, Mutations affecting the enzyme dynamics (Ser99Thr) or the chemical step (Arg55Lys) each drastically reduce kcat/KM 20 by reducing the bidirectional isomerization step on the enzyme (kcatisom) and not substrate affinity (KD) of CypA. b, 1H-1H NOE-exchange spectra at 0.2 s mixing time showing isomerization of the peptide A1A2P3F4 (1 mM) by catalytic amounts of wild-type (black), Ser99Thr (green) and Arg55Lys (blue) CypA. Assignments and dashed lines connecting exchange peaks are included for wild-type. Much higher concentrations of the Ser99Thr and Arg55Lys variants are needed relative to wild-type CypA to obtain similar exchange peaks, reflecting severely reduced catalytic activity. c, The cis-and trans- peaks coalesce (asterisk) for wild-type CypA at the same enzyme concentration as the mutant forms due to its much greater activity. The only remaining off-diagonal peak is a P3α-A2α NOE characteristic for a cis-prolyl peptide bond.
To independently quantify the mutational effects on the isomerization step (kcatisom), we measured catalysis of the cis/trans isomerization directly by ZZ exchange spectroscopy21 (Fig. 4b). These experiments detect the overall rate of converting the entire substrate pool from the cis- to the trans-Pro conformation and vice versa. The Ser99Thr and Arg55Lys mutations reduce overall turnover by similar amounts compared to wild-type CypA (Fig. 4, Supplementary Fig. 11). Quantitative fitting of these exchange data21 indicates that the Ser99Thr mutation severely reduces the rate of the bidirectional isomerization step on the enzyme (1.9×102+/−20 s−1 for Ser99Thr compared to 1.3×104+/−800 s−1 for wild-type CypA). Strikingly, this reduction (~70-fold) parallels the upper bound for the reduction in the rate constant for the major-to-minor conformational change for group I residues (~60-fold) in the free enzyme (Figs. 3c, d).
The similar rate reductions in the free enzyme and of substrate turnover strongly suggest that the slowed fluctuations due to the Ser99Thr mutation underlie the reduction of the catalytic rate. The effects of this “molecular traffic-jam” mutation remote from the active site support the idea that the coupled motions of the dynamic network tied to the rotation of Phe113 play a key role in turnover. The conformational transition of Phe113 observed here suggests that the side-chain rotation directly and distinctly impacts cis- and trans- forms of the proline-substrate during catalysis. Indeed, smaller22 or larger4 replacements for Phe113 substantially decrease CypA activity, suggesting the wild-type enzyme occupies a narrow structural optimum for catalysis. Both the interaction of the Arg55 side chain with the substrate8,9, which facilitates the chemistry, and side-chain motions throughout the dynamic network, which enable facile interconversion of conformational states, are necessary for catalysis (Fig. 4). Therefore, neither the dynamics nor active-site chemistry that lowers the transition-state energy is sufficient to promote efficient turnover.
Only by collecting X-ray diffraction data at ambient temperature did we observe agreement between the conformational substates detected using NMR and X-ray crystallography. This finding underscores previous studies demonstrating that the conformational distributions in crystals of myoglobin23,24 and ribonuclease16,25 are restricted at cryogenic temperatures. Unlike these classic studies, which accessed the manifold of crystal structures indirectly through reductions in protein volume, inhibition of ligand binding and reductions in crystallographic B-values at cryogenic temperatures, here we use Ringer13 to directly sample the electron density. This automated, systematic approach reveals alternate conformational substates. Our results emphasize that the common practices of crystal freezing and building models at the 1σ threshold, rather than capturing the conformational distribution in the crystal, can eliminate information about functionally critical conformations.
Our complementary results from NMR and X-ray crystallography for CypA demonstrate a general strategy to discover structural ensembles of interconverting substates, to identify which regions of enzymes couple to active sites, and to evaluate the roles of hidden, higher energy conformations in catalysis by other enzymes3,26. Particularly by identifying structurally remote residues that couple to the active site, defining the multiple conformations that contribute to function can afford a deeper understanding of the effects of sequence variation in disease and evolution27. Such knowledge may enable progress not only in understanding and manipulating the mechanisms of numerous macromolecular systems, but also in defining the manifold of conformations accessible to inhibitors and therapeutics28.
Methods Summary
CypA was purified as described4. Crystals were grown at pH 7.5 using PEG3350 or DL-malate as precipitants. All X-ray data sets derive from single crystals. For room-temperature data collection, we used the Rigaku free mounting device at ALS Beamline 12.3.117. Short exposures and crystal translation every 10 frames limited radiation damage. Ringer analysis was performed by real-space sampling of electron density around side-chain dihedral angles13. Inspection of plots of electron density versus dihedral angle revealed Ringer peaks above 0.3σ that guided placement of alternate conformers. This modeling process was performed iteratively using both conventional and simulated-annealing composite omit maps.
Constant-time 15N backbone CPMG TROSY relaxation-dispersion data18 were collected in an interleaved manner on Varian INOVA 500 and 600 spectrometers and a Bruker AVANCE-800 equipped with a cryoprobe. Dispersion curves were fit to the full Carver-Richards equation29, confirming the results for wild-type CypA where the exchange is in the intermediate to fast time regime4. In contrast for Ser99Thr CypA, global fits of dispersion of group-I residues at all temperatures, assuming temperature-independent ⊗⌉, unambiguously revealed exchange in the slow regime (REX ~ k1, rate constant for efflux from the major state). Numerical fitting and estimation of the dispersion amplitude gave the same rate constant k1 at 10 °C within experimental error.
Three-bond, aromatic J couplings were obtained at 25 °C as described30. Dissociation constants for AAPF were obtained by titrating the peptide into a solution of 0.2 mM CypA at 6 °C and fitting the resulting chemical shifts to single-site exchange. The enzyme-catalyzed cis/trans isomerization rates at 6 °C were measured using 1H-1H NOESY spectra of AAPF and fitting the data to equations for ZZ exchange21. The bidirectional kcatisom was calculated from the fitted exchange rate using the KD measured in titrations (see full methods).
Full Methods
Crystallography
The standard purification4 was augmented with a Superdex S75 column (GE Healthcare) equilibrated in 20 mM HEPES pH 7.5, 100 mM NaCl and 0.5 mM TCEP. Crystals (wild-type and Ser99Thr) with P212121 symmetry were grown by hanging-drop vapour diffusion by mixing an equal volume of well solution (100 mM HEPES pH 7.5, 22% PEG 3350, 5 mM TCEP) and protein (wild-type CypA at 60 mg/mL and mutants at 40 mg/mL). Trigonal (P3221) crystals of Ser99Thr and Arg55Lys CypA were grown similarly from 1.8 M DL-malic acid. For cryogenic data sets (collected at 100 K), crystals were flash frozen in liquid nitrogen with well solution plus 15% xylitol as a cryoprotectant for the orthorhombic crystals and 10% glycerol for the trigonal crystals. Wild-type CypA cryogenic x-ray data were collected at Beamline 9-1 at the Stanford Synchrotron Radiation Laboratory. Ser99Thr and Arg55Lys cryogenic data sets were collected at Beamline 8.3.1 at the Lawrence Berkeley National Laboratory Advanced Light Source (ALS).
Room-temperature X-ray data were collected at 15 °C with 96% humidity using a temperature- and humidity-controlled goniometer head17 (the Rigaku free-mounting device) at ALS Beamline 12.3.1. To limit the effects of radiation damage while maintaining high signal-to-noise, we collected short exposures (0.1 s) for 180° with 1° oscillations. A large crystal (1 mm × 0.5 mm × 0.3 mm) was translated halfway through this rapid collection protocol. Following this initial low-exposure pass, an additional complete data set of 90° with longer exposures (1 s) was collected while translating the crystal every 10 frames. During data processing, we ensured that radiation damage had not degraded the data quality by subdividing the data into subsets of 45 frames and confirming that unit cell parameters, scale factors and χ2 statistics were consistent throughout the dataset31. Due to the short total exposure time (108 s), the limited exposure of each spot on the crystal, the size of the crystal and the finite rate of crystal damage, the data were not significantly influenced by radiation damage.
Reflections were processed using HKL200032. Structures were solved using PHASER33 through phenix.automr34 with 2CPL chain A as the search model. To corroborate the features of residues 98, 99 and 113 observed in the initial electron density, reflections were processed using MOSFLM, through ELVES35, and XDS, through XIA236, and the structures were additionally solved using 1RMH chain A as the search model. The features described in the text were consistent across electron-density maps calculated from data processed with all three programs and both molecular-replacement search models.
Coordinates, atomic displacement parameters, and occupancy (where appropriate) were refined using phenix.refine34 with manual rebuilding using COOT37. Validation was performed using MOLPROBITY38 and PROCHECK39. Data collection, processing, refinement, and validation statistics are listed in Table S1. Structural figures were prepared using PyMol40.
Ringer
Ringer13 analyzes electron density in real space around the dihedral angles of the modeled side chains to discover unmodeled alternate conformers. For each residue, each side-chain dihedral angle is rotated in 10° increments, and the electron density value calculated using a cubic spline interpolation is tabulated at the position of the rotated atom. In sp3 geometry, for example, the sampled position is 1.53 Å away and oriented 111.1° from the preceding side-chain carbon atom. In regions of electron density that are not occupied by modeled atoms, Ringer peaks >0.3σ are significantly enriched at rotameric positions, and χ1 peaks are strongly correlated with rotameric χ2 peaks in long side chains, providing evidence that these features correspond to alternate conformers13. The contour level of 0.3σ provides an initial working cut-off to detect alternate rotamers in high-resolution electron density maps. Alternate conformers are modeled after inspection of the electron density maps for continuous or correlated electron density for the side chain and subsequently refined.
Comparison of crystal structures
Forty-eight CypA structures with 100% sequence identity (Supplementary Fig. 1) were superimposed using Theseus41 and analyzed for backbone root-mean-square-deviation (RMSD) to establish regions of conformational heterogeneity or flexibility.
NMR Methods
Unless otherwise specified, NMR samples consisted of 1 mM wild-type or mutant CypA in 50 mM Na2HPO4 pH 6.5, 0.02% NaN3, 1 mM DTT, 10% D2O. Constant-time relaxation dispersion spectra18,42 were collected in an interleaved manner, processed using NMRPipe43 software, and analyzed using NMRView44 and custom scripts. Errors in peak intensities were estimated using the variance for non-exchanging peaks, signal to noise, and duplicate points. Errors for the fits were estimated using 200 Monte-Carlo simulations. CPMG relaxation dispersion data collected at 25 °C at 500, 600, and 800 MHz not only constrained the fitted parameters but also provided α values for all residues19.
We note that in the slow exchange regime of Ser99Thr, CPMG relaxation data and fitting them to the Carver Richards equation is not robust to determine chemical shift differences or populations. The absence of the minor peak, in light of the estimated line-broadening, suggests an upper population limit for this species of about 10%. However, k1 is well determined in the slow time regime by the value of Rex.
Chemical shift differences between mutant and wild-type backbone amides or side-chain indoles (Trp121) were considered significant when Δ∂ was greater than 0.05 ppm in the 1H dimension or 0.25 ppm in the 15N dimension. Three-bond J couplings were assessed using difference spectra30.
NOESY experiments were performed at 6 °C in a buffer containing 860 μM AAPF and 100 % D2O using a standard gradient-filtered experiment45. Intensity curves were fit to previously described equations21 in Origin 6.0 using populations for the cis and trans isomer determined from the intensities of the peaks in a NOESY spectrum with a mixing time of zero. The fits of the data for all three enzyme forms yielded the corresponding exchange rates (kex), but the same R1 values for the auto-peaks within experimental error. The bidirectional kcatisom was calculated from this kex using the following equations:
where visom is the reaction velocity, KM is the Michaelis-Menten constant, and [S] and [E] are substrate and enzyme concentrations, respectively. For fast dissociation relative to the rate of the isomerization, KM ≈ KD, so
This yields a bidirectional kcatisom when one uses the KD averaged for the cis and trans isomers obtained from the titration experiments.
Coupled enzymatic assay
kcat/KM for the enzyme catalysed cis-to-trans isomerization of AAPF was measured at 10°C using the standard protease coupled assay20.
Supplementary Material
Acknowledgments
We thank S. Marqusee and B. Krantz for discussions; S. Classen, G. Meigs, J. Holton, A. Samelson, N. Echols, P. Afonine, and the Phenix team for technical support; J. Tainer for access to Rigaku free-mounting device at ALS Beamline 12.3.1; J. Pelton and D. Wemmer for providing essential help and access to NMR facilities. J.S.F. was supported by US NSF and Canadian NSERC fellowships. This work was funded by the US National Institutes of Health (to T.A.) and the US National Institutes of Health, the US Department of Energy Office of Basic Energy Sciences, and the Howard Hughes Medical Institute (to D.K.).
Footnotes
Supplementary Information accompanies the paper.
Atomic coordinates and structure factors for the reported crystal structures have been deposited in the PDB under accession codes (3K0M, 3K0N, 3K0O, 3K0P, 3K0Q, and 3K0R).
The authors declare no competing financial interest.
References
- 1.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 2.Mittermaier A, Kay LE. Review - New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 3.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 4.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 5.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 6.Schramm VL, Shi W. Atomic motion in enzymatic reaction coordinates. Curr. Opin. Struct. Biol. 2001;11:657–665. doi: 10.1016/s0959-440x(01)00269-x. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal PK. Cis/trans isomerization in HIV-1 capsid protein catalyzed by cyclophilin A: Insights from computational and theoretical studies. Proteins. 2004;56:449–463. doi: 10.1002/prot.20135. [DOI] [PubMed] [Google Scholar]
- 8.Hamelberg D, McCammon A. Mechanistic Insight into the Role of Transition-State Stabilization in Cyclophilin A. J. Am. Chem. Soc. 2009;131:147–152. doi: 10.1021/ja806146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li GH, Cui Q. What is so special about Arg 55 in the catalysis of cyclophilin A? Insights from hybrid QM/MM simulations. J. Am. Chem. Soc. 2003;125 doi: 10.1021/ja0367851. [DOI] [PubMed] [Google Scholar]
- 10.Trzesniak D, Van Gunsteren WF. Catalytic mechanism of cyclophilin as observed in molecular dynamics simulations: Pathway prediction and reconciliation of X-ray crystallographic and NMR solution data. Protein Sci. 2006;15:2544–2551. doi: 10.1110/ps.062356406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nat. Struct. Biol. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 12.Ke HM, Huai Q. Crystal structures of cyclophilin and its partners. Front. Biosci. 2004;9:2285–2296. doi: 10.2741/1396. [DOI] [PubMed] [Google Scholar]
- 13.Lang PT, et al. Automated electron-density sampling reveals widespread conformational polymorphism in proteins. submitted. [DOI] [PMC free article] [PubMed]
- 14.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 15.Halle B. Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4793–4798. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen BF, Stock AM, Ringe D, Petsko GA. Crystalline Ribonuclease-a Loses Function Below the Dynamic Transition at 220-K. Nature. 1992;357:423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- 17.Kiefersauer R, et al. A novel free-mounting system for protein crystals: transformation and improvement of diffraction power by accurately controlled humidity changes. J. Appl. Crystallogr. 2000;33:1223–1230. [Google Scholar]
- 18.Loria JP, Rance M, Palmer AG. A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J. Biomol. NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 19.Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J. Am. Chem. Soc. 2000;122:2867–2877. [Google Scholar]
- 20.Kofron JL, Kuzmic P, Kishore V, Colonbonilla E, Rich DH. Determination of Kinetic Constants for Peptidyl Prolyl Cis Trans Isomerases by an Improved Spectrophotometric Assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 21.Farrow NA, Zhang OW, Forman-Kay JD, Kay LE. A Heteronuclear Correlation Experiment for Simultaneous Determination of N-15 Longitudinal Decay and Chemical-Exchange Rates of Systems in Slow Equilibrium. J. Biomol. NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 22.Zydowsky LD, et al. Active-Site Mutants of Human Cyclophilin-a Separate Peptidyl-Prolyl Isomerase Activity from Cyclosporine-a Binding and Calcineurin Inhibition. Protein Science. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frauenfelder H, et al. Thermal-Expansion of a Protein. Biochemistry. 1987;26:254–261. doi: 10.1021/bi00375a035. [DOI] [PubMed] [Google Scholar]
- 24.Frauenfelder H, Petsko GA, Tsernoglou D. Temperature-Dependent X-Ray-Diffraction as a Probe of Protein Structural Dynamics. Nature. 1979;280:558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 25.Tilton RF, Dewan JC, Petsko GA. Effects of Temperature on Protein-Structure and Dynamics - X-Ray Crystallographic Studies of the Protein Ribonuclease-a at 9 Different Temperatures from 98-K to 320-K. Biochemistry. 1992;31:2469–2481. doi: 10.1021/bi00124a006. [DOI] [PubMed] [Google Scholar]
- 26.Beach H, Cole R, Gill ML, Loria JP. Conservation of us-ms enzyme motions in the apo- and substrate-mimicked state. J. Am. Chem. Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- 27.Tokuriki N, Tawfik DS. Protein Dynamism and Evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 28.Lee GM, Craik CS. Trapping Moving Targets with Small Molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis DG, Perlman ME, London RE. Direct Measurements of the Dissociation-Rate Constant for Inhibitor-Enzyme Complexes Via the T-1-Rho and T-2 (Cpmg) Methods. J. Magn. Reson. Ser. B. 1994;104:266–275. doi: 10.1006/jmrb.1994.1084. [DOI] [PubMed] [Google Scholar]
- 30.Hu JS, Grzesiek S, Bax A. Two-dimensional NMR methods for determining (chi 1) angles of aromatic residues in proteins from three-bond J(C’C gamma) and J(NC gamma) couplings. J. Am. Chem. Soc. 1997;119:1803–1804. [Google Scholar]
- 31.Southworth-Davies RJ, Medina MA, Carmichael I, Garman EF. Observation of decreased radiation damage at higher dose rates in room temperature protein crystallography. Structure. 2007;15:1531–1541. doi: 10.1016/j.str.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. Vol. 276 Methods in Enzymology. Academic Press Inc; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D-Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Delano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC, Palo Alto, CA, USA. 2008. http://www.pymol.org.
- 41.Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulder FAA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 43.Delaglio F, et al. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Johnson BA, Blevins RA. NMR View - a Computer-Program for the Visualization and Analysis of Nmr Data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 45.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of Exchange Processes by 2-Dimensional NMR-Spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.