Abstract
Although medulloblastoma is the most common pediatric malignant brain tumor, its molecular underpinnings are largely unknown. We have identified rare, recurrent homozygous deletions of Kruppel-like Factor 4 (KLF4) in medulloblastoma using high-resolution single nucleotide polymorphism arrays, digital karyotyping, and genomic real-time polymerase chain reaction (PCR). Furthermore, we show that there is loss of physiological KLF4 expression in more than 40% of primary medulloblastomas both at the RNA and protein levels. Medulloblastoma cell lines drastically increase the expression of KLF4 in response to the demethylating agent 5-azacytidine and demonstrate dense methylation of the promoter CpG island by bisulfite sequencing. Methylation-specific PCR targeting the KLF4 promoter demonstrates CpG methylation in approximately 16% of primary medulloblastomas. Reexpression of KLF4 in the D283 medulloblastoma cell line results in significant growth suppression both in vitro and in vivo. We conclude that KLF4 is inactivated by either genetic or epigenetic mechanisms in a large subset of medulloblastomas and that it likely functions as a tumor suppressor gene in the pathogenesis of medulloblastoma.
Introduction
Medulloblastoma is the most common malignant pediatric brain tumor. Current therapies including surgery, craniospinal radiotherapy, and chemotherapy result in modest 5-year survivals (60%–70%) and predispose to many long-term complications such as cognitive impairment, focal neurologic deficits, and secondary malignancies [1]. Development of novel targeted therapies for medulloblastoma has been hindered by modest levels of genetic and epigenetic data concerning its pathogenesis and, therefore, a paucity of targets for the development of novel therapies [2–10]. Owing to the relatively small number of children with medulloblastoma compared with adults with epithelial malignancies, it would be advantageous to identify medulloblastoma targets that are shared with the more common adult epithelial malignancies because compounds against these targets are more likely to be developed by the pharmaceutical industry. We demonstrate that the known tumor suppressor gene (TSG) KLF4 is inactivated in a substantial subset of medulloblastomas through either genetic or epigenetic mechanisms. KLF4 is a TSG that has previously been reported as epigenetically silenced in colonic, gastric, and pancreatic carcinoma, as well as in hematopoietic malignancies, and which is mutated in colon cancer [11–13]. KLF4 has also been reported to act as an oncogene in other histologic types of cancer [14–16]. Haploinsufficiency for Klf4 has been demonstrated to promote tumorigenesis in mouse models of colonic cancer [17]. Forced reexpression of KLF4 in a number of cancer cell lines diminishes tumorigenicity both in vitro and in vivo, supporting its role as a bona fide TSG [11,12].
In the current study, we demonstrate that KLF4 is either deleted or silenced by promoter CpG island methylation in a large subset of medulloblastomas. Whereas KLF4 is highly expressed in the normal human adult and fetal cerebella, there is no significant expression of KLF4 in approximately 46% of human medulloblastomas. Forced reexpression of KLF4 in the D283 medulloblastoma cell line results in decreased growth both in vitro and in vivo. Our results support the hypothesis that KLF4 functions as a TSG in the pathogenesis of medulloblastoma.
Materials and Methods
Cell Lines, Normal Cerebella, and Medulloblastoma Samples
ONS76 was obtained from the Institute for Fermentation (Osaka, Japan). UW228 and UW426 were obtained from J. Silber (University of Washington, Seattle, WA). D425, D458, and D384 were obtained from Darrell Bigner (Duke University, Durham, NC). MHH-MED1 and MED8a were obtained from Richard Gilbertson (St. Jude Children's Research Hospital, Memphis, TN). RES261 was obtained from Michael Bobola (University of Washington, Seattle, WA). Other cell lines were purchased from the American Type Culture Collection (Rockland, MD). Medulloblastoma samples were collected after institutional review board approval, and DNA and RNA were isolated as published [18]. Samples of normal adult and fetal cerebella were purchased from Biochain (Hayward, CA).
5-Azacytidine Treatment and Quantitative Reverse Transcription-Polymerase Chain Reaction
Cell lines were plated at 20% to 30% confluence in Dulbecco's modified Eagle medium with 10% fetal calf serum. Twenty-four hours later, the medium was replaced with fresh medium containing 5 mM 5-azacytidine (5-Aza; Sigma-Aldrich, Inc, St Louis, MO) or an equal volume of vehicle (PBS). Medium and drug or vehicle was replaced every 24 hours during a 72-hour period.
Copy Number Determination
Genotyping on the Affymetrix 100K single nucleotide polymorphism (SNP) arrays was performed as published [18]. Expression profiling of medulloblastoma specimens was performed on the Affymetrix Exon Array platform as published [19]. Digital karyotyping was performed as published [20].
Bisulfite Sequencing and Methylation-Specific PCR
Genomic DNA was treated with MethylEasy DNA Bisulphite Modification Kit (Human Genetic Signatures, North Ryde, Australia). For bisulfate sequencing, modified DNA was amplified using primers BSQ1: forward 5′-ttggaaaattattgattataaattaagg-3′ and reverse 5′-cttccctaaaaaataaccatatacc-3′; and BSQ2: forward 5′-gttygagtttttattattttttagtg-3′ and reverse 5′-attttactctcatcttcttaacaaaca-3′. Amplified products were cloned using the Topo-TA cloning system (Invitrogen, Carlsbad, CA) and Sanger sequenced at The Center for Applied Genomics (Toronto, Canada). Methylation-specific PCR (MSP) was carried out on bisulfate-treated DNA from 44 primary medulloblastomas. Unmethylated KLF4 forward 5′-gttaatttatgtttagaaagtgatattg-3′ and reverse 5′-tacacaccaaacccaccacaaac-3′. Methylated KLF4 forward 5′-gttaatttatgtttagaaagtgatattg-3′ and reverse 5′-tacacaccaaacccaccacaaac-3′. CPGenome universally methylated DNA (Chemicon, Temecula, CA) was used as a positive control.
Cell Proliferation Assay
MTS assay was performed as published [21].
Orthotopic Intracerebellar Xenografts
D283 medulloblastoma cells transfected with pcDNA-KLF4 or vector control were injected at a final concentration of 50,000 cells per milliliter into the cerebellum of Nu/Nu mice (aged 5–6 weeks, Charles River, Canada) as published [22]. Mice were killed at the time of development of a symptomatic intracranial mass such as a domed skull, incoordination, lethargy, or significant weight loss.
Human Medulloblastoma Tissue Microarray
Human medulloblastoma TMA was stained with anti-KLF4 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and interpreted as previously published [18].
Results
Recurrent Homozygous Deletion of KLF4 in Medulloblastoma
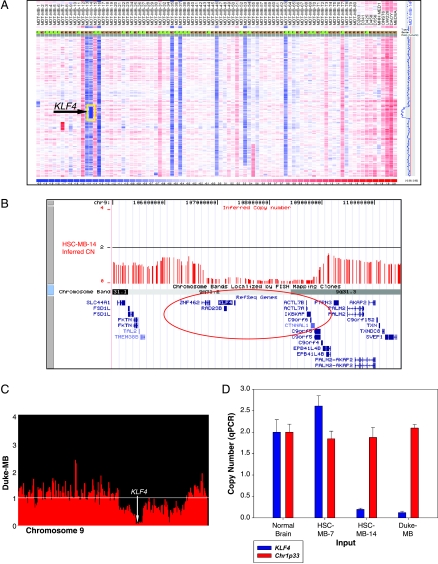
We used high-resolution SNP genotyping arrays to determine copy number aberrations in the genome of 201 primary medulloblastomas [18]. Loss of heterozygosity on chromosome 9q was observed in approximately 14% (11/79) of tumors, and one case harbored a region of homozygous deletion at 9q31.2-q31.3 that encompasses eight known genes (Figure 1A). This region of homozygous loss contains eight RefSeq genes: ZNF462, RAD23B, KLF4, ACTL7B, ACTL7A, IKBKAP, C9orf6, and CTNNAL1 (Figure 1B). In a separate study, on a nonoverlapping group of medulloblastomas, copy number aberrations in a series of 18 medulloblastomas were determined using digital karyotyping technology, and a second, overlapping but not identical homozygous deletion at 9q31.2 was identified (Figure 1C). Both of these homozygous deletions were validated using quantitative genomic PCR (Figure 1D). Recurrent homozygous deletions in primary tumors of a given tumor type are very significant, and indeed many of the best known TSGs have been discovered through the characterization of rare homozygous deletions [23–25].
Figure 1.
(A) Copy number analysis of 79 primary human medulloblastomas and 10 medulloblastoma cell lines profiled on the Affymetrix 100K SNP genotyping platform demonstrates that one primary tumor (sample MB-14) carries a 2.05-Mb homozygous deletion mapping to chromosome 9q31.2-q31.3 (highlighted in yellow box). Output is inferred log2 copy number data from dChip, with regions of gain shown in red and regions of loss shown in blue. Monosomy on chromosome 9q is observed in 8 (∼10.1%) of 79 samples. (B) UCSC output (NCBI Build 35; hg17) showing chromosome 9q inferred copy number data for HSC-MB-14, highlighting the region of homozygous deletion identified by SNP array. The homozygous deletion encompasses eight RefSeq genes (highlighted in red ellipsoid): ZNF462, RAD23B, KLF4, ACTL7B, ACTL7A, IKBKAP, C9orf6, and CTNNAL1. (C) Homozygous deletion encompassing the KLF4 locus as detected by digital karyotyping in 1 of 18 tumor samples. This region of homozygous deletion overlaps with the same region described in (B). Horizontal white line indicates a diploid copy number. Arrow points to the KLF4 locus. (D) Real-time genomic PCR confirming homozygous deletion of KLF4 in the two medulloblastoma samples described in (A, B) and (C). Template from normal brain and HSC-MB-7 were included as negative controls. An unrelated genomic locus on chromosome 1p33 (Chr1p33) was also included.
Loss of Expression of KLF4 in Medulloblastoma
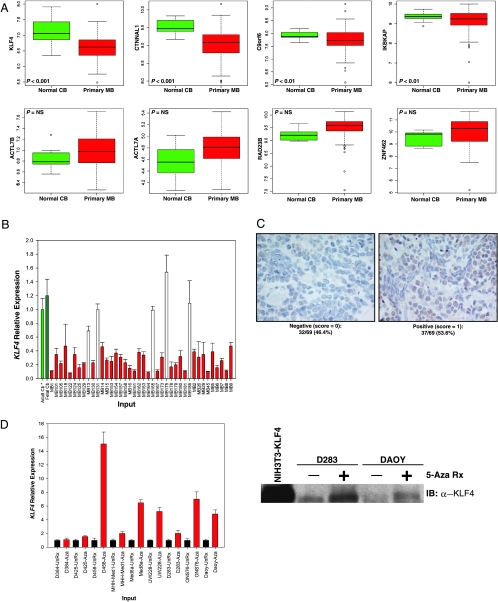
To gain insight into which gene(s) was acting as a “driver(s)” in the region of homozygous deletion on chromosome 9q31, we compared the expression pattern of the eight candidates in a cohort of 103 medulloblastomas compared with normal cerebellum (n = 10) using Affymetrix exon arrays [19]. Of these eight genes, only KLF4 and CTNNAL1 showed a significant decrease in expression in tumors compared with normal tissue control using exon array expression data (Figure 2A). KLF4 is a known TSG that is epigenetically silenced in colon cancer, gastric cancer, and leukemia [11–13,26]. Recurrent homozygous deletion and down-regulation of KLF4 in primary medulloblastoma suggests that it may function as a TSG in the pathogenesis of medulloblastoma.
Figure 2.
(A) Expression status of genes homozygously deleted on chromosome 9q31.2-q31.3 in normal cerebellum (n = 10) and primary medulloblastomas (n = 103) as determined by Affymetrix exon arrays. Box plots representing normalized log2-transformed expression array data for the eight RefSeq genes are shown. Of the eight candidate genes, KLF4 exhibits the greatest reduction in transcript expression level in medulloblastoma compared to normal controls. P values determined using t test statistics are shown. NS = not significant. y-axis = normalized signal intensity from exon array expression data. (B) Quantitative RT-PCR analysis of KLF4 in primary medulloblastomas (n = 40) demonstrates at least two-fold down-regulation of KLF4 expression compared with normal cerebellum in approximately 88% (35/40) of medulloblastomas. Samples with at least two-fold down-regulation are highlighted in blue. KLF4 expression values were determined by normalization to β-actin and calibration to normal adult cerebellum. (C) Staining of a human medulloblastoma tissue microarray with antibodies to KLF4 shows that KLF4 staining is absent in 32 (∼46%) of 69 cases. Representative cases are shown. (D) Right panel. Quantitative RT-PCR demonstrates robust reexpression of the KLF4 transcript in seven of nine medulloblastoma cell lines after treatment with the DNA methylation inhibitor 5-Aza (highlighted red) compared to mock-treated cells (highlighted black). Left panel. Western blot analysis demonstrating reexpression of KLF4 in the medulloblastoma cell lines D283 and DAOY after treatment with 5-Aza.
To confirm the expression array data, we used quantitative reverse transcription-polymerase chain reaction (RT-PCR) to compare the expression level of KLF4 in primary medulloblastomas to both adult and fetal human cerebellum. KLF4 expression was dramatically reduced in medulloblastomas compared with normal controls, with 35 (∼88%) of 40 cases exhibiting at least a two-fold reduction in transcript abundance compared with normal adult and fetal cerebellum when compared by quantitative RT-PCR (Figure 2B). To further investigate the status of KLF4 in primary medulloblastomas, we stained a medulloblastoma tissue microarray containing 69 primary cases with anti-KLF4 antibodies. KLF4 protein was not detected in approximately 46% (32/69) of tumors (Figure 2C). Lack of expression of KLF4 did not correlate with prognosis on our MB tissue microarray. Epigenetic silencing of TSGs through methylation of CpG dinucleotides within their promoter regions has become increasingly apparent in human cancers, including medulloblastomas [22,27,28]. To determine whether KLF4 deregulation may be attributable to promoter hypermethylation, we treated nine medulloblastoma cell lines with 5-Aza to reverse CpG methylation and subsequently checked for reexpression of KLF4 transcript. Indeed, treatment with 5-Aza resulted in robust reexpression of KLF4 in seven (∼78%) of nine medulloblastoma cell lines (Figure 2D, left panel). In addition, increased expression of the KLF4 protein is observed in the D283 and Daoy cell lines by immunoblot analysis after 5-Aza treatment (Figure 2D, right panel).
Methylation of the KLF4 Promoter in Medulloblastoma
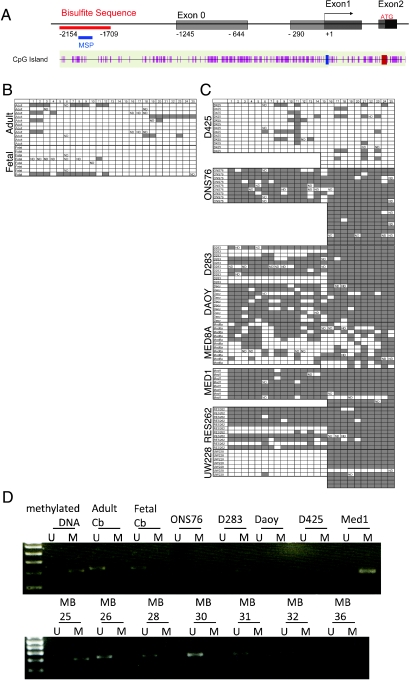
To elucidate whether the robust reexpression of KLF4 we observed in cell lines treated with 5-Aza was a direct result of CpG island hypermethylation, we next bisulfite-sequenced the KLF4 promoter region using genomic DNA templates isolated from normal human adult and fetal cerebella and eight medulloblastoma cell lines (Figure 3A). Profiling a region containing 25 CpG sites upstream of the KLF4 transcriptional start site demonstrated that there is only extremely rare CpG methylation in the normal human adult and fetal cerebellum (Figure 3B). In contrast, dense methylation of the KLF4 promoter is observed in seven of eight medulloblastoma cell lines (Figure 3C). Interestingly, of the cell lines profiled, only one, the D425 cell line, did not show significant CpG methylation in the KLF4 promoter (Figure 3C), which is significant in light of the fact that KLF4 expression remained unchanged in D425 cells treated with 5-Aza (Figure 2D). We performed MSP and demonstrated KLF4 promoter methylation in approximately 16% (7/44) of primary medulloblastomas (Figure 3D). We conclude that KLF4 promoter CpG hypermethylation is found in a subset of medulloblastomas, but not in normal cerebellum, and that this methylation is likely associated with transcriptional silencing. Additional medulloblastomas that fail to express KLF4, but for which we could not demonstrate promoter methylation using methylation sensitive PCR could carry other genetic or epigenetic events, including transcriptional repression to account for the lack of expression of KLF4.
Figure 3.
(A) Schematic representation of the CpG island found in the promoter region of the KLF4 genomic locus. Exons located within the CpG island are illustrated. Numbers indicate position relative to the transcriptional start site. The regions interrogated by bisulfite sequencing and MSP below are marked in red and blue, respectively. (B) Bisulfite-converted DNA from normal adult and fetal cerebellum were amplified at the KLF4 promoter, and fragments were TA-cloned and sequenced. CpG sites are represented as boxes, with shaded regions indicating methylation and unshaded regions indicating no methylation. The KLF4 promoter is hypomethylated in the normal adult and fetal cerebellum, with a minimal number of CpGs showing methylation. (C) Bisulfite sequencing of the KLF4 promoter in medulloblastoma cell lines shows densemethylation in ONS76, D283, Daoy, Med8a, Med1, RES262, and UW228. Notably, there is only sparse methylation in the D425 cell line, which also did not show an increase in KLF4 expression after treatment with 5-Aza. (D) MSP for a region of the KLF4 promoter. A total of 5 medulloblastoma cell lines and 44 primary medulloblastomas were included in the analysis. A methylated band was amplified in 7 (∼16%) of 44 primary samples. Universally methylated DNA and normal adult and fetal cerebellar samples were included as controls for the methylated (M) and unmethylated (U) primers, respectively.
KLF4 Functions as a TSG In Vitro and In Vivo
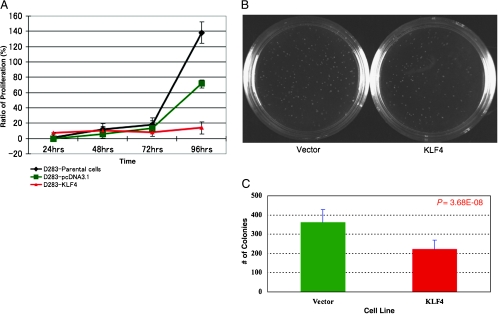
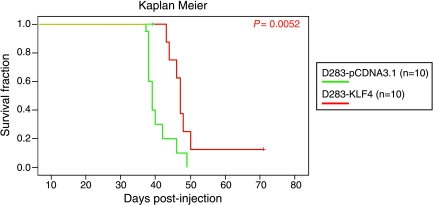
To functionally evaluate the role of KLF4 as a TSG in medulloblastoma, we stably reexpressed KLF4 in the D283 medulloblastoma cell line in which its expression is silenced. Stable reexpression of KLF4 in D283 cells resulted in nearly complete growth cessation compared with either parental or empty vector control cells by MTS assay (Figure 4A). This is very consistent with reexpression of a TSG in a malignant cell population. A similar pronounced effect was observed in the soft agar assay, with D283 cells stably expressing KLF4 exhibiting a significant reduction in colony-forming capacity compared to controls (Figure 4, B and C; t test, P = 3.68e - 08). Finally, intracerebellar xenografting of D283 medulloblastoma cells expressing KLF4 led to a clinically and statistically significantly increased survival compared with empty vector controls (Figure 5; log-rank test, P = .0052). That reexpression of just one gene—KLF4—can increase the survival of mice bearing medulloblastoma xenografts is quite striking, in that cancer genome resequencing projects are predicting that each tumor carries at least 10 and perhaps hundreds of driver mutations [29]. Our data demonstrate that KLF4 activity is abrogated by either genetic or epigenetic events in a significant subset of primary medulloblastomas and that restoration of KLF4 function in medulloblastoma results in decreased growth, decreased colony formation, and increased survival in an in vivo xenograft model. These data support our hypothesis that KLF4 functions as a TSG in medulloblastoma.
Figure 4.
(A) Stable reexpression of KLF4, but not controls, results in growth suppression of the D283medulloblastoma cell line as assessed by MTS assay. (B) D283 cells stably reexpressing KLF4 exhibit reduced colony-forming capacity compared to empty vector control cells as assessed by soft agar assay. (C) Quantification of colony formation data obtained for D283-empty vector and D283-KLF4 stable cell lines grown in soft agar. Values represent the mean colony number for three independent experiments performed in triplicate for each cell line. P value was calculated using t test statistics.
Figure 5.
Kaplan-Meier survival curve of nudemice with intracerebellar xenografts of D283 reexpressing KLF4 (n = 10) or vector control (n = 10). There is a statistically significant increase in survival of mice injected with the KLF4 expressing xenografts (log-rank test, P = .0052).
Discussion
KLF4 has been shown to interact with a number of pathways with well-documented links to medulloblastoma biology. KLF4 transactivates expression of the cell cycle arrest factor p27Kip1, which plays a critical role in the differentiation of granule neuron progenitor cells, one of the putative cells of origin for a subset of medulloblastomas, and a putative medulloblastoma TSG [30,31]. Klf4 has also been shown to bind to the promoter of Ccnd2 (encoding cyclin D2) and diminish its expression [32]. This is critical in that Ccnd2 knockout mice exhibit cerebellar hypoplasia, and CCND2 is frequently overexpressed in medulloblastoma [33,34]. Notch signaling has been shown to repress KLF4, which is pertinent in that Notch signaling plays an important role in the developing cerebellum, and NOTCH2 is amplified in a subset of medulloblastomas [35–37]. KLF4 has also been shown to repress the Wnt signaling pathway, which also has been shown to be hyperactivated in a subset of medulloblastomas [17,38,39]. As several of these signaling pathways are active in distinct subgroups of medulloblastoma [39], and as KLF4 has been shown to undergo loss of function in a variety of other cancer types, we suspect that KLF4 function in lost in more than one subgroup of medulloblastoma. To formally determine if this is true, the hypothesis will need to be tested on a clinically well-annotated series of medulloblastoma for which both high-quality genomic DNA and paraffin sections are available. Because KLF4 is targeted in a number of other neoplasms, we suspect that compounds targeting this event will be developed in the future, at which time they should be tested in medulloblastoma model systems as a prelude to future clinical trials in children with medulloblastoma.
Acknowledgment
The authors thank Susan Archer for editing.
Footnotes
These studies were supported with funds from the Canadian Cancer Society, the Pediatric Brain Tumor Foundation of the United States, The Sontag Foundation, and a CIHR Clinician-Scientist award (M.D.T.). P.A.N. was supported by a Restracomp salary award from the Hospital for Sick Children.
References
- 1.Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, Woo S, Wheeler G, Ahern V, Krasin MJ, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 2.De Smaele E, Fragomeli C, Ferretti E, Pelloni M, Po A, Canettieri G, Coni S, Di Marcotullio L, Greco A, Moretti M, et al. An integrated approach identifies Nhlh1 and Insm1 as Sonic Hedgehog-regulated genes in developing cerebellum and medulloblastoma. Neoplasia. 2008;10:89–98. doi: 10.1593/neo.07891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gershon TR, Oppenheimer O, Chin SS, Gerald WL. Temporally regulated neural crest transcription factors distinguish neuroectodermal tumors of varying malignancy and differentiation. Neoplasia. 2005;7:575–584. doi: 10.1593/neo.04637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DS, Hubbard SL, Peraud A, Salhia B, Sakai K, Rutka JT. Analysis of mammalian septin expression in human malignant brain tumors. Neoplasia. 2004;6:168–178. doi: 10.1593/neo.03310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michael LE, Westerman BA, Ermilov AN, Wang A, Ferris J, Liu J, Blom M, Ellison DW, van Lohuizen M, Dlugosz AA. Bmi1 is required for Hedgehog pathway-driven medulloblastoma expansion. Neoplasia. 2008;10:1343–1349. doi: 10.1593/neo.81078. 1345p following 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raffel C. Medulloblastoma: molecular genetics and animal models. Neoplasia. 2004;6:310–322. doi: 10.1593/neo.03454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao G, Pedone CA, Coffin CM, Holland EC, Fults DW. c-Myc enhances sonic hedgehog-induced medulloblastoma formation from nestin-expressing neural progenitors in mice. Neoplasia. 2003;5:198–204. doi: 10.1016/S1476-5586(03)80052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu LJ, Wu ML, Li H, Chen XY, Wang Q, Sun Y, Kong QY, Liu J. Inhibition of STAT3 expression and signaling in resveratrol-differentiated medulloblastoma cells. Neoplasia. 2008;10:736–744. doi: 10.1593/neo.08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Laterra J, Pomper MG. Hedgehog pathway inhibitor HhAntag691 is a potent inhibitor of ABCG2/BCRP and ABCB1/Pgp. Neoplasia. 2009;11:96–101. doi: 10.1593/neo.81264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamber MS, Bansal K, Liang ML, Mainprize TG, Salhia B, Northcott P, Taylor M, Rutka JT. Current concepts in the molecular genetics of pediatric brain tumors: implications for emerging therapies. Childs Nerv Syst. 2006;22:1379–1394. doi: 10.1007/s00381-006-0187-3. [DOI] [PubMed] [Google Scholar]
- 11.Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68:4631–4639. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasunaga J, Taniguchi Y, Nosaka K, Yoshida M, Satou Y, Sakai T, Mitsuya H, Matsuoka M. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64:6002–6009. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 15.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 16.Pandya AY, Talley LI, Frost AR, Fitzgerald TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA, Krontiras H, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- 17.Ghaleb AM, McConnell BB, Nandan MO, Katz JP, Kaestner KH, Yang VW. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 2007;67:7147–7154. doi: 10.1158/0008-5472.CAN-07-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, et al. The miR-17/92 polycistron is up-regulated in SonicHedgehog-driven medulloblastomas and induced by N-myc in Sonic Hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69(8):3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, Lengauer C, Cummins JM, Velculescu VE, Fults DW, et al. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- 21.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kongkham PN, Northcott PA, Ra YS, Nakahara Y, Mainprize TG, Croul SE, Smith CA, Taylor MD, Rutka JT. An epigenetic genome-wide screen identifies SPINT2 as a novel tumor suppressor gene in pediatric medulloblastoma. Cancer Res. 2008;68:9945–9953. doi: 10.1158/0008-5472.CAN-08-2169. [DOI] [PubMed] [Google Scholar]
- 23.Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, Jones S, Sjoblom T, Park BH, Parsons R, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varshavsky A. Targeting the absence: homozygous DNA deletions as immutable signposts for cancer therapy. Proc Natl Acad Sci USA. 2007;104:14935–14940. doi: 10.1073/pnas.0706546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox C, Bignell G, Greenman C, Stabenau A, Warren W, Stephens P, Davies H, Watt S, Teague J, Edkins S, et al. A survey of homozygous deletions in human cancer genomes. Proc Natl Acad Sci USA. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei D, Gong W, Kanai M, Schlunk C, Wan L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 27.Anderton JA, Lindsey JC, Lusher ME, Gilbertson RJ, Bailey S, Ellison DW, Clifford SC. Global analysis of the medulloblastoma epigenome identifies disease-subgroup-specific inactivation of COL1A2. Neuro Oncol. 2008;10:981–994. doi: 10.1215/15228517-2008-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsey JC, Lusher ME, Anderton JA, Gilbertson RJ, Ellison DW, Clifford SC. Epigenetic deregulation of multiple S100 gene family members by differential hypomethylation and hypermethylation events in medulloblastoma. Br J Cancer. 2007;97:267–274. doi: 10.1038/sj.bjc.6603852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayrault O, Zindy F, Rehg J, Sherr CJ, Roussel MF. Two tumor suppressors, p27Kip1 and patched-1, collaborate to prevent medulloblastoma. Mol Cancer Res. 2009;7:33–40. doi: 10.1158/1541-7786.MCR-08-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc andthe cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci USA. 2006;103:11579–11583. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klaewsongkram J, Yang Y, Golech S, Katz J, Kaestner KH, Weng NP. Kruppel-like factor 4 regulates B cell number and activation-induced B cell proliferation. J Immunol. 2007;179:4679–4684. doi: 10.4049/jimmunol.179.7.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huard JM, Forster CC, Carter ML, Sicinski P, Ross ME. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development. 1999;126:1927–1935. doi: 10.1242/dev.126.9.1927. [DOI] [PubMed] [Google Scholar]
- 34.Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, Ueda S, Dura W, Grajkowska W, Kuo JS, Rutka JT. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 35.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 36.Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 37.Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, Liu C, Ai W. KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol. 2009;296:G490–G498. doi: 10.1152/ajpgi.90393.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor MD, Zhang X, Liu L, Hui CC, Mainprize TG, Scherer SW, Wainwright B, Hogg D, Rutka JT. Failure of a medulloblastoma-derived mutant of SUFU to suppress WNT signaling. Oncogene. 2004;23:4577–4583. doi: 10.1038/sj.onc.1207605. [DOI] [PubMed] [Google Scholar]
- 39.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]