Abstract
Constitutive activation of signal transducer and activator of transcription 3 (STAT3) signaling is frequently detected in cancer, promoting its emergence as a promising target for cancer treatment. Inhibiting constitutive STAT3 signaling represents a potential therapeutic approach. We used structure-based design to develop a nonpeptide, cell-permeable, small molecule, termed as LLL12, which targets STAT3. LLL12 was found to inhibit STAT3 phosphorylation (tyrosine 705) and induce apoptosis as indicated by the increases of cleaved caspase-3 and poly (ADP-ribose) polymerase in various breast, pancreatic, and glioblastoma cancer cell lines expressing elevated levels of STAT3 phosphorylation. LLL12 could also inhibit STAT3 phosphorylation induced by interleukin-6 in MDA-MB-453 breast cancer cells. The inhibition of STAT3 by LLL12 was confirmed by the inhibition of STAT3 DNA binding activity and STAT3-dependent transcriptional luciferase activity. Downstream targets of STAT3, cyclin D1, Bcl-2, and survivin were also downregulated by LLL12 at both protein and messenger RNA levels. LLL12 is a potent inhibitor of cell viability, with half-maximal inhibitory concentrations values ranging between 0.16 and 3.09 µM, which are lower than the reported JAK2 inhibitor WP1066 and STAT3 inhibitor S3I-201 in six cancer cell lines expressing elevated levels of STAT3 phosphorylation. In addition, LLL12 inhibits colony formation and cell migration and works synergistically with doxorubicin and gemcitabine. Furthermore, LLL12 demonstrated a potent inhibitory activity on breast and glioblastoma tumor growth in a mouse xenograft model. Our results indicate that LLL12 may be a potential therapeutic agent for human cancer cells expressing constitutive STAT3 signaling.
Introduction
Cancer is the second leading cause of death in the United States, being attributed to one in four deaths. Approximately one in two men and one in three women will have their conditions diagnosed as an invasive cancer in their lifetime [1]. Breast cancer is the leading type of cancer affecting women. It is estimated that breast cancer accounts for just more than a quarter of all newly diagnosed cancer cases in women [1]. Pancreatic cancer is the fourth leading cause of cancer deaths in the United States. Diagnosis is followed by a poor prognosis, with a 5-year survival rate of only 5% [1]. Worldwide, the survival rate for pancreatic cancer is only 1% [2]. Gliomas, a type of brain cancer, account for more than 75% of all primary malignant brain tumors [3]. The most common type of glioma, glioblastoma, is also the most severe. It is a highly aggressive cancer and continues to have a poor survival rate, with most cases becoming fatal within 2 years of diagnosis [4,5]. The large number of cases and poor survival rates under current therapies necessitates the search for novel target therapies for cancer.
The signal transducer and activator of transcription (STAT) proteins are transcription factors that participate in relaying signals from cytokines and growth factors [6–8]. Constitutive activation of STATs has been found to contribute to oncogenesis [6,9]. STAT3, in particular, is constitutively active in a wide variety of human malignancies, including breast and pancreatic cancer and glioblastoma [6–8]. STAT3 is considered to be an oncogene owing to its ability to promote malignancy [7,9,10]. Experiments have shown that constitutively active STAT3 is sufficient for inducing cellular transformation [10]. Further support comes from a resistance to transformation seen in STAT3-deficient fibroblasts [11,12]. Constitutively active STAT3 has also been shown to have the potential to alter the phenotype of nonmalignant cells into one that is similar to malignant cells [13].
Persistent activation of STAT3 has been implicated in both the induction of cancer and the processes promoting the survival of cancer. STAT3 activation occurs when the tyrosine 705 (Tyr705) residue is phosphorylated, leading to dimerization and translocation from the cytoplasm to the nucleus [9,14,15]. In the nucleus, STAT3 binding to target genes induces the transcription and up-regulation of proliferation and antiapoptotic associated proteins [7,9,10,16]. Therefore, constitutive STAT3 signaling is involved in stimulating cell cycle progression and preventing apoptosis which contributes to malignant progression [7,9]. It has also been found to promote angiogenesis [6,17]. In addition, persistently activated STAT3 plays a role in impairing both innate and adaptive immune responses by enhancing immunologic tolerance and enabling cancer cells to evade immune surveillance [18]. Further, the survival of these tumors seems to depend on the presence of STAT3 signaling [6,11].
The implications of constitutive STAT3 signaling in tumors have presented it as a possible target for cancer treatment. Experiments aimed at blocking STAT3 signaling using dominant-negative STAT3, RNA interference, and STAT3 antisense oligonucleotides have provided further evidence of the potential of STAT3 as a target for treating cancer [6,8,19,20]. Inhibiting STAT3 using the stated approaches has been successful, resulting in an inhibition of growth and the induction of death in tumors. It was also determined that in normal cells, blocking STAT3 is neither harmful nor toxic to the cells [6,11]. Given the oncogenic functions of STAT3 and the promise of inhibiting it, directly targeting STAT3 signaling represents a potential therapeutic approach to treating cancer.
Using a structure-based drug design, we developed a novel STAT3 inhibitor, named LLL12. Computer models with docking simulation showed that LLL12 binds directly to the phosphoryl tyrosine 705 (pTyr705) binding site of the STAT3 monomer. We assessed the inhibitory effects of LLL12 in cancer cells. We demonstrated that LLL12 inhibits STAT3 phosphorylation (Tyr705) and STAT3 activities; downregulates STAT3 downstream targets; inhibits proliferation, colony formation, and cell migration; and induces apoptosis in various human breast and pancreatic cancer cells as well as in glioblastoma cells. We also demonstrated that LLL12 has minimal apoptotic effects on normal human cells.
Materials and Methods
Computational Binding Studies of LLL12
Computational docking program AutoDock4 was use to dock our designed nonpeptide small molecules and to predict their binding modes and approximate binding free energies to STAT3 SH2 dimerization site [21]. The small molecule LLL12 was docked using the Lamarckian Genetic Algorithm. The docking procedure involved the preparation of the ligand and macromolecule, the assignment of Gasteiger charges, and the identification of the torsional root and the three rotatable bonds of the ligand. An AutoGrid map was then precomputed for all atom types in the ligand set. After 10 million energy evaluations were completed, all the resulting conformations of the ligand in the binding pocket of the macromolecule were clustered into groups according to their conformations with a root mean square deviation threshold of 1.5 Å. A major lowest energy cluster was identified with 24% conformers and binding energy of -7.8 kcal/mol.
Synthesis of LLL12
Chemicals and reagents
Chemicals (except 3-hydroxy-2-pyrone, which was purchased from Tyger Scientific, Ewing, NJ) and silica gel were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI). The chemicals were checked for purity by thin layer chromatography and nuclear magnetic resonance (NMR). Melting points were determined on a Thomas Hoover capillary melting point apparatus and were uncorrected. Proton nuclear magnetic resonance spectra were obtained with a Bruker Avance 300 (300 MHz) spectrophotometer (Billerica, MA).
Synthesis of compound 2
Naphthalene sulfonyl chloride 1 (1 g, 4.41 mmol) was dissolved in acetone (52 ml) and was stirred at 0°C for 30 minutes. Ammonium hydroxide (52 ml) was cooled to 0°C and was added to the above mixture and stirred at room temperature for 3 hours. The acetone was then removed at reduced pressure. The residue was dissolved in dichloromethane (100 ml) and washed with water (2 x 100 ml). The organic layer was collected and evaporated under reduced pressure. The residue was purified by silica column chromatography (hexane-ethyl acetate, 3:1) yielding compound 2 (750 mg, 82.1%); melting point (m.p.) 147 to 149°C (literature 150°C) [22].
Synthesis of compound 3
Compound 2 (500 mg, 2.41 mmol) was dissolved in glacial acetic acid (5.0 ml). Chromium trioxide (1.08 g, 10.85 mmol) was dissolved in a mixture of water-glacial acetic acid (1:1, 2 ml) and added to the solution of compound 2 in glacial acetic acid and was stirred under reflux for 15 minutes. The solution was cooled to 0°C and water (25 ml) was added, and the resulting solution was stirred overnight at room temperature. The reaction mixture was diluted with water (500 ml) and extracted with ether (3 x 100 ml). The organic layer was collected, dried under reduced pressure, and purified with silica column chromatography ethyl acetate-hexane (2:3) to yield compound 3 (88 mg, 15.4%); m.p. (187–188°C); 1H NMR (300 MHz, dimethyl sulfoxide [DMSO]) δ 7.23 (2H, d, J = 9 Hz), 7.43 (2H, S), 8.11 (1H, t, J = 9 Hz), 8.34 (1H, d, J = 9 Hz), 8.515 (1H, d, J = 9 Hz). Mass spectrometry ([M + Na]+ 260.7).
Synthesis of LLL12
Asolution of compound 3 (200 mg, 0.843 mmol) in chloroform (14 ml) was stirred at -20°C for 10 minutes followed by the addition of triethylamine (0.01 ml) and stirring continued at -20°C for an additional 15 minutes. 3-Hydroxy-2-pyrone (86 mg, 0.767 mmol) dissolved in chloroform (1 ml) was added to the reaction mixture and stirred at room temperature for 1 hour. The solvent was removed under reduced pressure. The resulting residue was diluted with water (50 ml), and the aqueous solution was extracted with ethyl acetate (3 x 50 ml). The organic layer was separated, dried (brine), and evaporated. The crude product was purified by silica column chromatography (hexane-ethyl acetate, 4:1) yielding LLL12 (50 mg, 20%). m.p. (179–181°C); 1H NMR (300 MHz, DMSO) δ 7.42–7.85 (5H, m), 8.11 (1H, m), 8.56 (2H, m), 12.05 (1H, s). Mass spectrometry ([M + Na]+ 326.1).
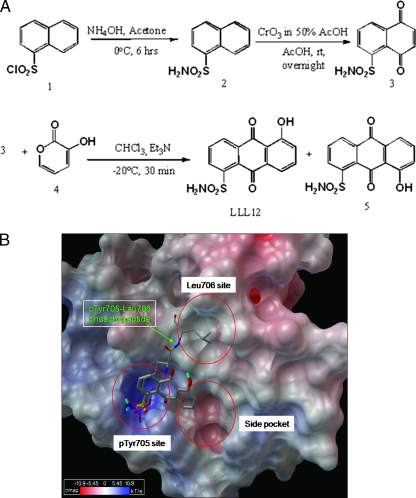
The synthesis of LLL12 began with the reaction of sulfonyl chloride 1 with ammonium hydroxide to form 2. Oxidation of 2 yielded the naphthoquinone 3 with chromium (VI) oxide. Base-catalyzed Diels-Alder reactions of 3-hydroxy-2-pyrone with compound 3 at -20°C yield LLL12 and are regioisomer in a ratio of 98:2.
Cell Lines
Human breast cancer cell lines (MDA-MB-231, MDA-MB-453, and SK-BR-3), human pancreatic cancer cell lines (HPAC and iPANC-1), glioblastoma cell line (U87), human hepatocytes (HHs), and normal human lung fibroblasts (WI-38) were purchased from the American Type Culture Collection (Manassas, VA). Human glioblastoma cell line (U373) was kindly provided by Dr. Sean Lawler (The Ohio State University). Human mammary epithelial cells (HMECs) were purchased from Lonza Walkersville, Inc. (Walkersville, MD), and maintained in Ham's F12 medium (Mediatech, Manassas, VA) supplemented with 5 µg/ml insulin, 1 µg/ml hydrocortisone, 10 µg/ml epidermal growth factor, 100 µg/ml cholera toxin, 5% fetal bovine serum (FBS). Immortalized human pancreatic duct epithelial (HPDE) cells were provided by Dr. Ming-Sound Tsao at the University of Toronto and maintained in CnT-07CF epidermal keratinocyte medium (CELLnTEC Advanced Cell Systems, Bern, Switzerland) supplemented with 0.07 mM CaCl2. The HHs were maintained in hepatocyte medium (ScienCell, Carlsbad, CA) plus hepatocyte growth supplement and 5% FBS. All other cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS, 4.5 g/L l-glutamine, sodium pyruvate, and 1% penicillin/streptomycin. All cell lines were stored in a humidified 37°C incubator with 5% CO2.
JAK2 and STAT3 Inhibitors
LLL12, a STAT3 inhibitor, and WP1066 [23], a JAK2 inhibitor, were synthesized in Dr. Pui-Kai Li's laboratory (College of Pharmacy, The Ohio State University). The powder was dissolved in sterile DMSO to make a 20-mM stock solution. Aliquots of the stock solution were stored at -20°C. S3I-201 [24], a STAT3 SH2 inhibitor, was purchased from Calbiochem (Gibbstown, NJ).
Cell Viability Assay
Human breast cancer cell lines (MDA-MB-231 and SK-BR-3), human pancreatic cancer cell lines (PANC-1 and HPAC), and glioblastoma cell lines (U87 and U373) were seeded in 96-well plates at a density of 3000 cells per well. Different concentrations of LLL12 (0.1–10 µM), WP1066 (1–10 µM), or S3I-201 (1–100 µM) were added in triplicate to the plates in the presence of 10% FBS. The cells were incubated at 37°C for a period of 72 hours. 3-(4,5-Dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT) viability assay was done according to manufacturer's protocol (Roche Diagnostics, Mannheim, Germany). The absorbance was read at 595 nm. Half-maximal inhibitory concentrations (IC50) were determined using Sigma Plot 9.0 Software (Systat Software, Inc., San Jose, CA).
Western Blot Analysis
Human breast cancer cell lines (MDA-MB-231 and SK-BR-3), human pancreatic cancer cell lines (HPAC and PANC-1), human glioblastoma cell lines (U87 and U373), and human normal cells lines (HPDE, HMEC, HH, and WI-38) were treated with LLL12 (5 or 10 µM) or DMSO at 60% to 80% confluence in the presence of 10% FBS for 24 hours, lysed in cold radio immunoprecipitation assay lysis buffer containing protease inhibitors, and subjected to SDS-PAGE. Membranes were probed with a 1:1000 dilution of antibodies (Cell Signaling Technology, Beverly, MA) against phosphospecific STAT3 (Tyr705), phosphospecific extracellular signal-regulated kinase (ERK)1/2 (threonine 202/tyrosine 204), phosphospecific Src (tyrosine 416), phosphospecific themammalian target of rapamycin (mTOR) (serine 2448), cleaved poly (ADP-ribose) polymerase (PARP), cleaved caspase-3, cyclin D, Bcl-2, surviving, and glyceraldehyde-3-phosphate dehydrogenase. Membranes were analyzed using Enhanced Chemiluminescence Plus reagents and scanned with the Storm Scanner (Amersham Pharmacia Biotech, Inc., Piscataway, NJ).
IL-6 Induction of STAT3 Phosphorylation
MDA-MB-453 breast cancer cells were seeded in 10-cm plates and allowed to adhere overnight. The following night, the cells were serum-starved. The cells were then left untreated or were treated with LLL12 (0.5–2 µM) or DMSO. After 2 hours, the untreated and LLL12-treated cells were stimulated by IL-6 (25 ng/ml). The cells were harvested at 30 minutes and analyzed by Western blot.
STAT3 and STAT1 DNA Binding Activity
MDA-MB-231, SK-BR-3, HPAC, and U87 cancer cells at 60% to 80% confluence were treated with LLL12 (5 or 10 µM) or DMSO in the presence of 10% FBS for 24 hours. A nuclear extract kit (Clontech, Inc., Mountain View, CA) was used to obtain nuclear extracts. The nuclear extracts were analyzed for STAT3 and/or STAT1 DNA binding activity using STAT3 or STAT1 Transcription Factor Kits (Clontech, Inc.), which provide an ELISA-based method to detect DNA binding by transcription factors.
STAT3-Dependent Transcriptional Luciferase Activity
STAT3-dependent transcriptional luciferase activity was measured using MDA-MB-231-cloned cells that stably integrate the STAT3-dependent luciferase reporter construct, pLucTKS3 [25]. The cells were grown in six-well plates until semiconfluent and treated in 5% FBS with LLL12 (1–10 µM) or DMSO for 24 hours. The luciferase assay (Promega, Madison, WI) was run according to the manufacturer's protocol. The STAT3 luciferase activity of the LLL12-treated cells is reported relative to pLucTKS3-transfected cells treated with DMSO arbitrarily set at 100%.
Reverse Transcription-Polymerase Chain Reaction
MDA-MB-231, HPAC, and U373 cells were treated with LLL12 (5 or 10 µM) or DMSO at 60% to 80% confluence in the presence of 10% FBS for 24 hours. RNA from the cells was then collected using RNeasy kits (Qiagen, Valencia, CA). Primer sequences and source information of STAT3 downstream target genes can be found in Table W1. Polymerase chain reaction (PCR) amplification was done under the following conditions: 5 minutes at 94°C followed by 25 cycles of 30 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C with a final extension of 5 minutes at 72°C.
Determination of Combinatorial Effects
MDA-MB-231 breast and HPAC pancreatic cancer cells were seeded in 96-well plates in triplicate at a density of 3000 cells per well and were treated with LLL12 (500 nM) and doxorubicin (100–400 nM, Sigma-Aldrich, St. Louis, MO) or with LLL12 (1000 nM) and gemcitabine (100–1000 nM, Sigma-Aldrich, St. Louis, MO) in the presence of 10% FBS. LLL12 and doxorubicin (Sigma-Aldrich, St. Louis, MO) or LLL12 and gemcitabine (Sigma-Aldrich, St. Louis, MO) synergy with regards to growth inhibition was determined as follows [26]. The log (fa/fu) was plotted against the concentration (D) for each compound alone or in combination, where fa is the fraction affected and fu is the fraction unaffected (1 - fa) of cells at each concentration. Calcusyn software (Biosoft, Ferguson, MO) was used to determine the combinational index (CI) for each drug and concentration combination. A CI value of less than 1 represents synergism. A CI value equal to 1 represents additive effects. A CI value greater than 1 represents antagonistic effects.
Wound Healing/Cell Migration Assay
MDA-MB-231 breast cancer cells (3 x 105 per well) were seeded in a six-well plate. Approximately 24 hours later, when the cells were 100% confluent, the monolayer was scratched using a 1-ml pipette tip and washed once to remove nonadherent cells. New medium in the presence of 10% FBS containing LLL12 (1–20 µM) or DMSO was added. The treatments were removed after 4 hours, and fresh medium was added. After an additional 20 hours without treatment, the cells were observed under the microscope. When the wound in the control was closed, the inhibition of migration was assessed by using the ImageJ software, available from the National Institutes of Health Web site (http://rsb.info.nih.gov/ij/). The percent of wound healed was calculated using the formula: 100 - [(final area / initial area) x 100%].
Colony Formation Assay
A base 0.6% agar gel with 10% FBS in DMEM was prepared and added to the wells of a six-well culture dish. MDA-MB-231 breast cancer cells were plated at a density of 5000 cells per well on top of the base agar for anchorage-independent growth analysis in 0.4% agar gel with 10% FBS in DMEM supplemented with LLL12 (1 or 5 µM) or DMSO. The cells were maintained at 37°C and allowed to grow for 2 weeks. The colonies were stained using MTT dye (100 µl per well). Pictures of the colonies were taken using a Leica MZ 16FA inverted microscope (Leica Microsystems, Bannockburn, IL) with a 7.4 Slider Camera (Diagnostic Instruments, Inc., Sterling Heights, MI). The colonies were scored by counting, and numbers were normalized as a percentage of colonies formed in DMSO.
Transfection with Constitutive STAT3
U87 glioblastoma cells were plated in 60-mm3 dishes or 96-well plates. The second day, the cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with a vector encoding the constitutive STAT3 (STAT3-C), which is tagged with the FLAG epitope [10]. Cells were treated with LLL12 (1–5 µM) or DMSO 24 hours after transfection. Twenty-four hours later, the cells in 60-mm3 dishes were harvested to run Western blot. Cell viability was determined by MTT assay in 96-well plates as previously described.
Mouse Xenograft Tumor Model
MDA-MB-231 breast cancer cells (1 x 107) and U87 glioblastoma cells (5 x 106) were injected (subcutaneously) into the right flank area of 4- to 5-week-old male athymic nude mice that were purchased from Harlan (Indianapolis, IN). After tumor development, mice were divided into three treatment groups consisting of five mice/group: DMSO vehicle control and 2.5 and 5 mg/kg of LLL12. Tumor growth was determined by measured the length (L) and width (W) of the tumor every other day with a caliper, and tumor volume was calculated on the basis of the following formula: volume = (π/6)LW2. After 14 days of treatment, tumors were harvested from killed mice, snap-frozen in liquid nitrogen, and stored at -80°C. Tumors tissue homogenates were lysed and separated by SDS-PAGE to examine the expression of STAT3 phosphorylation in vehicle- and LLL12-treated mice.
Results
LLL12, a Novel Small Molecule That Targets STAT3
pTyr705 is critical for the biologic function of STAT3, as it is critical for dimerization [14,15]. pTyr705 is located on a loop segment of the SH2 domain and binds together with several adjacent amino acid residues (leucine 706, threonine 708, and phenylalanine 710) to a cavity on the SH2 domain of the other STAT3 monomer. We designed a compound, LLL12, which binds to STAT3 SH2 domain. The structure and synthesis of LLL12 are shown in Figure 1A. To optimize potency and selectivity, the main scaffold of LLL12 contains fragments that directly contact the pTyr705 binding site of STAT3 (Figure 1B). A simulated docking model shows that the sulfonamide tail of LLL12 occupies the pTyr705 binding pocket of STAT3 with at least three hydrogen bonds. Simulated binding energy (-7.8 kcal/mol) of LLL12 to STAT3 predicts that it will be a potent inhibitor of the constitutive STAT3 pathway.
Figure 1.
(A) Synthesis of LLL12 (includes chemical structure). (B) Computer model of LLL12 binding to the STAT3 SH2 domain. The ball-and-stick model of pTyr705-Leu706 is the binding mode of the partnering SH2 during the STAT3 homodimerization. LLL12 effectively displaces its binding through stronger binding to pTyr705 binding site, indicating LLL12 can efficiently prevent STAT3 SH2 dimerization.
LLL12 Inhibits STAT3 Phosphorylation and Induces Apoptosis in Human Breast and Pancreatic Cancer Cells and Glioblastoma Cells
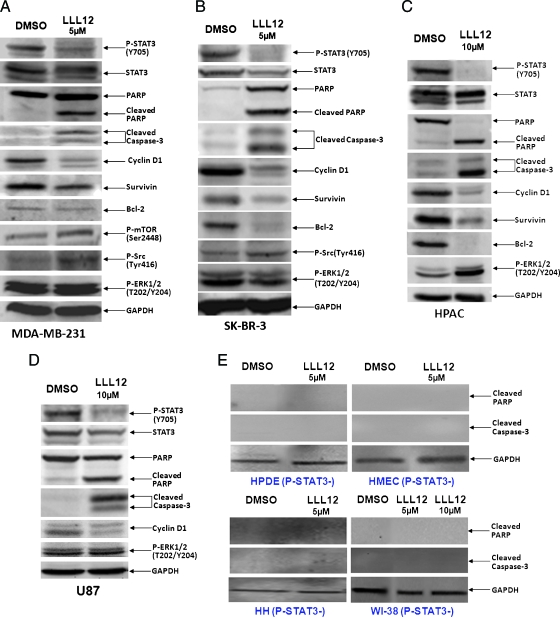
LLL12 was evaluated for its effect on breast cancer cells (MDA-MB-231 and SK-BR-3), pancreatic cancer cells (HPAC and PANC-1), and glioblastoma cells (U87 and U373) that express elevated levels of STAT3 phosphorylation. LLL12 inhibited STAT3 phosphorylation at Tyr705 in all six cancer cell lines (Figures 2, A–D, and W1). LLL12 was not found to inhibit phosphorylation of other kinase, such as ERK1/2, mTOR, and Src, indicating selectivity for STAT3. As shown in Figure 2, A–D, downstream targets of STAT3, such as cyclin D1, survivin, and Bcl-2 [10], were downregulated by LLL12. The inhibition of STAT3 phosphorylation by LLL12 seems to be consistent with the induction of apoptosis as evidence by the cleavages of PARP and caspase-3 (Figures 2, A–D, and W1). The effect of LLL12 was also examined in cells that do not express elevated levels of STAT3 phosphorylation (HPDE cells, HMECs, HHs, and WI-38 normal lung fibroblasts). LLL12 did not induce cleaved PARP or caspase-3 in any of these cells lines (Figure 2E). This indicates that LLL12 is selective for cancer cells expressing elevated levels of STAT3 phosphorylation.
Figure 2.
Western blot analysis of cells treated with LLL12. Cancer cell lines expressing constitutively active STAT3, (A) MDA-MB-231, (B) SK-BR-3, (C) HPAC, (D) U87, exhibit a decrease in the levels of expression of STAT3 phosphorylation after treatment with LLL12. Downstream targets of STAT3, cyclin D1, Bcl-2, and survivin, were inhibited. Apoptosis is also indicated by the induction of cleaved PARP and caspase-3. Normal cell lines that do not express elevated levels of STAT3 phosphorylation, (E) HPDE cells, HMEC, HHs, and normal human lung fibroblasts (WI-38), did not exhibit an induction of cleaved PARP or caspase-3 after treatment with LLL12.
LLL12 Inhibits STAT3 Phosphorylation Induced by IL-6
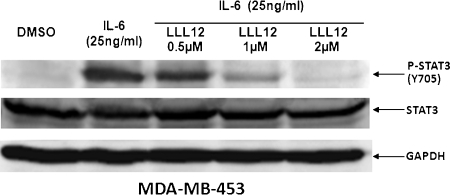
Activation of STAT3 can be induced by IL-6 [14,15]. MDA-MB-453 breast cancer cells, which do not express persistently phosphorylated STAT3, were used to determine if LLL12 is capable of inhibiting IL-6-induced STAT3 phosphorylation. We found that IL-6 stimulates STAT3 phosphorylation in MDA-MB-453 cells. This stimulation of STAT3 phosphorylation was blocked by LLL12 in a dose-dependent manner (Figure 3). These results support that LLL12 is a potent inhibitor of STAT3 phosphorylation in cancer cells.
Figure 3.
LLL12 inhibits STAT3 phosphorylation induced by IL-6 in MDA-MB-453 breast cancer cells. The cells were serum-starved overnight, then left untreated or were treated with LLL12 (0.5–2 µM) orDMSO. After 2 hours, the untreated and LLL12-treated cells were stimulated by IL-6 (25 ng/ml). The cells were harvested at 30 minutes and analyzed by Western blot.
LLL12 Inhibits STAT3 DNA Binding
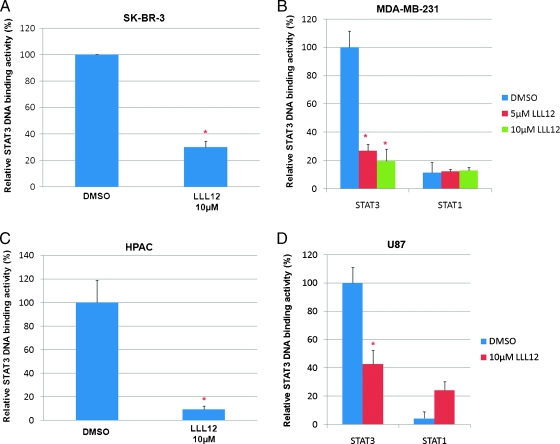
To confirm the inhibition of STAT3 signaling by LLL12, we examined the inhibition of STAT3 DNA binding activity. LLL12 caused a statistically significant inhibition of STAT3 DNA binding activity in breast cancer cell lines, SK-BR-3 (Figure 4A) and MDA-MB-231 (Figure 4B), pancreatic cancer cell line, HPAC (Figure 4C), and glioblastoma cell line, U87 (Figure 4D). LLL12 did not inhibit STAT1 DNA binding activity (Figure 4, B and D), indicating a specificity of LLL12 for STAT3 over STAT1.
Figure 4.
LLL12 has an inhibitory effect on STAT3 DNA binding activity and STAT3-dependent transcriptional activity. The nuclear extracts of (A) SK-BR-3, (B) MDA-MB-231, (C) HPAC, and (D) U87 cancer cells were analyzed for STAT3 DNA binding. STAT1 DNA binding was also looked at to demonstrate the specificity of LLL12 to STAT3 over STAT1 protein in (B) MDA-MB-231 and (D) U87 cancer cells. Statistical significance (P < .05) relative to the DMSO vehicle control is designated by an asterisk.
LLL12 Inhibits STAT3-Dependent Transcriptional Activities and Transcription of Downstream Targets of STAT3
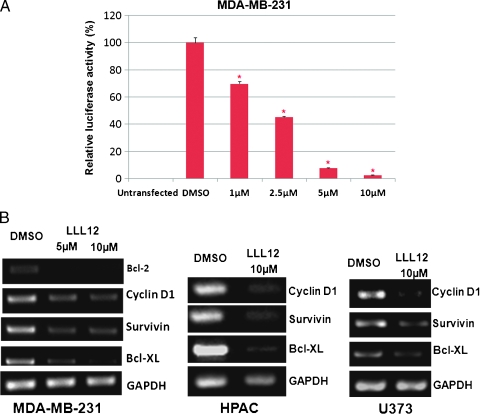
As previously mentioned, STAT3 binding to the promoters of the target genes induces the transcription of several proliferation- and antiapoptotic-associated proteins. STAT3-dependent transcriptional luciferase activity was then examined after treatment with LLL12 for 24 hours. As seen in the luciferase assay (Figure 5A), LLL12 also inhibited STAT3-dependent transcriptional activity in a dose-dependent manner. To further analyze the impact of LLL12 on the inhibition of STAT3, we looked at the transcription of downstream target genes of STAT3 by reverse transcription (RT)-PCR. We treated MDAMB-231 breast cancer cells, HPAC pancreatic cancer cells, and U373 glioblastoma cells with LLL12 (5 or 10 µM) or DMSO for 24 hours. RT-PCR was run for cyclin D1, survivin, and Bcl-XL. We found that treatment with LLL12 resulted in an inhibition of the transcription of STAT3-regulated genes (Figure 5B).
Figure 5.
(A) STAT3-dependent transcriptional activity was analyzed in a luciferase assay. MDA-MB-231 breast cancer cloned cells that stably integrate the STAT3-dependent luciferase reporter construct, pLucTKS3, were used. Results are reported relative to a pLucTKS3-transfected sample treated with DMSO set at 100%. Statistical significance (P < .05) relative to DMSO is designated by an asterisk. (B) Transcription of STAT3-regulated genes is inhibited by LLL12. RT-PCR reveals decreased expression of STAT3 target genes over a DMSO control after treatment with LLL12.
Inhibition of Cell Proliferation/Viability in Human Breast and Pancreatic Cancer Cells and Glioblastoma Cells by LLL12
STAT3 activation is important for cell proliferation and survival. Cell viability assays were run to examine the inhibitory affect of LLL12 on human breast and pancreatic cancer cells and glioblastoma cells. A dose-dependent inhibition in tumor cell proliferation/viability was seen after 72 hours of treatment. IC50 values were calculated for LLL12 and other previously characterized inhibitors (Table 1), namely, WP1066 [23], a JAK2/STAT3 inhibitor, and S3I-201 [24], a STAT3 inhibitor. The inhibitory efficacy of the three compounds was compared. LLL12 is substantially more potent in the inhibition of cell viability than the other available inhibitors in all the cell lines analyzed.
Table 1.
IC50 Obtained for STAT3 Inhibitors (µM) in Human Breast, Pancreatic Cancer and Glioblastoma.
| LLL12 | WP1066 | S3I-201 | |
| MDA-MB-231 | 0.97 | 7.48 | >100 |
| SK-BR-3 | 3.09 | 3.31 | >100 |
| PANC-1 | 0.29 | 5.12 | >100 |
| HPAC | 0.16 | 2.52 | >100 |
| U87 | 0.21 | 5.78 | 55.10 |
| U373 | 0.86 | 5.16 | 52.50 |
All values reflect concentrations calculated after 72 hours of treatment in an MTT viability assay.
Anchorage Independence and Cell Viability
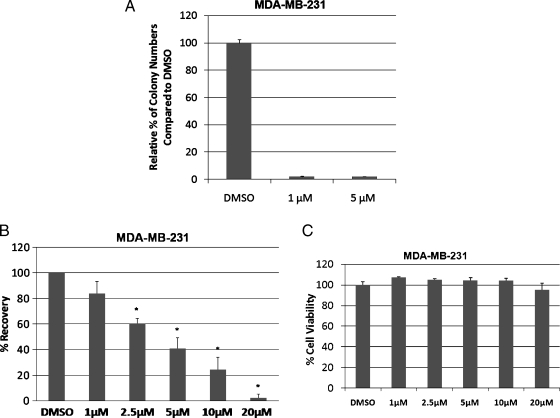
An indicator of transformation is the ability of cells to grow in the absence of substratum attachment [9]. Anchorage-independent growth is vitally important in the formation of the tumor [27]. The soft agar colony formation assay provides an assessment of tumor cells' susceptibility to a drug in an anchorage-independent environment. It is considered a more sensitive measure of toxicity, reflecting the efficacy of a drug, because it is analyzed when cells are in a proliferative state [28,29]. We examined the effect that LLL12's ability to inhibit STAT3 would have on colony formation of MDA-MB-231 cells in soft agar. Compared with the DMSO control, treatment with LLL12 led to a decrease of more than 95% in colony formation (Figure 6A). The results of this assay further confirm what was seen in the MTTassay: LLL12 is a potent inhibitor for cancer cell viability.
Figure 6.
(A) Colony formation of MDA-MB-231 cells in soft agar is inhibited by LLL12. The potency of LLL12 was assessed further in an anchorage-independent environment through a colony formation assay. Treatment with LLL12 greatly decreased the ability of MDA-MB-231 cells to form colonies in comparison to a DMSO control. (B) LLL12 inhibits cell migration in MDA-MB-231 breast cancer cells. A wound healing assay reveals that LLL12 has a significant impact on MDA-MB-231 cell migration. The ability of the cells to migrate is increasingly inhibited by an increase in dose of LLL12. Statistical significance (P < .05) relative to the DMSO control is designated by an asterisk. (C) A cell viability assay (MTT) was done to determine if the effect of LLL12 on MDA-MB-231 cell migration was due to its ability to inhibit cell proliferation. The time points of treatment (4 hours with LLL12) and incubation (additional 20 hours without LLL12) used in the wound healing assay was applied in the viability assay. The ability of LLL12 to inhibit cell migration does not seem to be due to an inhibition of cell proliferation.
LLL12 Inhibits Cell Migration in MDA-MB-231 Breast Cancer Cells
Cell migration is important in physiologic processes, such as wound healing and tumor metastasis. To assess the effect of LLL12 on cell migration, a wound healing assay was done. After the creation of a wound, cells were treated with various concentrations of LLL12. Treatment was removed after 4 hours. Cells were allowed to migrate into the denuded area for 24 hours. Treatment with LLL12 at a concentration of 2.5 µM or higher caused a significant decrease in cell migration (Figure 6B). The ability of LLL12 to inhibit cell migration may not be due to its ability to inhibit cell proliferation. MTT assay reveals that the dosages and time points used in the migration assay have minimal impact on cell viability (Figure 6C).
Quantitative Combinatorial Effects between LLL12 and Doxorubicin or Gemcitabine
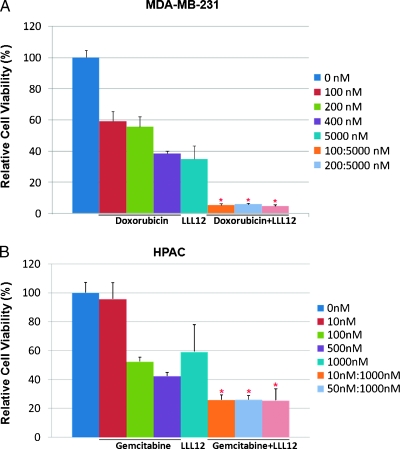
We evaluated the potential of LLL12 to act in a synergistic manner with doxorubicin or gemcitabine. MDA-MB-231 breast cancer cells were treated with doxorubicin or LLL12. HPAC pancreatic cancer cells were treated with gemcitabine or LLL12. The treatments lead to a dose-dependent decrease in cellular viability. To determine the combinatorial effects of the treatments, a constant concentration of LLL12 was used with varying concentrations of doxorubicin or gemcitabine. After 72 hours of treatment, a greater decrease in cell viability is seen in the combination treatments (Figure 7, A and B). The CI for each drug and concentration combination was calculated. The CI values of all the combinations of treatments were less than 1, indicating synergism between LLL12 and doxorubicin or gemcitabine. The synergistic effects seen with LLL12 and the currently used cancer therapeutic agents could prove useful in cancer therapy.
Figure 7.
The combinatorial effect of LLL12 and chemotherapy drugs, doxorubicin and gemcitabine. (A) MDA-MB-231 breast cancer cells were treated with LLL12 and doxorubicin individually and in combination. (B) HPAC pancreatic cancer cells were treated with LLL12 and gemcitabine individually and in combination. Cell viability was determined by MTT assay. A synergistic effect between LLL12 and doxorubicin or gemcitabine is indicated by an asterisk.
Effect of the Expression of Constitutively Active STAT3 Protein on LLL12-Mediated Inhibition
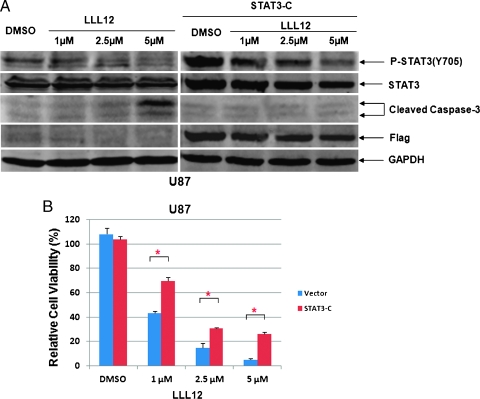
To confirm that LLL12 inhibition is indeed through the inhibition of STAT3, U87 glioblastoma cells were transfected with a constitutively active form of STAT3, STAT3-C (a murine STAT3). LLL12 (2.5 and 5 µM) inhibited STAT3 phosphorylation at Tyr705 and induced apoptosis that was indicated by caspase-3 cleavage in U87 cells (Figure 8A). However, LLL12 did not increase cleaved caspase-3 after the U87 cells were transfected with STAT3-C expression vector (Figure 8A). The expression of Flag-STAT3 was verified in STAT3-C-transfected U87 but not in nontransfected U87 cells (Figure 8A). The inhibition of cell viability of LLL12 in U87 cells was also partially reversed by the transfection with STAT3-C expression vector (Figure 8B). Our results show that STAT3-C can at least partially rescue LLL12-mediated inhibition. The fact that we did not observe a complete rescue by STAT3-C may be due to the transfection efficiency. Not 100% of U87 cells were transfected, and cells did not express that STAT3-C are still sensitive to LLL12 inhibition.
Figure 8.
The effect of STAT3-C expression on LLL12-mediated inhibition in U87 glioblastoma cells. Cells were transfected with a vector expressing constitutively active STAT3, STAT3-C for 24 hours, then treated with LLL12 for another 24 hours. (A) LLL12-induced caspase-3 cleavage was rescued in U87 cells when STAT3-C protein was expressed. (B) The inhibition of cell viability of LLL12 in U87 cells was also reduced in the presence of STAT3-C protein in MTT assay (*P < .05).
LLL12 Suppresses Tumor Growth in Mouse Model In Vivo
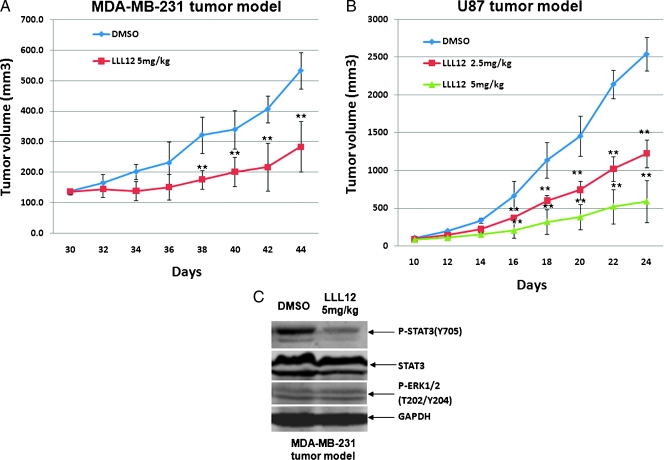
We further investigate whether LLL12 exhibits antitumor effect in vivo. Mouse xenograft experiments were performed by implanting MDA-MB-231 breast cancer cells or U87 glioblastoma cell line and then giving 2.5 and 5 mg/kg LLL12 or DMSO daily after tumor development. As shown in Figure 9, LLL12 significantly inhibited tumor growth compared with DMSO-treated controls in the MDAMB-231 (Figure 9A) and U87 xenografted mice (Figure 9B). STAT3 but not ERK1/2 phosphorylation of tumor tissue samples from these mice was also decreased by LLL12 (Figure 9C), suggesting that inhibition of STAT3 resulted in the suppression of tumor growth in mice.
Figure 9.
Effect of LLL12 on tumor growth in mouse xenografts with MDA-MB-231 breast cancer cells (A) or U87 glioblastoma cells (B). After tumor development, the mice were given daily intraperitoneal dosages of 2.5 to 5 mg/kg LLL12 or DMSO (*P < .05). STAT3 but not ERK1/2 phosphorylation of MDA-MB-231 tumor tissue samples from these mice was also decreased (C).
Compound Assessments for Drug-Likeness
Drug-likeness characteristics of LLL12 were evaluated using QikProp (Schrodinger LLC, Portland, OR). The absorption, distribution, metabolism, excretion, and toxicity of LLL12 were computed. Fifty “druglikeness” parameters were evaluated, including molecular weight, polarity, solubility, cell permeability, blood-brain barrier, Human ether-a-gogo related gene K+ blockage, human serum albumin binding, metabolic stability, and more. LLL12 showed decent “druglike” properties. Selected highlights are listed here: (1) possible in vivo metabolic reactions range only from 1 to 3; (2) composite log P values range from -2 to 2; (3) predicted IC50 values for HERG K+ channels are around -3, well above -5 for any concern; (4) predicted Caco-2 and Madin-Darby canine kidney cell permeability values are acceptable; (5) predicted brain-blood partition coefficients are above -3; (6) predicted index of binding to human serum albumin ranges from -0.5 to -0.8, well within the recommended range of -1.5 to 1.5; (7) predicted human oral absorption percentage is around 60%. Compared with existing drugs, LLL12 is 90% similar to sulfacytine and chlorthalidone. Overall, LLL12 is worthy of medicinal chemistry research effort for further optimization.
Discussion
STAT3, a member of the STAT family of transcription factors, is an oncogenic protein that is frequently activated in many types of cancer [6–8]. STAT3 activation results in the expression of downstream genes that promote cell proliferation and provide resistance to apoptosis, such as cyclin D1 and Bcl-2 respectively [7,9,10,16]. STAT3 has been classified as an oncogene because activated STAT3 can mediate oncogenic transformation in cultured cells and tumor formation in nude mice [10]. In contrast, STAT3-deficient fibroblasts were shown to be resistant to transformation by a variety of oncogenes [11,12]. Constitutive STAT3 signaling participates in oncogenesis by stimulating cell proliferation, mediating immune evasion, promoting angiogenesis, and conferring resistance to apoptosis induced by conventional therapies [6,16,18,30]. The crucial role of STAT3 in cancer progression and tumorigenesis has allowed STAT3 to emerge as a promising molecular target for treating cancer [6,8,9]. Previous methods aimed at blocking STAT3 have included the use of RNA interference, STAT3 antisense oligonucleotides, and dominant-negative STAT3 [14,19,31]. Although the stated approaches have been successful, limitations apply to their delivery and stability [32,33]. Peptide-based inhibitors of STAT3 have also been reported, but their use is confined by poor cell permeability and in vivo stability [34,35]. In addition, nonpeptide small-molecule inhibitors have been developed, which inhibit STAT3, including WP1066 [23], S3I-201 [24], STA-21 [36], Stattic [37], and SD-1029 [38].
We developed a novel small-molecule STAT3 inhibitor using structure-based design, named LLL12. We evaluated the inhibitory efficacy of LLL12 in breast and pancreatic cancer cells and glioblastoma cells with constitutively active STAT3. The results demonstrate that LLL12 is able to inhibit STAT3 phosphorylation. The inhibitory effects of LLL12 for STAT3 downstream target genes result in an inhibition of cell viability and the induction of apoptosis. Further, LLL12 is more potent than STAT3 inhibitors that have previously been reported, namely S3I-201 and WP1066, in the inhibition of cancer cell viability. We also show that LLL12 works synergistically with doxorubicin and gemcitabine in MDA-MB-231 breast and HPAC pancreatic cells, respectively. We also found that LLL12 did not induce detectable cleavage of caspase-3 and PARP in normal cells, in which STAT3 is not constitutively activated. In addition, the phosphorylation of other kinases, such as ERK1/2, mTOR, and Src, were not found to be inhibited by LLL12, suggesting the selectivity of LLL12 to these kinases. LLL12 inhibited STAT3 DNA binding activity, but STAT1 DNA binding activity was not inhibited further indicating its selectivity on STAT3 over STAT1. All these results indicate the potency and selectivity of LLL12 for STAT3, which is derived from its direct interaction with the pTyr705 binding site of STAT3 as predicted in our computer-generated docking model.
On the basis of our findings, treatment with LLL12 produces both antiproliferative and proapoptotic effects. LLL12 should be a suitable agent for targeting cancer cells with constitutively activated STAT3 owing to its ability to inhibit STAT3 phosphorylation and its potent growth-suppressive activity. Thus, LLL12 has the potential to be a therapeutic agent for the treatment of human breast and pancreatic cancer as well as glioblastoma with constitutively activated STAT3. We also observed that LLL12 is a potent inhibitor of STAT3 phosphorylation and DNA binding activity in cancer cells from other cancer types such as colorectal and liver cancers (data not shown). Therefore, LLL12 should be capable to have extended application to inhibit other types of cancer cells that have constitutively activated STAT3. LLL12 shows potential as a cancer therapeutic and is deserved for further exploration of its use as a potential agent in the treatment of cancer.
Supplementary Material
Acknowledgments
Chenglong Li acknowledges the CPU time allocated by the Ohio Super-computer Center.
Abbreviations
- DMEM
Dulbecco's modified Eagle medium
- DMSO
dimethyl sulfoxide
- PARP
poly (ADP-ribose) polymerase
- STAT
signal transducer and activator of transcription
- Tyr705
tyrosine 705
Footnotes
This research was partly funded by The Susan G. Komen Breast Cancer Foundation, The James S. McDonnell Foundation, The National Foundation for Cancer Research, and NIHR21 grant (R21CA133652-01) to Jiayuh Lin.
This article refers to supplementary materials, which are designated by Table W1 and Figure W1 and are available online at www.neoplasia.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. p 1. [DOI] [PubMed] [Google Scholar]
- 3.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. quiz 1 p following 516. [DOI] [PubMed] [Google Scholar]
- 4.Brantley EC, Benveniste EN. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res. 2008;6:675–684. doi: 10.1158/1541-7786.MCR-07-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 7.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 8.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 9.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 10.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 11.Inghirami G, Chiarle R, Simmons WJ, Piva R, Schlessinger K, Levy DE. New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle. 2005;4:1131–1133. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- 12.Schlessinger K, Levy DE. Malignant transformation but not normal cell growth depends on signal transducer and activator of transcription 3. Cancer Res. 2005;65:5828–5834. doi: 10.1158/0008-5472.CAN-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang HF, Murphy TF, Shu P, Barton AB, Barton BE. Stable expression of constitutively-activated STAT3 in benign prostatic epithelial cells changes their phenotype to that resembling malignant cells. Mol Cancer. 2005;4:2. doi: 10.1186/1476-4598-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 15.Faruqi T, Gomez D, Bustelo X, Bar-Sagi D, Reich N. Rac1 mediates STAT3 activation by autocrine IL-6. Proc Natl Acad Sci USA. 2001;98:9014–9019. doi: 10.1073/pnas.161281298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 17.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. NatMed. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 19.Ling X, Arlinghaus RB. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 2005;65:2532–2536. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 20.Xiong H, Zhang Z, Tian X, Sun D, Liang Q, Zhang Y, Lu R, Chen Y, Fang J. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huey R, Morris GM, Olson AJ, Goodsell DS. An empirical free energy force field with charge-based desolvation and intramolecular evaluation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 22.Khorgami MH. Preparation of sulfonamides by reduction of N-hydroxysulfonamides. Synthesis. 1972;10:574–575. [Google Scholar]
- 23.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 24.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 28.Hashimura T, Yoshida O. Soft agar colony formation of mouse epidermal cells during the early phase of two-stage chemically induced carcinogenesis. Jpn J Cancer Res. 1985;76:321–323. [PubMed] [Google Scholar]
- 29.Wylie PG, Bowen WP. Determination of cell colony formation in a high-content screening assay. Clin Lab Med. 2007;27:193–199. doi: 10.1016/j.cll.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Shen Y, Devgan G, Darnell JE, Jr, Bromberg JF. Constitutively activated Stat3 protects fibroblasts from serum withdrawal and UV-induced apoptosis and antagonizes the proapoptotic effects of activated Stat1. Proc Natl Acad Sci USA. 2001;98:1543–1548. doi: 10.1073/pnas.041588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton BE, Karras JG, Murphy TF, Barton A, Huang HF. Signal transducer and activator of transcription 3 (STAT3) activation in prostate cancer: direct STAT3 inhibition induces apoptosis in prostate cancer lines. Mol Cancer Ther. 2004;3:11–20. [PubMed] [Google Scholar]
- 32.Sledz CA, Williams BR. RNA interference in biology and disease. Blood. 2005;106:787–794. doi: 10.1182/blood-2004-12-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein CA. The experimental use of antisense oligonucleotides: a guide for the perplexed. J Clin Invest. 2001;108:641–644. doi: 10.1172/JCI13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton AD, Jove R. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 35.Burke TR, Jr, Yao ZJ, Liu DG, Voigt J, Gao Y. Phosphoryltyrosyl mimetics in the design of peptide-based signal transduction inhibitors. Biopolymers. 2001;60:32–44. doi: 10.1002/1097-0282(2001)60:1<32::AID-BIP1002>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 36.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a smallmolecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Duan Z, Bradner JE, Greenberg E, Levine R, Foster R, Mahoney J, Seiden MV. SD-1029 inhibits signal transducer and activator of transcription 3 nuclear translocation. Clin Cancer Res. 2006;12:6844–6852. doi: 10.1158/1078-0432.CCR-06-1330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.