Summary
Cancer initiating translocations such as those associated with lymphomas require the formation of paired DNA double-strand breaks (DSBs). Activation-induced cytidine deaminase (AID) produces widespread somatic mutation in mature B cells; however the extent of “off-target” DSB formation and its role in translocation-associated malignancy is unknown. Here we show that deregulated expression of AID causes widespread genome instability, which alone is insufficient to induce B cell lymphoma; transformation requires concomitant loss of the tumor suppressor p53. Mature B cell lymphomas arising as a result of deregulated AID expression are phenotypically diverse and harbor clonal reciprocal translocations involving a group of Immunoglobulin (Ig) and non-Ig genes which are direct targets of AID: this group includes miR-142, a previously unknown micro-RNA target which is translocated in human B cell malignancy. We conclude that AID produces DSBs throughout the genome, which can lead to lymphoma associated chromosome translocations in mature B cells.
Introduction
Chromosome translocations are characteristic features of several different forms of B cell cancers in mice and humans, including lymphomas and myelomas (Kuppers, 2005; Kuppers and Dalla-Favera, 2001; Potter, 2003). These karyotypic abnormalities are etiologically important in malignancy because they either deregulate oncogene expression, or create novel oncogenes by bringing together disparate transcription units (Rabbitts, 2009). Chromosome translocations are believed to be particularly frequent in mature B lymphocytes because, in addition to V(D)J rearrangements early in development, these cells undergo two forms of programmed DNA damage in their immunoglobulin (Ig) loci during immune responses: somatic hypermutation and class switch recombination (Peled et al., 2008; Stavnezer et al., 2008; Teng and Papavasiliou, 2007).
Somatic hypermutation introduces non-templated point mutations in antibody variable genes and class switch recombination replaces one constant region for another by a deletional recombination reaction (Peled et al., 2008; Stavnezer et al., 2008; Teng and Papavasiliou, 2007). Although the two reactions are mechanistically distinct, both are initiated by a single enzyme, AID, which is believed to act by deaminating cytosine to produce U:G mismatches in target DNA (Di Noia and Neuberger, 2007; Honjo et al., 2002; Muramatsu et al., 2000). Diverse and overlapping DNA repair pathways process these lesions to produce somatic mutations, or DNA double-strand breaks (DSB), which are obligate intermediates in the class switch reaction (Peled et al., 2008). These same DSBs can also become substrates for chromosome translocations (Jankovic et al., 2007; Ramiro et al., 2007; Ramiro et al., 2006b). However, a single DSB at Ig alone is not sufficient for translocation (Robbiani et al., 2008). To account for the DSBs in non-Ig genes it has been proposed that the enzymes that create the breaks in Ig loci, RAG1/2 and AID, can also cause damage in non-Ig genes (Lieber et al., 2008; Tsai et al., 2008). Consistent with this idea we found that the oncogene c-myc is a target of AID, although the frequency of DSB formation at c-myc is significantly lower than at IgH (Robbiani et al., 2008).
Burkitt’s lymphoma, diffuse large B cell lymphoma, and multiple myeloma represent different types of mature B cells that carry clonal translocations and may have expressed AID (Kuppers, 2005; Shaffer et al., 2002). A role for AID in inducing translocations in these cancers has been suggested by studies of activated B cells in vitro and plasmacytosis in IL-6 transgenic and pristane-treated mice, all of which develop AID dependent c-myc/IgH translocations (Dorsett et al., 2007; Ramiro et al., 2006a; Ramiro et al., 2004; Unniraman et al., 2004). In addition, AID expression accelerates the rate of tumor development in Bcl6 transgenic mice (Pasqualucci et al., 2008). Finally, transgenic AID produces mutations in c-myc and T cell receptor genes as well as epithelial and T cell malignancies (Okazaki et al., 2003).
Curiously, however, deregulated AID does not cause malignancy or translocation-associated cancer in B cells (Muto et al., 2006; Okazaki et al., 2003; Shen et al., 2008). In addition, despite its obligate role in c-myc/IgH translocation, AID is not required for the development of plasmacytosis or plasmacytoma in IL-6 transgenic or pristane-treated mice, respectively (Kovalchuk et al., 2007; Ramiro et al., 2004). Finally, most human B cell lymphoma associated translocations do not involve c-myc and many do not involve Ig genes (Kuppers, 2005). Thus, the issue of whether AID can induce a sufficiently varied genomic damage to account for Ig and non-Ig gene translocation associated B lymphocyte malignancy in vivo remains to be determined.
Results
Increased switching, mutation and c-myc/IgH translocation in AID transgenic mice
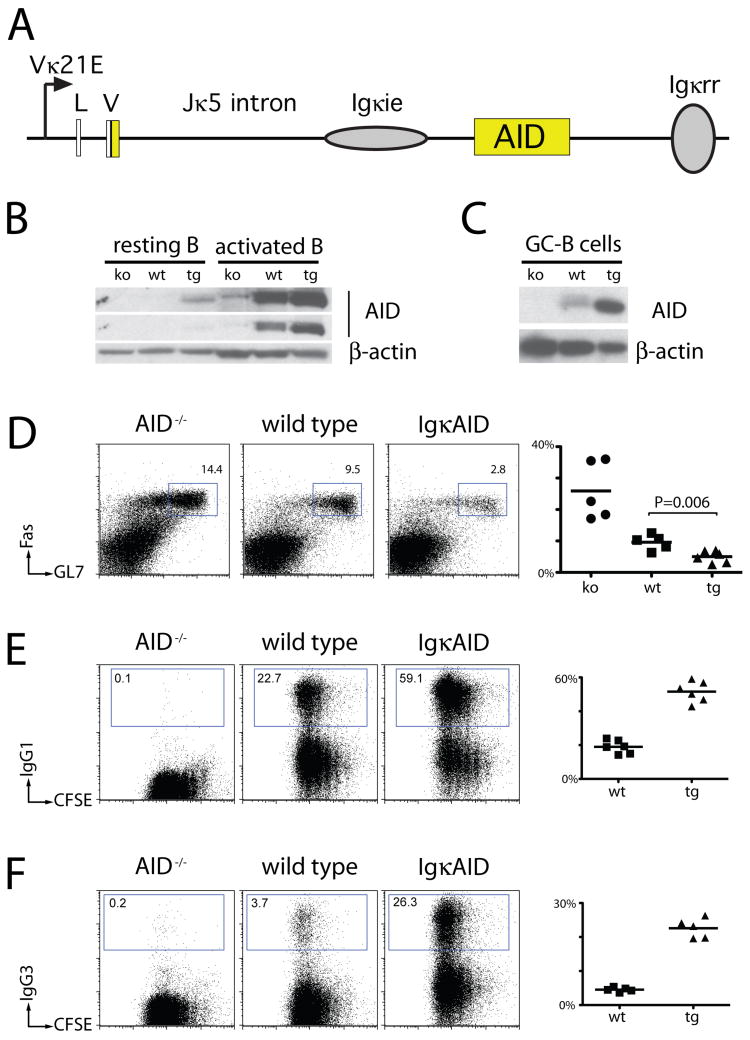
To determine whether deregulated AID expression can destabilize the genome and induce B cell malignancy in vivo, we produced transgenic mice that express AID under the control of Igκ regulatory elements (IgκAID, Figure 1A). AID is expressed at low levels in resting B cells in IgκAID mice, and increases in activated and germinal center B cells (Figure 1B and 1C and Supplemental Figure S1). Nevertheless, with the exception of the expected decrease in germinal center cells (Figure 1D and (Dorsett et al., 2008; Muramatsu et al., 2000)), B cell development and distribution is normal in IgκAID mice and serum Ig isotypes are not increased (Supplemental Figures S2 and S3). Consistent with the increase in AID expression in stimulated B cells in vitro, IgκAID B cells switched at higher frequency to IgG1 and IgG3 (Figure 1E and 1F). Thus, although deregulated AID expression does not significantly alter B cell development in vivo, it does lead to increased levels of class switch recombination in vitro. This effect is consistent with the observation that AID concentrations are limiting for this reaction (Dorsett et al., 2008; McBride et al., 2006; Takizawa et al., 2008).
Figure 1. Increased class switching in Igκ AID B cells.
(A) Schematic representation of the IgκAID transgene. AID cDNA (yellow) is embedded in Igκ regulatory elements (Vκ21E promoter, non-coding leader (L) and V gene exons, intronic sequence with enhancer (ie), and 3′ regulatory region (rr)). (B) and (C) Western Blot for AID protein in IgκAID B cells. (B) 1 million cells of the indicated genotypes were assayed in each lane. Top and middle are different exposures of the same blot. (C) 0.5 million sorted germinal center (GC) B cells were assayed in each lane. (D) Flow cytometric analysis of Peyer’s patch germinal center B cells from matched 6–10 week old mice. Graph (right) shows a summary of two independent experiments with 2–3 mice each, the bar indicates the mean value for each group (25.9% ko, 9.6% wt, 5.0% tg), P value was calculated by unpaired T-test. (E) and (F) Flow cytometric analysis for class switching to IgG1 and IgG3 and cell division. Graph (right) shows a summary of two independent experiments with 2–3 mice each, the bar indicates the mean value for each group (IgG1: 18.9% wt, 51.6% tg; IgG3: 3.8% wt, 22.6% tg).
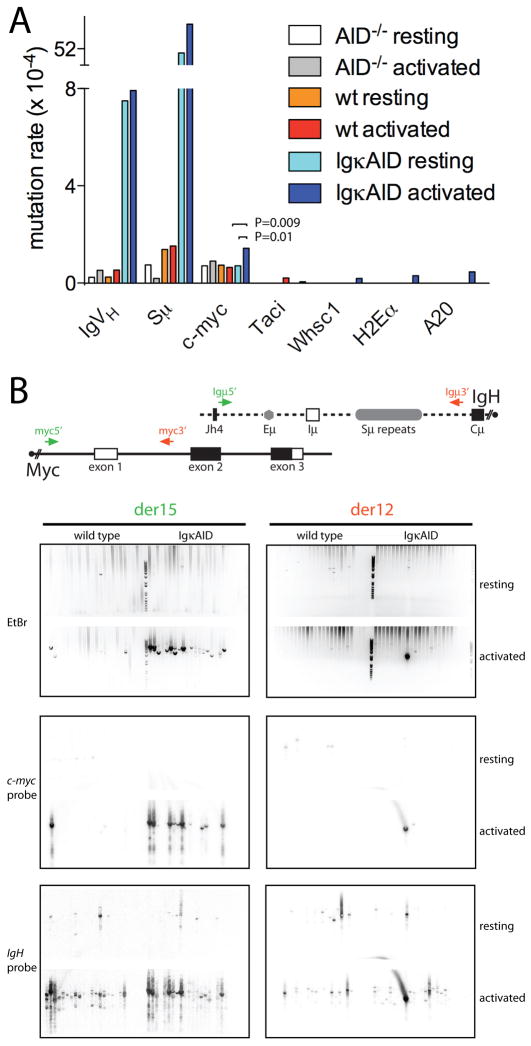
To determine whether transgenic AID expression might alter its targeting, we examined the extent of somatic mutation in documented AID target (IgVH, Sμ, c-myc) and non-target genes (Taci, Whsc1, H2Eα, A20), all of which are expressed in stimulated B cells (Supplemental Figure S4 and (Liu et al., 2008)). IgVH mutation was measured by combining the B1-8hi IgVH knock-in allele (Casellas et al., 2001) with IgκAID (IgκAID/B1-8hi) or AID−/− controls (AID−/−/B1-8hi). Consistent with constitutive AID expression in naïve B cells, resting IgκAID/B1-8hi B cells showed high levels of mutation in IgVH and Sμ, and only a modest increase upon in vitro activation with LPS and IL-4 (Figure 2A). IgVH and Sμ mutation was not significantly detectable in controls, but was found at rates of 7.5×10−4 and 51×10−4, respectively in naïve AID transgenic B cells. This rate of mutation is one order of magnitude lower than the rates normally found in germinal center B cells (Liu et al., 2008; McKean et al., 1984). In contrast, the increased levels of AID did not result in hypermutation in any of the non-target genes tested (Figure 2A). Also consistent with a physiologic pattern of target selection, c-myc, which has a propensity to error-free DNA repair (Liu et al., 2008), shows only a modest increase in mutation rate in activated IgκAID B cells (1.43×10−4 vs. 0.64×10−4). We conclude that deregulated expression of AID induces hypermutation in resting B cells at levels close to those normally found in germinal center cells; however the specificity of target selection is maintained since there is no significant DNA damage in non-target genes.
Figure 2. Somatic mutations and c-myc/IgH translocations.
(A) Frequency of somatic mutations in indicated loci of resting and LPS and IL-4 stimulated B cells. (B) C-myc/IgH translocations in resting B cells or B cells activated with LPS and IL-4 for four days. Top: Schematic representation of the Myc and IgH alleles with the PCR primers for detecting der15 and der12 c-myc/IgH translocations. Bottom: ethidium bromide (EtBr) stained gel with PCR bands indicative of translocations was blotted and probed for c-myc and IgH, as indicated, to verify translocations. Translocation frequency: wild type 0.4×10−6 (der15) and 0.1×10−6 (der12); IgκAID 4.6×10−6 (der15) and 1.3×10−6 (der12). One of two independent experiments is shown.
Even though c-myc is a bona fide target of AID, DSBs at c-myc, and c-myc/IgH translocations are difficult to detect under physiological conditions (Robbiani et al., 2008). Despite constitutive AID expression in IgκAID B cells, we found no c-myc/IgH translocations in resting transgenic B cells (Figure 2B). In contrast, IgκAID B cells stimulated with LPS and IL-4 showed an increase in translocation frequency when compared to controls (Figure 2B). Therefore, transgenic B cell expression increases the frequency of c-myc/IgH translocations in activated B cells.
B cell lymphoma
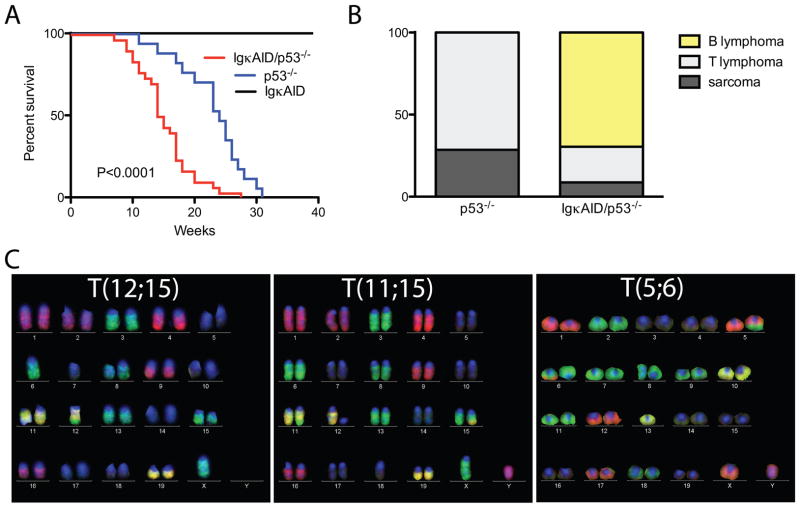
Despite high levels of c-myc/IgH translocations in stimulated B cells expressing deregulated AID, B cell lymphomas did not develop in IgκAID or in three other previously reported strains of AID transgenic mice (Figure 3A and (Muto et al., 2006; Okazaki et al., 2003; Shen et al., 2008)). All eight of the IgκAID mice survived beyond 40 weeks (Figure 3A). One potential explanation for this unexpected result is that B cells are protected from AID mediated DNA damage induced transformation by gatekeepers such as p53, which trigger cell cycle arrest or apoptosis (Bassing et al., 2003; Green and Kroemer, 2009; Kruse and Gu, 2009; Lowe et al., 2004; Ramiro et al., 2006a). Consistent with this idea, gatekeeper genes, or genes that control them such as Bcl6 (Phan and Dalla-Favera, 2004), are frequently deleted or mutated in mature human B cell lymphomas (Kuppers, 2005).
Figure 3. IgκAID/p53−/− mice succumb to B cell lymphomas with reciprocal translocations.
(A) Survival curve for n=8 IgκAID, n=17 p53−/− and n=30 IgκAID/p53−/− mice. The P value refers to the difference between IgκAID/p53−/− and p53−/− (log-Rank test). The median survival for IgκAID/p53−/− is 14.5 weeks, for p53−/− it is 24 weeks. (B) Phenotype of tumors at necropsy (n=7 p53−/− and n=23 IgκAID/p53−/−). (C) Representative M-FISH images of reciprocal translocations found in B cell lymphomas of IgκAID/p53−/− mice.
To determine whether p53 is protective against AID-induced B cell lymphomas in vivo we produced IgκAID/p53−/− mice. In contrast to IgκAID mice, all 30 of the IgκAID/p53−/− mice died within 28 weeks and they succumbed more rapidly to malignancy than p53−/− controls (P<0.0001, Figure 3A and 3B). In addition, tumors that developed in IgκAID/p53−/− mice differed from those in the p53−/− controls: the former were predominantly mature B cell lymphomas, whereas the p53−/− controls developed thymic lymphomas and sarcomas (Figure 3B and (Jacks et al., 1994)). The mature B cell lymphomas in IgκAID/p53−/− mice represent a broad spectrum of the different stages of B cell development; from naïve B cells that express high levels of IgM and little or no IgD to plasmacytomas that express CD138 and no surface Ig (Table 1 and Supplemental Figure S5). We conclude that deregulated AID expression in a p53 deficient background leads to the emergence of a broad spectrum of mature B cell lymphomas.
Table 1.
Karyotype and phenotype of B lymphomas in IgkAID/p53−/−.
| Tumor ID | Clonal translocation | Additional changes in at least one metaphase | Tumor phenotype |
|---|---|---|---|
| #1461 | T(11;15) | Dic(13;13) | CD19+ IgM+ IgD− |
| #1535 | T(11;15) | −12, +13, −19 | CD19+ IgMdim IgD+/− |
| #1536 | T(5;6) | −13, T(6;19) | CD19+ IgM+ IgDdim |
| #1550 | T(11;15) | −13, −16, −17, −18, T(16;17), T(16;Y) | CD19+ IgM+ IgDdim/+ CD138+/− |
| #1561 | T(11;15) | +3,−12, −Y, T(12;16), T(12;17) | CD19+ IgM+ IgD− CD138dim |
| #1583 | T(11;15) | −12, −13, −14 | CD19+ IgM+/++ IgD+/− CD138dim |
| #1584 | T(11;15) | −12, −18 | CD19+ IgM+/++ IgD+/− CD138dim |
| #1589 | none | −5, −14, −18, −X, T(3;4), T(3;11), T(3;19) | CD19+ IgM− IgD− Igκ− CD138+/− |
| #1592 | T(11;15) | −7, +16 | CD19+ IgM+ IgD+/− CD138+/− |
| #1593 | T(12;15) | −7 | CD138++ CD19+ IgM− IgD− Igκ− |
| #1594 | T(11;15) T(12;17) |
T(2;11), T(9;15), T(11;16), T(13;5) | CD138++ CD19+ IgM− IgD− Igκ− |
| #1595 | T(11;15) | −12, −13, −19, T(13;16) | CD19+ IgM+ IgD− CD138+/− |
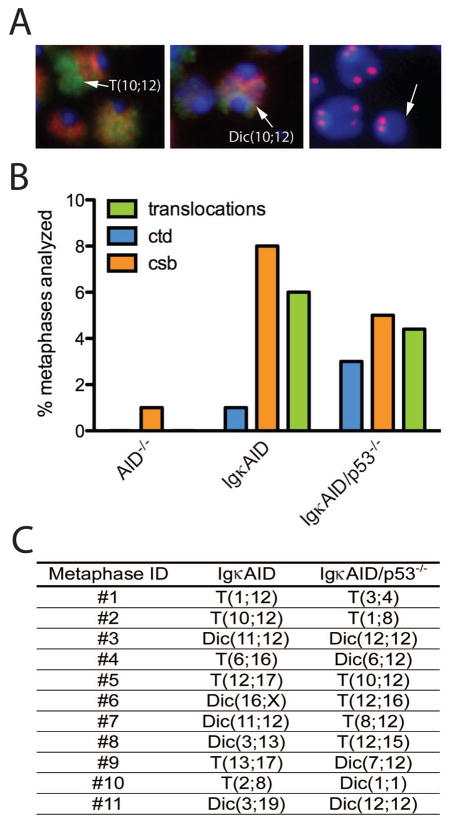
To determine whether IgκAID/p53−/− lymphomas were associated with chromosome translocations we performed multicolor-FISH (M-FISH) analysis on tumor metaphase spreads. We found that 11 out of the 12 IgκAID/p53−/− B cell lymphomas tested contained a clonal reciprocal translocation. In all cases additional non-clonal chromosome aberrations were present (Figure 2C and Table 1). In contrast, clonal reciprocal translocations were not observed in the p53−/− control T cell lymphomas (data not shown and (Liao et al., 1998)). Thus, in the absence of p53, deregulated AID can produce B cell lymphomas that harbor clonal reciprocal chromosome translocations, similar to those that are characteristic of human B cell lymphomas.
Tumor associated translocations induced by AID in vivo
Nine of the eleven IgκAID/p53−/− B cell lymphomas analyzed by M-FISH showed a clonal T(11;15) translocation; the other two exhibited a T(12;15), or T(5;6) translocation, with a variety of additional clonal and non-clonal chromosomal abnormalities (Table 1). The tumor with T(12;15) was a typical mouse plasmacytoma that expressed CD19 and high levels of CD138 but not surface Ig (Table 1 and Supplemental Figure S5). PCR analysis was used to determine that the translocation involved c-myc and IgH and sequence analysis revealed that the breakpoint on c-myc was in the promoter region, and that the breakpoint at the IgH locus was in the IgH switch μ region (Supplemental Table 1). Such reciprocal translocation is similar to those observed in human Burkitt’s lymphomas and mouse plasmacytomas, and are consistent with IgH and c-myc being targets of AID activity (Robbiani et al., 2008).
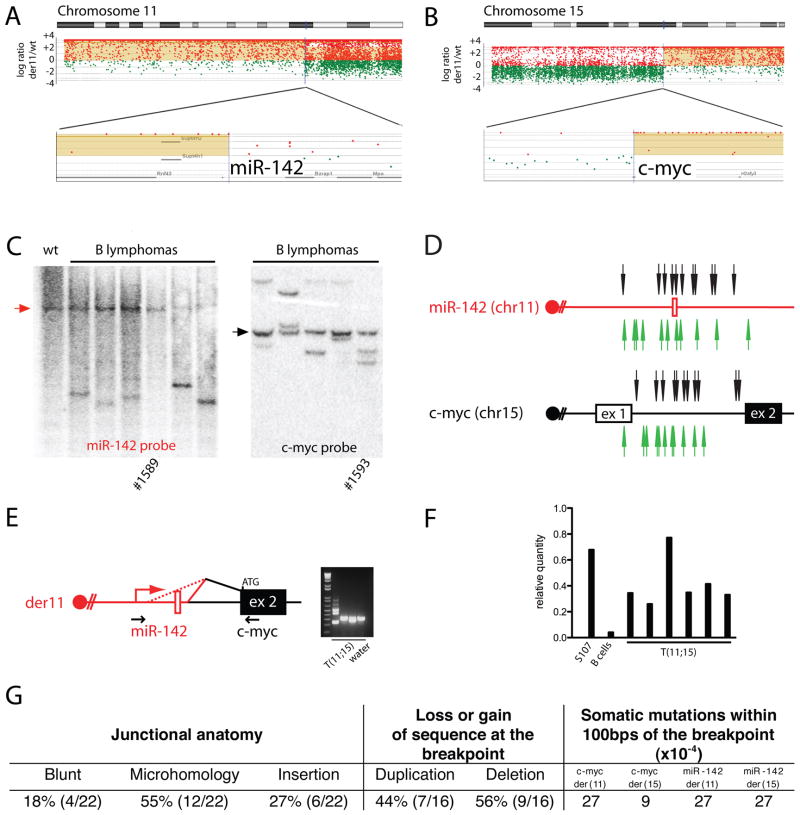
To identify the translocation breakpoints in tumors containing T(11;15) and T(5;6), we performed high resolution array painting. Because the translocations in IgκAID/p53−/− B cell lymphomas appeared to be balanced as determined by M-FISH (Figure 3C), we sorted derivative chromosomes from the tumor samples to compare with normal reference DNA (Gribble et al., 2007). Translocation breakpoints and copy number changes were revealed as discontinuities (high-low or low-high ratio) in the hybridization ratio of sorted derivative chromosomes compared to unsorted reference DNA. We found that the breakpoints in the T(11;15) translocation were in the region of micro-RNA 142 (miR-142) and c-myc and in T(5;6) at Anxa4 and Wdfy3 (Figure 4A and 4B and Supplemental Figure S6). The apposition of miR-142 on chr11 with c-myc on chr15 was confirmed by Southern blotting (Figure 4C). The high resolution mapping by array painting allowed for the design of PCR primers to characterize fourteen T(11;15) translocations molecularly. The miR-142 breakpoints were clustered in 1.8kbps surrounding the micro-RNA, while the breakpoints at c-myc were in a 1.5kbps region centered in intron 1 (Figure 4D). The resulting miR-142/c-myc fusion genes on der11 chromosome contained the miR-142 promoter and the coding region of c-myc. These genes were associated with the production of fusion transcripts between miR-142 and c-myc (der11), leading to the over-expression of c-myc at levels similar to those found in the S107 plasmacytoma, which carries a c-myc/IgH translocation (Figure 4E and 4F). Importantly, a closely related clonal translocation between miR-142 and c-myc has been reported in human B cell leukemia (Gauwerky et al., 1989). We conclude that the T(11;15) translocation found in the vast majority of IgκAID/p53−/− B cell cancers results in deregulated expression of c-myc by juxtaposition to regulatory elements from miR-142, one of the most highly expressed micro-RNAs in hemaetopoetic cells (Chen et al., 2004; Landgraf et al., 2007). Based on the B cell developmental stages as well as karyotype and high resolution analysis of the T(12;15) and T(11;15) translocations, it appears that the IgκAID/p53−/− mouse provides a relevant model for studying the development of human B cell lymphoma in vivo.
Figure 4. Identification of the T(11;15) translocation partners.
(A) and (B) aCGH analysis of sorted chromosomes identifies the breakpoint in proximity of miR-142 (Chr11) and c-myc (Chr15). (C) Southern blotting with c-myc and miR142 probes. Unless otherwise noted, tumors have T(11;15) by M-FISH. Arrows point to the unrearranged germline bands. (D) Map of translocation breakpoints in T(11;15) B cell lymphomas from IgκAID/p53−/− mice (n=14: in addition to the 9 T(11;15) identified by M-FISH, an additional 5 were identified by PCR). For some tumors the breakpoint could be identified in one only derivative (see Supplemental Table S1). Arrows point to translocation breakpoints for der11 (black) or der15 (green). (E) RT-PCR for hybrid transcripts between miR-142 and c-myc. (F) qPCR for c-myc expression in T(11;15) tumors. S107 is a plasmacytoma cell line with c-myc/IgH translocation (positive control). Negative control is activated B cells. (G) Features of the T(11;15) breakpoints.
AID might induce lymphoma-associated translocations by producing direct genomic damage or indirectly by compromising the genes that maintain genomic stability (Jankovic et al., 2007). The T(11;15) translocation junction sequences revealed either blunt end joining or small amounts of microhomology (Figure 4G and Supplemental Table S1). In eight cases, we were able to characterize both derivative chromosomes. Surprisingly, six of these showed duplications in one or both of the translocated genes. In agreement with this observation, duplications were also present in the lymphomas with T(12;15) and T(5;6) translocation. In this setting, duplication could only be produced by filling in a break created by two staggered single-strand nicks, one on each DNA strand. This type of DSB is consistent with a staggered pair of lesions introduced by AID (Figure 4G, Supplemental Table S2, and Supplemental Figure S7).
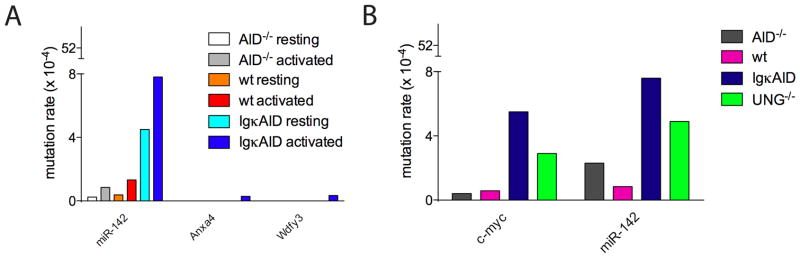
To determine if any of the translocated genes are direct targets of AID, we obtained DNA sequences from primary resting and activated B cells (Figure 5A). We found a high rate of mutation in miR-142 in resting (4.5×10−4) and activated (7.8×10−4) IgκAID B cells (P=0.0004 and P=0.0007, respectively, Figure 5A and Supplemental Figure S8), but not in Anxa4 or Wdfy3. Hypermutation indicative of AID activity at miR-142 was also detected in the region surrounding the translocation breakpoints as well as, more significantly, in the non-translocated allele of T(11;15) lymphomas (Figure 4G, Supplemental Table 1 and Supplemental Figure S9). Finally, we confirmed the in vitro results by analyzing c-myc and miR-142 mutations in germinal center B cells. Both c-myc and miR-142 were hypermutated in IgκAID germinal center B cells at levels similar to those found in activated B cells in vitro (Figure 5B). In addition, both c-myc and miR-142 mutations were enhanced in the absence of UNG in cells with endogenous levels of AID (Figure 5B). Therefore, miR-142 resembles c-myc in that AID-induced lesions have a propensity to error free repair, which is UNG dependent (Figure 5B and (Liu et al., 2008)). Thus, the majority of the B cell lymphomas induced by deregulated AID expression involves direct AID target genes (a total of 15 T(12;15) and T(11;15)) while a minority (1 T(5;6)) may result indirectly from other genome destabilizing mutations.
Figure 5. miR-142 is an AID target.
(A) Frequency of somatic mutations in miR-142, Anxa4 and Wdfy3 in resting and LPS and IL-4 stimulated B cells. (B) Frequency of somatic mutations in miR-142 and c-myc in purified germinal center B cells from AID−/−, wild type, IgκAID, and UNG−/− mice.
Genomic DNA damage
To examine the spectrum of genomic DNA damage induced by AID deregulation in primary non-transformed B cells, we measured chromosome translocations by M-FISH (Figure 6A and 6B). We found that 4.4% of all IgκAID/p53−/− B cells showed translocations in LPS, IL-4 and RP105 stimulated B cells (n= 250; Figure 6B). Metaphases containing translocations were rare in wild type B cells (3/500 metaphases, Supplemental Table S3 and (Callen et al., 2007; Ramiro et al., 2006a; Wang et al., 2009)). Interestingly, IgκAID, although tumor-free, showed a similar frequency of translocation to IgκAID/p53−/− (6%, n=500 metaphases; Figure 6B and Supplemental Table S3). Thus, p53 does not impact on the formation of translocations, and this factor must act downstream of translocation to prevent malignancy. Significantly, out of 500 metaphases from AID−/− mice none showed chromosome aberrations, indicating that the translocations in IgκAID mice are completely AID-dependent (Figure 6B). Every chromosome (except Chr5, Chr14, Chr18 and ChrY) was found translocated in at least one metaphase from IgκAID B cells. The distribution of translocations was similar between IgκAID/p53−/− and IgκAID mice (Figure 6C). Remarkably some of the chromosomes were found at an extremely high frequency including Chr12 (21/45 translocations), Chr16 (9/45 translocations), Chr 11 and Chr13 (both 7/45 translocations)(Figure 6C and Supplemental Table S3). We conclude that in addition to c-myc/IgH translocations, AID induces a large number of translocations in B cells and that loss of p53 does not increase the frequency of translocations.
Figure 6. AID-dependent chromosome instability in stimulated primary B cells.
(A) Representative M-FISH and telomere FISH images of B cells activated in vitro for 4 days with LPS, IL-4 and RP105. The arrows indicate (left-to-right) a translocation, a dicentric chromosome and a chromosome break. (B) Genomic instability and translocations in IgκAID primary B cells. Frequencies of ctd (chromatid breaks) and csb (chromosome breaks) were determined by analysis of metaphases hybridized with a telomere probe and counterstained with DAPI (n=100). Frequency of translocations (including dicentrics) was determined by analysis of M-FISH images (n=250). (C) Table of chromosome aberrations detected in IgκAID and IgκAID/p53−/− primary B cells. See also Supplemental Table S3.
Translocations are products of DNA breaks. To directly determine the frequency of DNA breaks we examined DAPI-stained metaphase spreads that were hybridized with a telomere-repeat specific peptide nucleic acid probe marking chromosome ends (Figure 6A and 6B and (Callen et al., 2007)). The level of chromosome and chromatid breaks was similar in IgκAID (9%) and IgκAID/p53−/− (8%) B cells, and this level of DNA damage was 8–9 fold higher than found in AID−/− mice (Figure 6A and 6B). Thus, deregulated AID expression produces widespread DNA damage which is independent of p53 and which promotes chromosomal translocations in mature B cells.
Discussion
Lymphoid malignancies account for 5% of all human cancers; the majority of these involve mature B cells at various stages of development. Chromosome translocations between Ig and non-Ig genes feature prominently in these cancers; however, they can also occur between two non-Ig genes. For example, Burkitt’s lymphomas typically contain c-myc/IgH translocations; whereas in MALT lymphomas the translocation commonly involves API2 and MALT (Inagaki, 2007). Multiple origins have been suggested for the paired DNA lesions that are obligate intermediates in lymphoid translocations, including the RAG1/2 V(D)J recombinase (Bassing et al., 2002; Callen et al., 2009; Dudley et al., 2005; Schatz, 2004; Schlissel et al., 2006; Tonegawa, 1983), AID (Ramiro et al., 2007; Ramiro et al., 2006a; Ramiro et al., 2004), reactive oxygen species, fragile sites and combinations of the above (Callen et al., 2007; Greaves and Wiemels, 2003; Lieber et al., 2008; Lieber et al., 2006; Tsai et al., 2008; Wang et al., 2009). However, none of these is directly demonstrated to produce the necessary initiating lesions on multiple chromosomes in lymphoid cells. Our experiments show that transgenic over-expression of AID in mice produces DSBs on most chromosomes and indicate that expression of this enzyme may be sufficient to account for many of the lesions that lead to mature B cell lymphoma.
AID is known to produce translocations between c-myc and IgH in B cells in vitro as well as in IL-6 transgenic plasmacytes and pristane-induced plasmacytomas (Dorsett et al., 2007; Ramiro et al., 2006a; Ramiro et al., 2004; Unniraman et al., 2004). However, a number of laboratories have shown that deregulated AID expression is not sufficient to induce B cell lymphoma in vivo (Muto et al., 2006; Okazaki et al., 2003; Shen et al., 2008). Our data resolve this apparent disparity and reveal that AID is indeed a cancer susceptibility gene that causes a broad spectrum of B cell malignancies. In contrast to gatekeeper genes that control cell proliferation and cell death, or caretakers that regulate DNA repair, AID is a tumor promoter that destabilizes the genome by inducing DNA breaks and chromosome translocations.
AID creates DNA lesions by cytosine deamination (Petersen-Mahrt et al., 2002). Like other DSBs, AID breaks are recognized by proteins that mediate the DNA damage response including H2AX, Nbs1 and 53BP1 (Manis et al., 2004; Petersen et al., 2001; Reina-San-Martin et al., 2005; Ward et al., 2004), and ATM (Difilippantonio et al. 2005; Reina-San-Martin et al., 2004), which in turn phosphorylates and recruits a canonical cascade of additional factors to the breaks (Jankovic et al., 2007). Although AID’s activity is focused on antibody genes, where it produces high rates of mutation, its specificity is not absolute and its mechanism of targeting is still to be defined. The lack of absolute specificity results in low frequency off-target mutation of oncogenes such as Bcl6 and c-myc (Pasqualucci et al., 1998; Shen et al., 1998). However, the rate of AID-induced mutations in genes like c-myc is reduced by error free repair by a yet to be determined UNG-dependent pathway resulting in mutation rates that are difficult to detect (Liu et al., 2008). Therefore the complete list of genes that can be mutated by AID is not known.
Even less is understood about the extent to which the genome might be susceptible to AID-induced DSBs. Our experiments demonstrate that although deregulating AID expression is not sufficient to overcome its targeting specificity, it induces a wide spectrum of chromosome aberrations, including reciprocal and non-reciprocal translocations, dicentric chromosomes as well as chromosome deletions in primary B cells.
AID deregulation leads to increased somatic mutation and increases genomic instability, but is not sufficient to cause malignant transformation. Conversely, loss of p53 was required for malignancy but it did not significantly increase the level of translocation or DSBs. Thus, the presence of p53 did not prevent translocation per se, but p53 was required for transformation of cells bearing translocations. This would be in keeping with p53’s ability to respond to oncogenic stress and induce the death of cells that over-express oncogenes (Green and Kroemer, 2009; Hoffman and Liebermann, 2008; Kruse and Gu, 2009; Lowe et al., 2004). For example, p53 dependent cell death is triggered when c-myc is deregulated by the Ig enhancer in the c-myc/IgH translocation or by the miR-142 promoter in the c-myc/miR-142 translocation. Of note, p53 or its regulators are frequently mutated in human B cell malignancies including diffuse large-B cell lymphoma, Burkitt’s lymphoma and chronic lymphocytic leukemia (reviewed in (Kuppers, 2005)). Finally, finding that there was no difference in the number of DSBs or chromosome abnormalities in the presence or absence of p53 indicates that the majority of the damage created by deregulated AID involves genes that do not activate p53. Based on our observations in mice, we speculate that p53’s role in human lymphomas is to eliminate cells bearing AID induced translocations that activate oncogenes.
Human B cell lymphomas can carry reciprocal translocations between two non-Ig genes; however, whether AID produces the lesions that lead to the malignancies associated with this class of translocations was not known. We found one example of a translocation where neither of the two aberrantly joined non-Ig genes was an AID target as measured by somatic hypermutation (Anxa4/Wdfy3). However, the remaining non-Ig translocations in the B cell malignancies that arose in IgκAID/p53−/− mice (n=14) were associated with c-myc/miR-142 translocation, which is also found in human B cell leukemia (Gauwerky et al., 1989). Both of these genes are AID targets. Consistent with bilateral initiation of these non-Ig translocations by AID, most showed junctional duplications that can only arise when the initiating lesions are staggered nicks on two DNA strands including the Anxa4/Wdfy3 translocation. Therefore, in addition to translocations involving Ig, AID can also induce cancer-associated translocations between two non-Ig genes. It is important to note that somatic mutation as currently measured is not a sensitive indicator of whether or not a gene is an AID target simply because the rates of mutation sometimes approach the error rates of the polymerases used for PCR amplification. Thus, the lesions that lead to Anxa4/Wdfy3 translocation, which show junctional duplication but no evidence of somatic mutation may still be due to AID. Given its destabilizing effects on the genome and oncogenic potential in vivo, it is not surprising that the expression of this enzyme and its residence in the nucleus are tightly regulated by a combination of cytoplasmic retention signals (Patenaude et al., 2009), nuclear export (McBride et al., 2004), phosphorylation (Cheng et al., 2009; McBride et al., 2006; McBride et al., 2008), micro-RNAs (Dorsett et al., 2008; Teng et al., 2008) and ubiquitylation (Aoufouchi et al., 2008).
In conclusion, AID is a potent endogenous tumor promoter as it mutates oncogenes like Bcl6, and creates DSBs in Ig and non-Ig genes that can serve as substrates for chromosome translocation. These lesions may account for a significant fraction of the mature B cell lymphomas that frequently arise in humans.
Experimental Procedures
Mice
The IgκAID transgene is based on VkMYC (Robbiani et al., 2005) and includes the coding sequence of mouse AID (split over two exons) embedded in regulatory elements of the κ light chain gene. The plasmid backbone was removed with MluI and NotI prior to pronuclear injection into C57BL/6 x (C57BL/6 x CBA) oocytes. Transgenic males were backcrossed to C57BL/6 for at least ten generations. AID−/−, B1-8hi and p53−/− mice were backcrossed to C57BL/6 (Casellas et al., 2001; Jacks et al., 1994; Muramatsu et al., 2000). UNG−/− mice were previously described (Endres et al., 2004). All experiments were performed in accordance with protocols approved by the Rockefeller University and National Institutes of Health (NIH) Institutional Animal Care and Use Committee.
B cell cultures
Resting B lymphocytes were isolated from mouse spleens by immunomagnetic depletion with anti-CD43 MicroBeads (Miltenyi Biotech), and cultured at 0.5x106 cells/ml in R10 medium as previously described (Robbiani et al., 2008). For class switch recombination to IgG3, cells were stimulated with LPS (25μg/ml, Sigma) and RP105 (0.5 μg/ml, Pharmingen). For class switch recombination to IgG1, IL-4 (5 ng/ml, Sigma) was supplemented in addition to LPS and RP105. Cells were analyzed after three days of culture.
Flow cytometry and cell sorting
For flow cytometric analysis, single-cell suspensions were stained with the indicated fluorochrome-conjugated antibodies (all from Pharmingen). CFSE-labeling for cell division was at 37C for ten minutes in 5μM carboxyfluorescein succinimidyl ester (CFSE, Molecular Probes). Data were acquired on a FACSCalibur or LSRII instrument (Becton Dickinson) and analyzed with FlowJo software.
Lymphoid populations were sorted to >95% purity with a FACSVantage SE with DIVA option or FACSAria instruments (Becton Dickinson). Follicular B cells (CD19+CD23+CD21+) from naïve spleens and bone marrow pre-B cells (B220+CD43low/intIgM−IgD−) were enriched with anti-CD19 microbeads (Miltenyi Biotech). Splenic germinal center B cells (CD19+GL7+CD95+) from P. chabaudi infected mice (day 14) were enriched with anti-CD19 microbeads prior to sorting.
Germinal center induction
Plasmodium chabaudi chabaudi clone AS (MRA 429) was obtained through the MR4 (Peters and Robinson, 2000). Parasites were maintained as frozen stocks and passaged in mice as described previously (Meding et al., 1990). For experiments, mice were injected intra-peritoneally with 105 infected red blood cells. Infections were monitored by microscopic examination of Giemsa-stained thin blood smears.
Western Blot
AID expression was analyzed as previously described (McBride et al., 2008). Lymphoid cells were purified as described above from AID−/−, wild type or IgκAID mice. Activated splenic B cells were stimulated in vitro with LPS and IL-4 for four days.
PCR assay for translocations
DNA was extracted from resting splenic B cells or B cells that were stimulated in vitro for four days with LPS and IL-4. PCR reactions were performed on genomic DNA from 105 cells, using the Expand Long Template PCR System (Roche). Nested reactions were performed as detailed and confirmed by Southern Blot as described (Robbiani et al., 2008).
Mutational analysis
The following primers were used to amplify the indicated genes with Pfu polymerase (Stratagene) and 25 cycles of amplification, prior to cloning in TOPO vector (Invitrogen) and sequencing. VH(B1-8hi): 5-CCATGGGATGGAGCTGTATCATCC-3 and 5-GAGGAGACTGTGAGAGTGGTGCC-3. Sμ: 5-GACCCAGGCTAAGAAGGCAATC-3 and 5-GCGGCCCGGCTCATTCCAGTTCATTACAG-3. c-myc: 5-TGGTCTTTCCCTGTGTTCTTTCTG-3 and 5-GACACCTCCCTTCTACACTCTAAACCG-3. Taci: 5-GTCAGGTCAGACAACTCAGGAAGG-3 and 5-GTTTGCCACCCACATCAAGC-3. Whsc1: 5-ACGACGAAACGGTATGAAATCG-3 and 5-AAAAATGAAGGCTGCTGGGC-3. H2Eα: 5-CCAGAGACCAGGATGCCGC-3 and 5-TGGGCACCTTAGCACCGTAGTTAC-3. A20: 5-GGACCATGGCTGAACAACTT-3 and 5-ATCTGGCCGTTTGAGACAAC-3. miR-142: 5-CGTTGGATTCAAGACTGTGGGTC-3 and 5-AATGAGGGCGTGTGAGAGATGCTC-3. Anxa4: 5-ACGGCACCATCTTCTGCTGTC-3 and 5-TCCTCCACACCTTGTTCTCTTGAG-3. Wdfy3: 5-AGAGGAGCCTGGTTTATGTAGCAG-3 and 5-TGGGAGGCTTATTGATTAGGCTG-3. Activated B cells were stimulated for 4 days with LPS and IL-4. Single B cell preparations from matched mice were compared. For additional details see Supplemental Table S4. P-values were determined with the Student’s-t test using a two-tailed distribution.
Southern Blot
Rearrangements at the c-myc and miR-142 loci were determined by digesting DNA with SspI (c-myc) and PstI (miR-142) and probing with PCR products of 5-CATTCTGACTCCTTTTGCCC-3 and 5-TCAGAGGTGGCTATTCAGTTGC-3 (c-myc); or 5-CCCAGGCATTTTTTCCACG-3 and 5-TTGAATCCAACGGAGGCAGC-3 (miR-142).
qPCR
Reverse-transcribed RNA from tumors and controls were amplified for c-myc (5-GCCCCTAGTGCTGCATGAG-3 and 5-CCACAGACACCACATCAATTTCTT-3) and normalized with Gapdh (5-TGAAGCAGGCATCTGAGG-3 and 5-CGAAGGTGGAAGAGTGGGAG-3) using SYBR Green (Applied Biosystems).
RT-PCR and PCR for T(11;15) breakpoints
Der11 c-myc/miR-142 fusion transcripts were amplified with 5-AGTCGGCAAGAAAAGCAGGTG-3 and 5-TCGTCGCAGATGAAATAGGGC-3 and verified by direct sequencing. Der11 breakpoints were amplified with 5-TCGCTCTGCTGTTGCTGGTGATAG-3 and 5-TTGTCGCTGGTTTCCTGTCAG-3. Der15 breakpoints were amplified with 5-GTGAAAACCGACTGTGGCCCTGGAA-3 and 5- CACAACCCCAATAACAGAGTCAGAC-3. The breakpoint was determined by direct sequencing of the gel-extracted PCR products.
Array painting
Tumor cells growing in culture were arrested by overnight treatment with colcemid. The cells were treated with hypotonic KCl solution and 0.25% TX-100, before incubation with Chromomycin A3 (final concentration 40 pg/ml) and Hoechst 33258 (final concentration 2 pg/ml). Sorting was performed on a FACStar Plus flow sorter (Becton Dickinson) equipped with two argon ion lasers. 5000 chromosomes were sorted. Amplification of sorted chromosomes was achieved using WGA4 (Sigma). Oligonucleotide array CGH (aCGH) using mouse 244K tiling arrays was performed according to the manufacturer’s protocol (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis, version 4.0, June 2006). Unsorted liver genomic DNA from sex-matched littermates was used as reference DNA. Slides were scanned using a G2565BA scanner, version 9.1. Feature Extraction software was applied and the data were visualized using CGH Analytics 3.4.40 (Agilent).
M-FISH and analysis of genomic instability
For analysis of chromosome and chromatid breaks, metaphase spreads were prepared from tumor cells and hybridized with telomere repeat-specific peptide nucleic acid (PNA) labeled with Cy3 (Applied Biosystems; (Callen et al., 2007)). Metaphases were counterstained with DAPI. For M-FISH, metaphases were hybridized with the 21x mouse probe cocktail (Metasystems). FISH-labeled metaphases were imaged using a Zeiss AxioImager M1, equipped with a motorized scanning stage and analyzed using Isis software.
Supplementary Material
Acknowledgments
All members of the Nussenzweig labs for discussions. Klara Velinzon for FACSorting, and David Bosque for help in managing the mouse colonies. The work was supported in part by a Fondazione Ettore e Valeria Rossi grant to D.F.R. and by an NIH grant to M.C.N. (AI037526). A.N. and S.B. were supported by the Intramural research Program of the NIH, National Cancer Institute and the Center for Cancer Research and I.A.K. by NIH MSTP grant GM07739. D.F.R. was and N.F. is a Fellow of the Leukemia and Lymphoma Society, A.B. is a predoctoral Fellow of the Cancer Research Institute, and M.C.N. is an HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoufouchi S, Faili A, Zober C, D’Orlando O, Weller S, Weill JC, Reynaud CA. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Callen E, Bunting S, Huang CY, Difilippantonio MJ, Wong N, Khor B, Mahowald G, Kruhlak MJ, Ried T, Sleckman BP, et al. Chimeric IgH-TCR/translocations in T lymphocytes mediated by RAG. Cell Cycle. 2009:8. doi: 10.4161/cc.8.15.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Casellas R, Shih TA, Kleinewietfeld M, Rakonjac J, Nemazee D, Rajewsky K, Nussenzweig MC. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, et al. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci U S A. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Robbiani DF, Jankovic M, Reina-San-Martin B, Eisenreich TR, Nussenzweig MC. A role for AID in chromosome translocations between c-myc and the IgH variable region. J Exp Med. 2007;204:2225–2232. doi: 10.1084/jem.20070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Endres M, Biniszkiewicz D, Sobol RW, Harms C, Ahmadi M, Lipski A, Katchanov J, Mergenthaler P, Dirnagl U, Wilson SH, et al. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J Clin Invest. 2004;113:1711–1721. doi: 10.1172/JCI20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauwerky CE, Huebner K, Isobe M, Nowell PC, Croce CM. Activation of MYC in a masked t(8;17) translocation results in an aggressive B-cell leukemia. Proc Natl Acad Sci U S A. 1989;86:8867–8871. doi: 10.1073/pnas.86.22.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble SM, Kalaitzopoulos D, Burford DC, Prigmore E, Selzer RR, Ng BL, Matthews NS, Porter KM, Curley R, Lindsay SJ, et al. Ultra-high resolution array painting facilitates breakpoint sequencing. J Med Genet. 2007;44:51–58. doi: 10.1136/jmg.2006.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Inagaki H. Mucosa-associated lymphoid tissue lymphoma: molecular pathogenesis and clinicopathological significance. Pathol Int. 2007;57:474–484. doi: 10.1111/j.1440-1827.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jankovic M, Nussenzweig A, Nussenzweig MC. Antigen receptor diversification and chromosome translocations. Nat Immunol. 2007;8:801–808. doi: 10.1038/ni1498. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, duBois W, Mushinski E, McNeil NE, Hirt C, Qi CF, Li Z, Janz S, Honjo T, Muramatsu M, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J Exp Med. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao MJ, Zhang XX, Hill R, Gao J, Qumsiyeh MB, Nichols W, Van Dyke T. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol Cell Biol. 1998;18:3495–3501. doi: 10.1128/mcb.18.6.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Raghavan SC, Yu K. Mechanistic aspects of lymphoid chromosomal translocations. J Natl Cancer Inst Monogr. 2008:8–11. doi: 10.1093/jncimonographs/lgn012. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meding SJ, Cheng SC, Simon-Haarhaus B, Langhorne J. Role of gamma interferon during infection with Plasmodium chabaudi chabaudi. Infect Immun. 1990;58:3671–3678. doi: 10.1128/iai.58.11.3671-3678.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muto T, Okazaki IM, Yamada S, Tanaka Y, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Negative regulation of activation-induced cytidine deaminase in B cells. Proc Natl Acad Sci U S A. 2006;103:2752–2757. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, Di Noia JM. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The Biochemistry of Somatic Hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Peters W, Robinson BL. The chemotherapy of rodent malaria. LVIII. Drug combinations to impede the selection of drug resistance, Part. 2: The new generation--artemisinin or artesunate with long-acting blood schizontocides. Ann Trop Med Parasitol. 2000;94:23–35. doi: 10.1080/00034980057581. [DOI] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. Commonality but diversity in cancer gene fusions. Cell. 2009;137:391–395. doi: 10.1016/j.cell.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Ramiro A, San-Martin BR, McBride K, Jankovic M, Barreto V, Nussenzweig A, Nussenzweig MC. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv Immunol. 2007;94:75–107. doi: 10.1016/S0065-2776(06)94003-6. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006a;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Nussenzweig MC, Nussenzweig A. Switching on chromosomal translocations. Cancer Res. 2006b;66:7837–7839. doi: 10.1158/0008-5472.CAN-06-0863. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc Natl Acad Sci U S A. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Colon K, Affer M, Chesi M, Bergsagel PL. Maintained rules of development in a mouse B-cell tumor. Leukemia. 2005;19:1278–1280. doi: 10.1038/sj.leu.2403774. [DOI] [PubMed] [Google Scholar]
- Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Kaffer CR, Curry JD. Leukemia and lymphoma: a cost of doing business for adaptive immunity. Genes Dev. 2006;20:1539–1544. doi: 10.1101/gad.1446506. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920–932. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- Shen HM, Bozek G, Pinkert CA, McBride K, Wang L, Kenter A, Storb U. Expression of AID transgene is regulated in activated B cells but not in resting B cells and kidney. Mol Immunol. 2008;45:1883–1892. doi: 10.1016/j.molimm.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Papavasiliou FN. Immunoglobulin somatic hypermutation. Annu Rev Genet. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nat Immunol. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.