Abstract
Targeting of IP3R (inositol 1,4,5-trisphosphate receptors) to membranes of the ER (endoplasmic reticulum) and their retention within ER or trafficking to other membranes underlies their ability to generate spatially organized Ca2+ signals. N-terminal fragments of IP3R1 (type 1 IP3R) were tagged with enhanced green fluorescent protein, expressed in COS-7 cells and their distribution was determined by confocal microscopy and subcellular fractionation. Localization of IP3R1 in the ER requires translation of between 26 and 34 residues beyond the end of the first transmembrane domain (TMD1), a region that includes TMD2 (second transmembrane domain). Replacement of these post-TMD1 residues with unrelated sequences of similar length (24–36 residues) partially mimicked the native residues. We conclude that for IP3R approx. 30 residues after TMD1 must be translated to allow a signal sequence within TMD1 to be extruded from the ribosome and mediate co-translational targeting to the ER. Hydrophobic residues within TMD1 and TMD2 then ensure stable association with the ER membrane.
Keywords: calcium signalling; endoplasmic reticulum; inositol 1,4,5-trisphosphate receptor (IP3R); protein targeting; signal sequence
Abbreviations: ER, endoplasmic reticulum; EYFP, enhanced yellow fluorescent protein; GFP, green fluorescent protein; EGFP, enhanced GFP; HRP, horseradish peroxidase; IP3, inositol 1,4,5-trisphosphate; IP3R, IP3 receptor(s); PBS-T, PBS containing 0.1% Tween 20; SRP, signal recognition particle; TMD, transmembrane domain(s); the abbreviations used for fragments of IP3R1 are defined in Figure 1(C)
INTRODUCTION
IP3R [IP3 (inositol 1,4,5-trisphosphate) receptors] are the intracellular Ca2+ channels that both initiate and regeneratively propagate the cytosolic Ca2+ signals evoked by the many receptors that stimulate IP3 formation [1]. All IP3R are tetramers, each with an IP3-binding site lying close to the N-terminus and six TMD (transmembrane domains) lying close to the C-terminus (Figure 1A). The last pair of TMD from each subunit together with their intervening luminal loop form the pore [2,3]. In most animal cells, IP3R are expressed mainly within the membranes of the ER (endoplasmic reticulum), but they are also expressed within the nuclear envelope [4], nucleoplasmic reticulum [5], Golgi apparatus [6], plasma membrane [7], and perhaps also in secretory vesicles [8], although the latter is contentious [9]. Within these membranes, IP3R are not uniformly distributed and different subtypes may differ in their distributions [10,11]. The subcellular distribution of IP3R accounts for their ability to generate cytosolic Ca2+ signals that are spatially organized, thereby allowing Ca2+ to regulate specifically a diverse array of cellular processes [1]. The versatility of Ca2+ as a ubiquitous intracellular messenger thus depends upon precise targeting of IP3R to specific subcellular compartments.
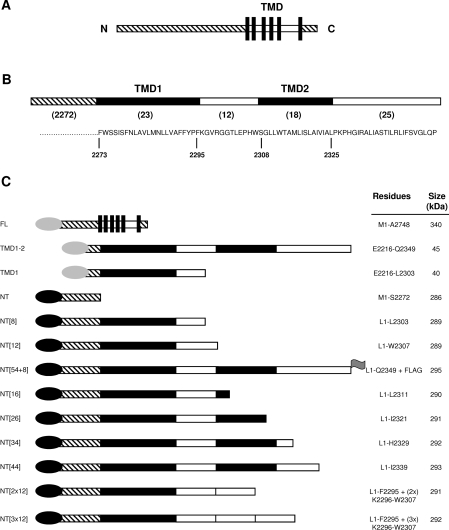
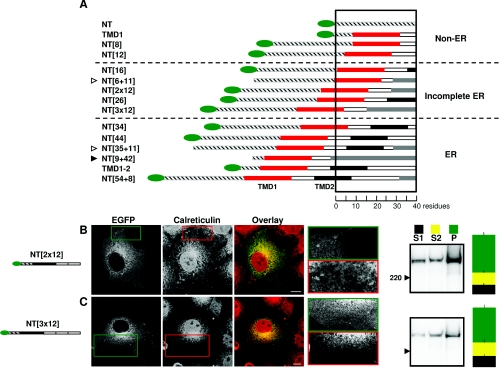
Figure 1. Fusion proteins used.
(A) Key regions of IP3R1. (B) Sequence of the TMD1-2 region. Numbers in parentheses denote the number of residues within each region. (C) The proteins used, and their abbreviations, are shown with N-terminal EGFP (black) or EYFP (grey) tags represented as ovals, and the C-terminal FLAG epitope as a flag. TMD are shown by black bars, linking loops by white bars, and the N- and C-termini by hatching.
Whatever the final destination of an IP3R, it must first be directed to the ER, where it may either be retained (the fate of most IP3R) or be allowed to move on to other membranes via the Golgi apparatus. Targeting of proteins to the ER is mediated by a short stretch of amino acid residues, the signal sequence, which may be either an N-terminal sequence that is later cleaved, or an internal non-cleavable sequence [12]. The latter, signal-anchor sequences, serve the dual purpose of directing the protein to the ER and anchoring it within the membrane. Signal sequences vary widely in primary sequence, but in both prokaryotes and eukaryotes they share a hydrophobic core of 8–12 residues for cleavable signals and of 20–30 residues for internal signals [13]. The diversity of signal sequences allows them to function with different efficiencies and also provides a mechanism that allows proteins that might become terminally misfolded to be directed away from the ER for degradation during ER stress [14].
The signal sequence, whether N-terminal or internal, is recognized by the SRP (signal recognition particle). For most eukaryotic secretory or membrane proteins, this occurs co-translationally [15], but a minority of proteins (those with a C-terminal signal sequence) are post-translationally targeted [16]. Co-translational targeting is initiated when SRP binds simultaneously to the exposed signal sequence and the ribosome, forming the SRP–ribosome nascent chain complex [15]. SRP may also recognize a conformation of the ribosome within which a signal-anchor sequence is still concealed and so pre-associate with the ribosome before binding tightly to the emerging signal sequence [17]. The SRP–ribosome nascent chain complex then binds to the SRP receptor within the ER membrane [18], SRP dissociates, protein synthesis resumes and the growing protein is directed into the ER through the open translocon. The latter is a channel formed largely from the Sec61 complex that allows proteins to pass into either the lumen of the ER or laterally into the ER membrane [19,20].
After incorporation into the ER membrane, proteins may either remain there or move on. Proteins remain because they express signals that prevent them from leaving the ER or promote their retrieval from the Golgi apparatus. Luminal ER proteins are retrieved by a C-terminal KDEL motif, whereas integral membrane proteins are retrieved from post-ER compartments by cytosolic motifs such as the C-terminal di-lysine or N-terminal di-arginine motif [21,22]. TMD can also mediate ER retention [23–25]. None of the cytosolic motifs known to mediate ER retrieval are present in IP3R, but our earlier work demonstrated that any pair of TMD with a linking luminal loop can retain an IP3R fragment or a plasma membrane protein within the ER [26].
IP3R lack an N-terminal signal sequence, but, as with other ER membrane proteins, hydrophobic residues within the TMD can provide internal signal sequences. RyR1 (type 1 ryanodine receptor), for example, is targeted to the ER by its first TMD [27], and a sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1) is targeted and retrieved by its first pair of TMD [28]. For IP3R1, the TMD region is sufficient for ER targeting [29,30], with later work suggesting that the first pair of TMD is essential [3,26]. Our analysis of fragments comprising individual TMD or pairs of TMD demonstrated that any pair of TMD linked by their luminal loops is sufficient to localize IP3R1 to the ER [26]. These results, derived from short fragments of IP3R1 lacking the N-terminus, suggest that IP3R1 is targeted to the ER only after translation of the first and second TMD and the following loops. This highlights residues lying between the beginning of TMD1 and the end of the cytosolic loop following TMD2 (Figure 1B) as the most likely determinants of IP3R1 targeting to the ER. However, the relative roles of residues within this region when IP3R1 has its normal large cytosolic N-terminus are not clear. In the present study, we address this issue by systematically examining the subcellular distribution of IP3R1 fragments progressively truncated from the C-terminus.
EXPERIMENTAL
Expression constructs
All constructs are based on the full-length rat IP3R1 [31] (GenBank® accession number GQ233032) lacking the S1 splice region [32]. Most proteins were N-terminally tagged with EGFP [enhanced GFP (green fluorescent protein)] (Figure 1C). Molecular masses were calculated without the S1 splice region, but the numbering of residues includes the S1 region. All constructs containing the N-terminal region of IP3R1 have a TTG (Leu) codon instead of ATG (Met) at the beginning of the open reading frame to prevent internal initiation.
The plasmid encoding NT[8] was constructed by ligating the EcoRI/SalI fragment of IP3R1 generated from pCMVI-9-IP3R1 [31] by PCR into pEGFP-C2 (Clontech). The same fragment was ligated into the pENTR 1A vector (Invitrogen) to provide the key plasmid from which all other constructs were assembled. In brief, PCR fragments were generated using the primers listed in Supplementary Table S1 (available at http://www.BiochemJ.org/bj/425/bj4250061add.htm), they were digested with SalI/BstBI and ligated to the SalI/BstBI-digested key plasmid. EcoRI/SalI was used to excise the inserts which were then ligated into pEGFP-C2 vector, resulting in the desired constructs. Expression plasmids for FL, TMD1 and TMD1-2 were prepared as described previously [26] using the primers listed in Supplementary Table S1. NT was prepared by PCR from FL and ligated into pEGFP-C2. The coding sequences of all constructs were confirmed by sequencing.
Cell culture and transfection
COS-7 cells were maintained in minimum essential medium supplemented with 10% (v/v) foetal bovine serum and 2 mM L-glutamine at 37 °C in 5% CO2. One day before transfection, cells were plated on to coverslips (13-mm-diameter, No. 1 thickness) coated with 0.01% poly-L-lysine in 24-well plates for confocal imaging (8×104 cells/well) or directly into six-well plates (3×105 cells/well) for subcellular fractionation. Cells at 80–90% confluence were transfected using Lipofectamine™ 2000 reagent (Invitrogen) during an overnight incubation with (per well) 1 μg of DNA and 1 μl of Lipofectamine™ 2000 for confocal imaging, or 2–4 μg of DNA and 2–4 μl of Lipofectamine™ 2000 for subcellular fractionation.
Confocal microscopy
Cells were used 24 h after transfection. They were washed twice with PBS and fixed with 3.5% paraformaldehyde at pH 7.4 in PBS for 5–20 min at 20 °C. Similar results were obtained after fixation with methanol/acetone (1:1, v/v) for 10 min at −20 °C (see Supplementary Figure S1 available at http://www.BiochemJ.org/bj/425/bj4250061add.htm). Cells were then permeabilized with 0.1% Triton X-100 in PBS for 5–10 min. Non-specific antibody binding was blocked by incubation with 5% (w/v) BSA in PBS for 10 min at 20 °C. To identify calreticulin, an ER luminal protein, cells were incubated with rabbit anti-calreticulin antibody [Calbiochem; 1:100 dilution in PBS with 5% (w/v) BSA] for 2.5 h at 20 °C and then for 12 h at 4 °C. We and others [33] also find calreticulin in the nucleoplasm of some cells. There was no consistent relationship between this nucleoplasmic expression of calreticulin and either the fixation method or the transfected construct. The benefits of using an antibody to an endogenous protein to identify the ER outweigh the drawbacks of it occasionally also staining the nucleoplasm. Cells were washed (3×5 min) with PBS and then incubated with goat anti-rabbit Alexa Fluor® 633 secondary antibody [Molecular Probes; 1:500 dilution in PBS with 5% (w/v) BSA] for 1 h at 20 °C. Cells were washed (3×5 min) with PBS and once with water, and the coverslips were mounted in Prolong anti-fade mounting medium (Molecular Probes). To identify mitochondria, cells were either co-transfected with the IP3R construct and DsRed-Mito (Clontech), or with the IP3R construct alone, and then incubated in normal culture medium supplemented with 100 nM MitoTracker Red CMXRos (Molecular Probes) for 30 min at 37 °C. Fixation and mounting were as described above. Slides were stored at −20 °C before confocal imaging.
All imaging used a Leica TCS SP2 AOBS confocal microscope with a ×63, 1.4 numerical aperture oil-immersion objective. EGFP, EYFP (enhanced yellow fluorescent protein), DsRed, MitoTracker Red and Alexa Fluor® 633 were excited with the 488 nm, 514 nm, 561 nm, 594 nm and 633 nm lines respectively. Emitted signals were collected using emission filters with detection bands of 500–565 nm, 520–600 nm (520–560 nm when imaged together with MitoTracker and 520–540 nm with DsRed), 565–675 nm, 600–650 nm and 640–750 nm respectively. In all dual-labelling analyses, we confirmed that there was no bleed-through between the two wavelengths. All images were exported as tiff files and processed using Adobe Photoshop.
Most confocal images are shown to highlight a single typical cell. Views of fields of cells are shown in Supplementary Figure S2 (available at http://www.BiochemJ.org/bj/425/bj4250061add.htm). Because the cytosol and ER are entwined, the distinction between them is not always immediately obvious in confocal images. The difference between reticulate and cytosolic distributions is more clearly discernible at the cell boundary. The ER extends to the periphery without clear boundaries, while the cytosol clearly defines the cell boundary. Images of the cell periphery are shown at higher magnification in each figure and in Supplementary Figure S3 (available at http://www.BiochemJ.org/bj/425/bj4250061add.htm). To allow a more quantitative analysis of the co-localization of IP3R fragments and calreticulin immunostaining (see Table 1), we adopted the method shown in Supplementary Figure S4 (available at http://www.BiochemJ.org/bj/425/bj4250061add.htm). Briefly, three lines were drawn across each cell to exclude the nucleus, and the Pearson correlation coefficient (r) was computed for the relationship between the intensities of the two fluorescence channels (green for IP3R fragments and red for calreticulin). For an IP3R construct known to be expressed primarily within membranes of the ER (FL) r was 0.70±0.07, and for a construct known to be cytosolic (NT) r was 0.37±0.06 (see Table 1). These values of r, for an ER and cytosolic protein, are those against which all other fragments are compared statistically in Table 1.
Table 1. Co-localization of IP3R fragments and calreticulin.
Pearson's correlation coefficient (r) was calculated for the fluorescence intensities for the wavelengths corresponding to each IP3R fragment and calreticulin as described in Supplementary Figure S4. Results are means±S.E.M. for three to six cells (n is shown for each fragment), with three lines analysed in each cell. *Values significantly different (P<0.05) from NT using one-way ANOVA with Dunnett's post-hoc test.
| IP3R fragment FL | TMD1-2 | NT | NT[8] | NT[12] | NT[54+8] | NT[16] | NT[26] | NT[34] | NT[44] | NT[2×12] | NT[3×12] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | 0.70±0.07* | 0.64±0.03 | 0.37±0.06 | 0.39±0.12 | 0.46±0.04 | 0.74±0.07* | 0.61±0.06* | 0.61±0.06 | 0.70±0.05* | 0.75±0.03* | 0.50±0.07 | 0.45±0.08 |
| (n) | (3) | (3) | (6) | (3) | (5) | (3) | (6) | (3) | (3) | (5) | (5) | (6) |
Subcellular fractionation and Western blot analysis
Cells were harvested 24 h after transfection, washed with PBS, scraped into 250 μl of ice-cold PBS containing protease inhibitors (1 tablet/10 ml; Roche) and disrupted by 30 passages through a 25-gauge needle. After centrifugation (30000 g for 30 min), the supernatant (S1), containing cytosolic proteins, was saved. The pellet was resuspended in 250 μl of sodium carbonate (0.1 M, pH 11.5), incubated on ice for 45 min to dissociate peripheral membrane proteins and, after further centrifugation (30000 g for 30 min), the second supernatant (S2) containing peripheral membrane proteins was saved. The pellet (P) containing integral membrane proteins was resuspended in 250 μl of ice-cold PBS containing protease inhibitors and 1% Triton X-100.
Samples (corresponding to equivalent numbers of cells for each fraction) were loaded on to pre-cast NuPAGE 3–8% Tris-acetate or 4–12% Bis-Tris gels (Invitrogen). SDS/PAGE (XCell SureLock Mini-Cell; Invitrogen) and transfer on to PVDF membrane (XCell II Blot Module or iBlot dry gel system; Invitrogen) were performed according to the manufacturer's instructions. Membranes were blocked overnight in PBS-T (PBS containing 0.1% Tween 20) supplemented with 1% (w/v) BSA, incubated for 1 h with a rabbit polyclonal antibody to GFP [AbCam; 1:1000 dilution in PBS-T with 1% (w/v) BSA], washed (3×10 min) with PBS-T, and then incubated with a secondary donkey anti-rabbit antibody coupled to HRP (horseradish peroxidase) [Santa Cruz Laboratories; 1:5000 dilution in PBS-T with 1% (w/v) BSA]. Membranes were washed (3×10 min) with PBS-T, and HRP was detected using Supersignal West Pico Chemiluminescent substrate (Pierce). Immunoreactive bands were quantified using GeneTools software (Syngene).
RESULTS AND DISCUSSSION
ER localization of N-terminally truncated IP3R requires more than TMD1
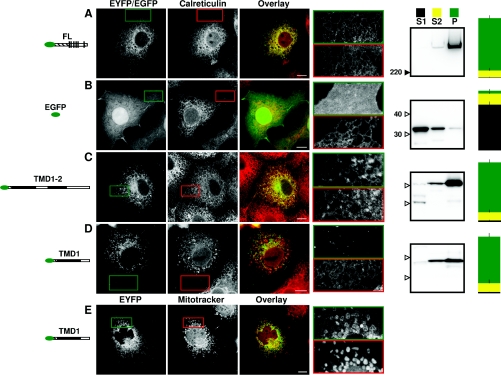
Full-length IP3R1 tagged at its N-terminus with EYFP (FL; Figure 1C) was localized in the ER of COS-7 cells. It exhibited strong perinuclear fluorescence that extended towards the periphery of the cell in a tubular network and it co-localized with calreticulin, an ER luminal protein (Figure 2A, Table 1 and Supplementary Figure S2). After subcellular fractionation, FL was found mostly (88±4%) in the P fraction (integral membrane proteins; see the Experimental section). EYFP (results not shown) or EGFP alone was diffusely spread throughout the cell, including the nucleus, and did not co-localize with calreticulin (Figure 2B). EGFP was found largely (77±5%) in the S1 fraction (cytosolic proteins; Figure 2B). An EYFP-tagged fragment of IP3R1 (TMD1-2) that includes only the last 58 residues of the N-terminal cytosolic region and extends to the end of the cytosolic loop following TMD2 had a distribution similar to that of FL and it co-localized with calreticulin (Table 1). TMD1-2 was present largely (84±5%) in the P fraction (Figure 2C). A similar, but shorter, IP3R1 fragment (TMD1) truncated after eight of the 12 residues linking TMD1 to TMD2 did not co-localize with calreticulin (Figure 2D), but instead co-localized with MitoTracker Red (Figure 2E) [26]. As expected [26], there was no such co-localization of TMD1-2 with mitochondria (Supplementary Figure S5 available at http://www.BiochemJ.org/bj/425/bj4250061add.htm). TMD1 was also found largely in the P fraction (82±6%). The presence of TMD1 in the P fraction highlights the limitations of using simple fractionation methods alone to resolve the targeting of IP3R fragments to the ER. A fragment (TMD2) comprising only the last eight residues of the loop linking TMD1 to TMD2 and terminating 12 residues after the end of TMD2 was also expressed in mitochondria, but not in the ER (results not shown) [26]. Western blotting with an anti-GFP antibody confirmed that the expressed proteins had the expected sizes (Figures 2A–2D).
Figure 2. N-terminally truncated IP3R1 fragments are targeted to the ER by the first TMD pair.
COS-7 cells transiently transfected with the indicated constructs are shown in the left column and stained for calreticulin [MitoTracker in (E)] in the second column. The third column shows the first two columns overlaid with the construct in green and calreticulin (or MitoTracker) in red. Bars, 10 μm. The fourth column shows enlargements of the highlighted boundaries, with green and red borders enclosing the construct and organelle marker respectively. Here, and in all subsequent Figures, images are representative of at least three independent transfections. The fifth column shows Western blots (with an antibody to GFP) of the three fractions derived from subcellular fractionation of the cells: S1 (first supernatant; cytosolic proteins), S2 (second supernatant; peripheral membrane proteins) and P (pellet; integral membrane proteins). For each gel, the three lanes were loaded with material from an equivalent number of cells. Molecular-mass markers are shown in kDa. The final column summarizes the results obtained from the subcellular fractionation (values are means±S.E.M., n≥3; see the Experimental section).
These results confirm earlier work showing that, in these minimal C-terminal fragments of the IP3R1, neither of the first two TMD is alone sufficient to allow expression in the ER, while together they mediate effective ER localization [26]. Subsequent experiments aim first to establish whether there is a similar requirement for localization of full-length IP3R in the ER, and secondly to define the role of these residues in ER targeting. To identify the first ER-targeting signal in native IP3R1, we examined the distribution of fragments of IP3R1 truncated only at the C-terminus (Figure 1C). For simplicity, the truncated IP3R constructs used to address these issues, all of which have the same N-terminus, are abbreviated by reference to the number of residues after TMD1 (Figure 1C).
Localization of IP3R1 in the ER requires translation of TMD1 and TMD2
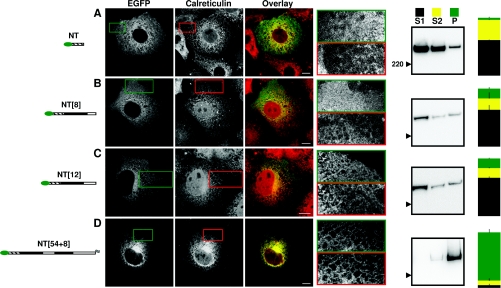
In keeping with previous reports [29,30], the large N-terminal fragment of the IP3R1 preceding TMD1 (NT) was cytosolic. It was diffusively distributed throughout the cytosol, excluded from the nucleus and it did not co-localize with calreticulin (Table 1 and Figure 3A). After subcellular fractionation, very little of the NT (4±1%) was detected in the P fraction (integral membrane proteins; Figure 3A). The latter is consistent with an earlier conclusion, although in that study there was more contamination of the membrane fraction with the N-terminal fragment [3].
Figure 3. Localization of IP3R1 in the ER requires translation of the first TMD pair.
Cells transiently transfected with the indicated constructs are shown in the same format as in Figure 2.
A fragment of IP3R1 truncated part way through the loop following TMD1 (NT[8]) had a similar distribution to NT. It was diffusely spread from around the nucleus to the plasma membrane, uniformly defined the boundaries of the cell, it was not co-localized with calreticulin and very little of the protein (17±2%) was detected in the P fraction after subcellular fractionation (Figure 3B and Table 1). It is noteworthy that while neither the long (NT[8]) nor short (TMD1) fragment truncated after TMD1 is localized to the ER, the former remains cytosolic, while the latter is expressed in mitochondria (Figure 2E) [26]. We speculate that TMD1 is released from the ribosome before the signal sequence can be recognized by SRP (see later discussion) and the basic residues flanking TMD1 then favour post-translational targeting to mitochondria [34].
Addition to NT[8] of the four remaining residues from the loop following the first TMD (to give NT[12]; Figure 1C) had no effect on the distribution; NT[12] was cytosolic (Figure 3C and Table 1). Extending the N-terminal fragment further to include TMD2 and the following loop (NT[54+8]) caused the protein to be localized in the ER. The distribution of NT[54+8] was indistinguishable from that of FL, it defined the cell boundary in a reticulate fashion, it co-localized with calreticulin (Table 1) and most protein (87±6%) was found in the P fraction (Figure 3D). NT[54+8], which includes the entire N-terminal region of IP3R1, has the same native C-terminal residues as the much smaller fragment (TMD1-2), although NT[54+8] has an additional C-terminal FLAG tag comprising eight residues. Both fragments were similarly expressed in the ER (Figures 2C and 3D). These results suggest that for both short fragments and native IP3R1, translation of TMD1 and TMD2 are required for effective localization in the ER.
The minimal requirement for localization of IP3R in the ER is translation of 26–34 residues after TMD1
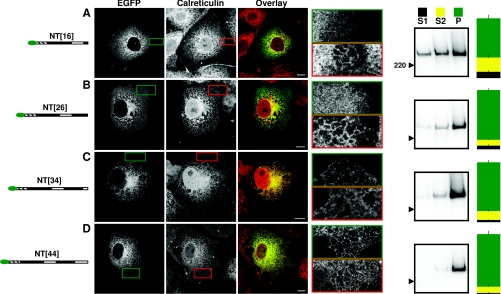
To define more specifically the minimal requirements for ER localization, we used constructs truncated within TMD2 or the succeeding loop. IP3R1 truncated just four residues into TMD2 (NT[16]) was diffusively expressed. It did not obviously co-localize with calreticulin (Figure 4A), with the quantitative analysis (r=0.61±0.16) suggesting a distribution intermediate between that of an ER protein (FL, r=0.70±0.07) and cytosolic protein (NT, r=0.37±0.06) (Table 1). Its definition of the cell boundary was similar to that of cytosolic fragments (Supplementary Figure S3). After subcellular fractionation, however, most NT[16] was in the P fraction (66±6%).
Figure 4. Localization of IP3R1 in the ER requires translation of 26–34 residues after TMD1.
Cells transiently transfected with the indicated constructs are shown in the same format as in Figure 2.
Results from confocal and fractionation assays concur where proteins are entirely cytosolic (e.g. NT; Figure 3A) or entirely localized in ER (e.g. FL; Figure 2A), but quantitative analysis of cell fractions seems better able to resolve incomplete targeting than is the more qualitative assessment of confocal images (Figures 3 and 4).
A fragment (NT[26]) that extends 14 residues into TMD2 appeared to be cytosolic in most cells (Figure 4B and Table 1), but in a minority of cells (~20%) the distribution was reticulate and similar to that of FL (Supplementary Figure S6 available at http://www.BiochemJ.org/bj/425/bj4250061add.htm); 84±3% of NT[26] was in the P fraction. A slightly longer fragment (NT[34]) that includes TMD2 and four residues from the following loop was localized in the ER, as was a fragment extended by a further ten residues (NT[44]). For both fragments, the distribution was reticulate, they co-localized with calreticulin (Figures 4C and 4D, and Table 1) and they were predominantly found in the P fraction (81±4% and 88±5% for NT[34] and NT[44] respectively). These results define the minimal number of residues beyond TMD1 that must be translated to allow effective localization of IP3R1 in the ER: 12 residues are not sufficient (Figure 3C), 26 residues are partially effective (Figure 4B and Supplementary Figure S6) and 34 residues after TMD1 allow the protein (Figure 4C) to be targeted to the ER as effectively as FL (Figure 2A and Table 1). We conclude that between 26 and 34 residues beyond the end of TMD1 must be translated for native IP3R1 to be effectively localized in the ER (Figure 5A).
Figure 5. Non-native sequences after TMD1 facilitate ER localization.
(A) Summary of the present results and those from previous analyses of expression in COS-1 cells [3] (open triangles) or in vitro [35] (filled triangle). The native IP3R sequence is shown in red (TMD1), black (TMD2), white (non-TMD) or hatched (N-terminal domain); EGFP/EYFP tags are shown in green, and C-terminal tags or non-native sequence in grey. All sequences from TMD1 towards the C-terminal (including C-terminal tags) are drawn to scale (see scale bar). The boxed area shows the ~40 residues concealed within the ribosome tunnel. As more of TMD2 and the following loop are translated, the putative signal sequence in TMD1 emerges from the ribosome, allowing it to bind SRP. (B, C) Cells transiently transfected with the indicated constructs are shown in the same format as in Figure 2.
Another analysis of similar IP3R1 fragments truncated at the C-terminus but with a C-terminal tag (11 residues) demonstrated that, when these were expressed in COS-1 cells, a fragment truncated six residues after TMD1 (NT[6+11]) was equally distributed between cytosolic and membrane fractions, whereas all constructs longer than NT[35+11] were entirely in the membrane fraction (Figure 5A) [3]. Using an in vitro translation system, a construct that included 21 residues before TMD1, nine native residues after it and a C-terminal tag of 42 residues (NT[9+42]) was effectively co-translationally inserted into microsomal membranes [35]. NT[9+42] and NT[35+11] have similar numbers of residues after TMD1 (51 and 46 respectively). Although the results from these analyses in vitro (suggesting a requirement for only TMD1) [35] and cells (suggesting a requirement for TMD1 and TMD2) [3] were thought to be contradictory, they are each consistent with a need for ≤51 residues after TMD1 to be translated to allow ER localization of IP3R1. We can now refine that requirement and conclude that between 26 and 34 residues beyond the end of TMD1 must be translated for IP3R1 to be targeted to the ER.
Residues after TMD1 mediate both exposure of the signal sequence and membrane-anchoring of IP3R
Signal sequences bind to SRP only after about ten residues have emerged from the ribosomal tunnel [36], which is ~10 nm long and can accommodate 30–40 unfolded residues [37]. The requirement for translation of 26–34 residues after TMD1, which includes TMD2 and some of the following loop (Figures 1B and 1C), may therefore reflect a need for these specific residues to contribute to ER targeting or they may be required only to allow exposure of a signal sequence in TMD1 (Figure 5A). The latter would be consistent with the analysis in vitro showing that TMD1 with the following loop mediates co-translational targeting when it is followed by a sequence of 42 residues unrelated to the IP3R [35].
If the post-TMD1 residues serve only to expel the signal sequence from the ribosome tunnel, any sequence of 30–40 residues after the signal sequence within TMD1 would be sufficient to allow ER targeting. We therefore expressed IP3R1 with the loop following TMD1 duplicated to provide 24 residues beyond the end of TMD1 (NT[2×12]) or with three repeats to provide 36 residues (NT[3×12]) (Figure 1C). These proteins are similar in length to the shortest fragments that were partially (NT[26]; Figure 4B) or completely (NT[34]; Figure 4C) localized in the ER. Both NT[2×12] and NT[3×12] were found largely in the P fraction (63±4% and 58±5% respectively). Their integration into membranes is therefore much greater than for NT[12] (17±1%), but clearly less than for NT[26] (84±3%) and NT[34] (81±4%). These results suggest that a major role of the post-TMD1 residues is to allow a hydrophobic signal sequence within TMD1 to be pushed out of the ribosome tunnel and allow its recognition by SRP.
However, the replicated (hydrophilic) residues from the post-TMD1 loop were not as effective as the native (hydrophobic) residues of TMD2 in causing IP3R1 fragments to associate with the P fraction (~60% compared with ~80%), and nor were the fragments with replicated loops clearly co-localized with the ER in confocal images (Figures 5B and 5C, and Table 1). We suggest that, in addition to providing residues that allow the signal sequence to be extruded from the ribosome, hydrophobic residues within TMD2 also contribute to anchoring the large IP3R fragments in the ER membrane.
Conclusions
Co-translational targeting of IP3R to the ER [35] is the first step in the sequence of events that leads to IP3R being precisely located within intracellular membranes and thereby placed to generate spatially organized Ca2+ signals [1]. The large N-terminal cytosolic region of the IP3R (2272 residues) is translated and folds to include a functional IP3-binding site before the protein is directed to the ER. This ensures the final cytosolic disposition of this region and dictates the transmembrane topology of the complete IP3R. Evidence derived from expression in vitro and in cells of IP3R fragments with and without the complete N-terminus indicates that the sequence that includes TMD1 and TMD2 is sufficient for ER targeting [3,26,35] (Figure 2). Effective targeting occurs after translation of between 26 and 34 residues beyond the end of TMD1, a region that includes TMD2 (Figure 5A). Replacement of this post-TMD1 region with a similar number of hydrophilic residues can partially substitute for the native residues in mediating ER targeting (Figures 5B and 5C), suggesting that a major role is to facilitate extrusion from the ribosome of a signal recognition sequence within TMD1 allowing its recognition by SRP. Once targeted to the ER, the TMD provide the hydrophobic anchors that retain IP3R within the ER, with TMD1 and TMD2 together sufficient to ensure complete retention within the ER [26].
Online data
AUTHOR CONTRIBUTION
Evangelia Pantazaka completed the experiments. Colin Taylor directed the study. Both authors contributed to the design and analysis of experiments, and to writing the paper.
ACKNOWLEDGEMENTS
We thank Skarlatos Dedos (Department of Pharmacology, University of Cambridge, Cambridge, U.K.) for help with molecular biology.
FUNDING
This work was supported by the Medical Research Council UK [grant number G0700843]; the Wellcome Trust [grant number 085295]; and by a studentship from the Propondis Foundation (to E.P.).
References
- 1.Berridge M. J., Lipp P., Bootman M. D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Foskett J. K., White C., Cheung K. H., Mak D. O. Inositol trisphosphate receptor Ca2+ release channels. Physiol. Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galvan D. L., Borrego-Diaz E., Perez P. J., Mignery G. A. Subunit oligomerization, and topology of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 1999;274:29483–29492. doi: 10.1074/jbc.274.41.29483. [DOI] [PubMed] [Google Scholar]
- 4.Cardenas C., Liberona J. L., Molgo J., Colasante C., Mignery G. A., Jaimovich E. Nuclear inositol 1,4,5-trisphosphate receptors regulate local Ca2+ transients and modulate cAMP response element binding protein phosphorylation. J. Cell Sci. 2005;118:3131–3140. doi: 10.1242/jcs.02446. [DOI] [PubMed] [Google Scholar]
- 5.Echevarria W., Leite M. F., Guerra M. T., Zipfel W. R., Nathanson M. H. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat. Cell Biol. 2003;5:440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Missiaen L., Van Acker K., Parys J. B., De Smedt H., Van Baelen K., Weidema A. F., Vanoevelen J., Raeymaekers L., Renders J., Callewaert G., et al. Baseline cytosolic Ca2+ oscillations derived from a non-endoplasmic reticulum Ca2+ store. J. Biol. Chem. 2001;276:39161–39170. doi: 10.1074/jbc.M104044200. [DOI] [PubMed] [Google Scholar]
- 7.Dellis O., Dedos S., Tovey S. C., Rahman T.-U.-, Dubel S. J., Taylor C. W. Ca2+ entry through plasma membrane IP3 receptors. Science. 2006;313:229–233. doi: 10.1126/science.1125203. [DOI] [PubMed] [Google Scholar]
- 8.Gerasimenko O. V., Gerasimenko J. V., Belan P. V., Petersen O. H. Inositol trisphosphate and cyclic ADP-ribose-mediated release of Ca2+ from single isolated pancreatic zymogen granules. Cell. 1996;84:473–480. doi: 10.1016/s0092-8674(00)81292-1. [DOI] [PubMed] [Google Scholar]
- 9.Yule D. I., Ernst S. A., Ohnishi H., Wojcikiewicz R. J. H. Evidence that zymogen granules are not a physiologically relevant calcium pool: defining the distribution of inositol 1,4,5-trisphosphate receptors in pancreatic acinar cells. J. Biol. Chem. 1997;272:9093–9098. doi: 10.1074/jbc.272.14.9093. [DOI] [PubMed] [Google Scholar]
- 10.Vermassen E., Parys J. B., Mauger J.-P. Subcellular distribution of the inositol 1,4,5-trisphosphate receptors: functional relevance and molecular determinants. Biol. Cell. 2004;96:3–17. doi: 10.1016/j.biolcel.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Taylor C. W., Genazzani A. A., Morris S. A. Expression of inositol trisphosphate receptors. Cell Calcium. 1999;26:237–251. doi: 10.1054/ceca.1999.0090. [DOI] [PubMed] [Google Scholar]
- 12.Hegde R. S., Bernstein H. D. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 13.von Heijne G. Signal sequences: the limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 14.Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egea P. F., Stroud R. M., Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Abell B. M., Pool M. R., Schelenker O., Sinning I., High S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004;23:2755–2764. doi: 10.1038/sj.emboj.7600281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berndt U., Oellerer S., Zhang Y., Johnson A. E., Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halic M., Gartmann M., Schlenker O., Mielke T., Pool M. R., Sinning I., Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- 19.Alder N. N., Johnson A. E. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- 20.Rapoport T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 21.Teasdale R. D., Jackson M. R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 22.Michelsen K., Yuan H., Schwappach B. Hide and run: arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 2005;6:717–722. doi: 10.1038/sj.embor.7400480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczesna-Skorupa E., Kemper B. Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P450 2C1. J. Biol. Chem. 2000;275:19409–19415. doi: 10.1074/jbc.M002394200. [DOI] [PubMed] [Google Scholar]
- 24.Honsho M., Mitoma J. Y., Ito A. Retention of cytochrome b5 in the endoplasmic reticulum is transmembrane and luminal domain-dependent. J. Biol. Chem. 1998;273:20860–20866. doi: 10.1074/jbc.273.33.20860. [DOI] [PubMed] [Google Scholar]
- 25.Cocquerel L., Duvet S., Meunier J. C., Pillez A., Cacan R., Wychowski C., Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 1999;73:2641–2649. doi: 10.1128/jvi.73.4.2641-2649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker A. K. T., Gergely F. V., Taylor C. W. Targeting of inositol 1,4,5-trisphosphate receptors to the endoplasmic reticulum by multiple signals within their transmembrane domains. J. Biol. Chem. 2004;279:23797–23805. doi: 10.1074/jbc.M402098200. [DOI] [PubMed] [Google Scholar]
- 27.Meur G., Parker A. K. T., Gergely F. V., Taylor C. W. Targeting and retention of type 1 ryanodine receptors to the endoplasmic reticulum. J. Biol. Chem. 2007;282:23096–23103. doi: 10.1074/jbc.M702457200. [DOI] [PubMed] [Google Scholar]
- 28.Newton T., Black J. P. J., Butler J., Lee A. G., Chad J., East J. M. Sarco/endoplasmic-reticulum calcium ATPase SERCA1 is maintained in the endoplasmic reticulum by a retrieval signal located between residues 1 and 221. Biochem. J. 2003;371:775–782. doi: 10.1042/BJ20021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayers L. G., Miyawaki A., Muto A., Takeshita H., Yamamoto A., Michikawa T., Furuichi T., Mikoshiba K. Intracellular targeting and homotetramer formation of a truncated inositol 1,4,5-trisphosphate–green fluorescent protein chimaera in Xenopus laevis oocytes: evidence for the involvement of transmembrane spanning domain in endoplasmic reticulum targeting and homotetramer formation. Biochem. J. 1997;323:273–280. doi: 10.1042/bj3230273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mignery G. A., Südhof T. C. The ligand binding site and transduction mechanism in the inositol-1,4,5-trisphosphate receptor. EMBO J. 1990;9:3893–3898. doi: 10.1002/j.1460-2075.1990.tb07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mignery G. A., Newton C. L., Archer B. T., Südhof T. C. Structure and expression of the rat inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 1990;265:12679–12685. [PubMed] [Google Scholar]
- 32.Cardy T. J. A., Taylor C. W. A novel role for calmodulin: Ca2+-independent inhibition of type-1 inositol trisphosphate receptors. Biochem. J. 1998;334:447–455. doi: 10.1042/bj3340447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem. J. 2009;417:651–666. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- 34.Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., Mikhara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseph S. K., Boehning D., Pierson S., Nicchitta C. V. Membrane insertion, glycosylation, and oligomerization of inositol trisphosphate receptors in a cell-free translation system. J. Biol. Chem. 1997;272:1579–1588. doi: 10.1074/jbc.272.3.1579. [DOI] [PubMed] [Google Scholar]
- 36.Okun M. M., Eskridge E. M., Shields D. Truncations of a secretory protein define minimum lengths required for binding to signal recognition particle and translocation across the endoplasmic reticulum membrane. J. Biol. Chem. 1990;265:7478–7484. [PubMed] [Google Scholar]
- 37.Jenni S., Ban N. The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr. Opin. Struct. Biol. 2003;13:212–219. doi: 10.1016/s0959-440x(03)00034-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.