Abstract
The caspase-3 zymogen has essentially zero activity until it is cleaved by initiator caspases during apoptosis. However, a mutation of V266E in the dimer interface activates the protease in the absence of chain cleavage. We show that low concentrations of the pseudo-activated procaspase-3 kill mammalian cells rapidly and, importantly, this protein is not cleaved nor is it inhibited efficiently by the endogenous regulator XIAP (X-linked inhibitor of apoptosis). The 1.63 Å (1 Å = 0.1 nm) structure of the variant demonstrates that the mutation is accommodated at the dimer interface to generate an enzyme with substantially the same activity and specificity as wild-type caspase-3. Structural modelling predicts that the interface mutation prevents the intersubunit linker from binding in the dimer interface, allowing the active sites to form in the procaspase in the absence of cleavage. The direct activation of procaspase-3 through a conformational switch rather than by chain cleavage may lead to novel therapeutic strategies for inducing cell death.
Keywords: apoptosis, cancer therapy, conformational switch, co-operative dimer interface, procaspase activation
Abbreviations: Ac-DEVD-AFC, acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethylcoumarin; Ac-DEVD-CMK, acetyl-Asp-Glu-Val-Asp-chloromethyl ketone; bEVD-AOMK, biotinylhexanoyl-Asp-Glu-Val-acyloxymethane; D3A, procaspase-3(D9A,D28A,D175A); D3A,V266E, procaspase-3(D9A,D28A,D175A,V266E); HEK-293A cell, human embryonic kidney-293A cell; IL, intersubunit linker; PARP, poly(ADP-ribose) polymerase; V266E, procaspase-3(V266E); WT, wild-type procaspase-3; XIAP, X-linked inhibitor of apoptosis; Z-VAD-FMK, benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone
INTRODUCTION
Caspase activation, more than any other event, defines a cellular response to apoptosis. Synthesized initially as zymogens (procaspases) (Figure 1A), the cytosolic pool of inactive zymogen is converted rapidly into active protease upon the induction of apoptosis. A fundamental difference exists in the caspase subfamilies regarding maturation, and this difference is a key aspect for regulating apoptosis. Initiator procaspases are stable monomers in the cell, and dimerization is facilitated following recruitment to activation platforms [1]. Importantly, once dimerized, the initiator procaspases have high enzymatic activity, and subsequent chain cleavage simply stabilizes the active site [2,3]. In contrast, the effector procaspase-3 is a stable dimer, but has very low enzymatic activity (< 0.4% of that of the active protease) [4,5]. In this case, full activation occurs after cleavage of the IL (intersubunit linker) by initiator caspases, resulting in ordering of the active sites due to the release of two active site loops (L2 and L2′) from the IL and subsequent formation of the substrate-binding pocket (active site loop 3) (Figure 1B, and see Supplementary Movie S1 at http://www.BiochemJ.org/bj/424/0335/bj4240335add.htm). In short, the cell maintains a cytosolic pool of inactive procaspase-3 that is poised to carry out cell death.
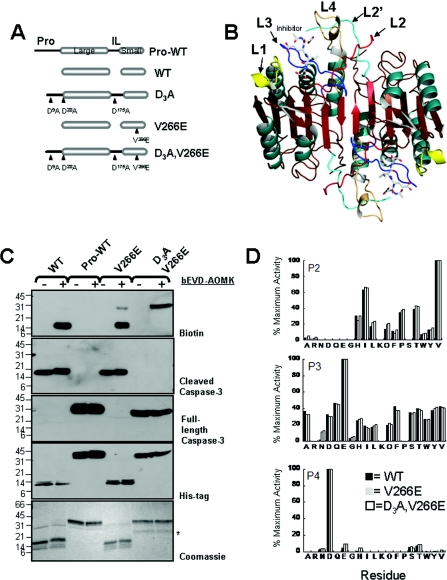
Figure 1. Procaspase-3(D3A,V266E) is enzymatically active without cleavage of the IL.
(A) The interface mutation V266E was designed in the context of wild-type caspase-3 (WT) and the uncleavable procaspase-3(D9A,D28A,D175A), called D3A. Low expression generates ‘one-chain’ procaspase-3 (Pro-WT). Overexpression generates the ‘two-chain’ caspase-3 (WT or V266E) by automaturation. ‘Pro’ refers to the pro-domain. (B) Structure of WT caspase-3 (PDB code 2J30) highlighting the active site loops L1 (yellow), L2 (red), L3 (blue), L4 (brown) and L2′ (cyan). The prime (′) indicates residues from the second monomer. For clarity, only one active site is labelled. (C) Labelling the V266E mutants by affinity-based probes. Proteins labelled with bEVD-AOMK were probed by Western blot analysis with anti-biotin, anti-cleaved caspase-3, anti-full-length caspase-3 or anti-His-tag antibodies, or subjected to trichloroacetic acid precipitation and stained with Coomassie Blue. The positive control was WT, and the negative control was Pro-WT. The asterisk indicates a contaminating protein from E. coli expression. Molecular masses are shown to the left in kDa. (D) Determining the substrate specificity of the recombinant caspase-3 mutants. Activity of the caspase-3 mutants (10 nM) was measured using a tetrapeptide positional scanning library, with P1 fixed as an aspartate residue and 7-amino-4-carbamoylmethylcoumarin as the fluorophore group. Hydrolysis rates are presented as a percentage of the maximum rate for each subset (P2, P3 and P4).
Structural and mutational studies show that the packing of amino acid side chains in the dimer interface is intimately connected to active site formation (reviewed in [6–9]). The importance of the dimer interface was described by Wells and co-workers, who showed that allosteric inhibitors bind in the interface of the mature caspase and stabilize a form of the protein with a disorganized active site, similar to that of the procaspase [10].
In addition to small molecule binding, mutations in the allosteric site of the dimer interface were shown to affect active site formation in procaspase-3 [11]. In one case, a V266E mutation increased the activity by 60-fold, representing a pseudo-activation of the zymogen. Notably, the increase in activity did not require cleavage of the IL but rather occurred via conformational changes in the intact zymogen. This is an important consideration, because the results demonstrated that procaspase-3 can indeed gain a substantial amount of catalytic activity without cleavage of the polypeptide chain.
An understanding of procaspase dimerization and how it relates to activation will be an important step toward effective therapies for a variety of diseases that involve the dysregulation of apoptosis. Although a number of cancer treatments target proteins in the apoptotic pathways [12], most of the current therapies are upstream of caspase activation and often require combined treatments to be effective [13]. Ultimately, however, these therapies indirectly induce the activation of caspase-3. Because there is a larger pool of quiescent procaspase-3 in most cancer cells compared with normal cells [14–16], directly targeting procaspase-3 could lead to more effective therapy, since effector caspases are the terminal proteases in the cell death cascade. Overall, our results from the present study show that the dimer interface should be considered a potential target for cancer treatment, where the identification of small molecules that bind to the interface of procaspase-3, resulting in its activation, may be a viable alternative to current therapies.
EXPERIMENTAL
Materials
Ac-DEVD-AFC (acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethylcoumarin) was prepared in the Salvesen laboratory, bEVD-AOMK (biotinylhexanoyl-Asp-Glu-Val-acyloxymethane; KMB01) was generously provided by Dr Matthew Bogyo (Stanford Comprehensive Cancer Center, Stanford, CA, U.S.A.), Z-VAD-FMK (benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone) was from Enzyme System Products, and Ac-DEVD-CMK (acetyl-Asp-Glu-Val-Asp-chloromethyl ketone) was from Calbiochem. Isopropyl β-D-thiogalactoside was from BioVectra.
Constructs
Plasmids expressing C-terminal His-tagged proteins (Figure 1A) have been described previously [11]. The following (pro)caspase-3 mutants were cloned into pcDNA3 (Invitrogen) as C-terminus FLAG-tag constructs: wild-type procaspase-3, procaspase-3(D9A,D28A,D175A), procaspase-3(V266E) and procaspase-3(D9A,D28A,D175A,V266E). These constructs are referred to as WT, D3A, V266E and ‘D3A,V266E’ respectively. pcDNA3 containing the gene for Bax and pcDNA/myc-XIAP (X-linked inhibitor of apoptosis) were gifts from Dr John Reed (The Burnham Institute for Medical Research, La Jolla, CA, U.S.A.). C-terminal His-tagged caspase-3 mutants were expressed and purified as described previously [11]. Full-length XIAP (N-terminal His tag) [17] and baculovirus protein p35 [18] were expressed and purified as described previously.
Labelling with affinity-based probes
Proteins were diluted in buffer [50 mM Hepes, pH 7.8, 100 mM NaCl and 1 mM DTT (dithiothreitol)] at 250 nM final concentration and incubated with bEVD-AOMK (2.5 μM) for 30 min at room temperature (~25 °C) in a total volume of 500 μl. Half the samples were immediately subjected to Western blot analysis, and the other half were concentrated ten times by precipitation with trichloroacetic acid (10%) and examined by SDS/PAGE (8–18% gradient gel), followed by Coomassie Blue staining.
Positional scanning libraries
Positional scanning substrate combinatorial libraries (P4, P3 and P2 positions) were synthesized similarly to the previously published methods [19–21], as summarized in the Supplementary online data (at http://www.BiochemJ.org/bj/424/bj4240335add.htm).
Cell culture and transfection
HEK-293A (human embryonic kidney-293A) cells were cultivated in Dulbecco's modified Eagle's medium. Jurkat and MCF-7 cells were cultivated in RPMI 1640 medium. The media were supplemented with 10% heat-inactivated bovine serum (Irvine Scientific), 2 mM L-glutamine and penicillin/streptomycin (Invitrogen). For transfection, the adherent cells, at 40–60% confluence, were grown either in 6- or 12-well plates and were transfected using GeneJuice® (Novagen), as suggested by the manufacturer, with 3 μl of transfectant reagent per 1.0–2.0 μg of total DNA in 0.1 ml of serum-free medium. The control or the dilution vector for transfected DNA was pcDNA3. At 2 h post-transfection, Z-VAD-FMK (100 μM) or DMSO was added to the cultured cells. The final concentration of DMSO was 0.2%. Cells were treated and harvested after 12–24 h, as described in the Figure legends. For flow cytometry experiments, cells were trypsinized on the plate, resuspended in the culture medium, washed with PBS and stained with Annexin V-PE using the Apoptosis Detection Kit (BioVision). Positive Annexin V-PE cells were analysed by FACS on a Becton Dickinson FACSort. Individual experiments were normalized by dividing each sample by the highest value (by Annexin V-PE staining) and multiplying by 100 to give ‘percentage maximum apoptosis’. Statistical analysis was performed using the paired Student's t test with two-tailed distribution. Untreated duplicate samples were processed for immunoblotting and caspase activity assays. Preparation of cell lysates and caspase activity measurements were performed as summarized in the Supplementary online data.
Western blot analysis and antisera
Cell lysates, balanced for total protein concentration, were examined by SDS/PAGE (8–18% polyacrylamide gradient gel). Proteins were transferred on to a PVDF membrane, blocked with 5% (v/v) non-fat dried skimmed milk powder in TBS-T (20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 0.1% Tween 20) and subjected to immunoblotting overnight at 4 °C against the primary antibodies [in TBS-T with 5% (v/v) non-fat dried skimmed milk powder]. The blot was washed with TBS-T followed by incubation for 1 h with the appropriate secondary antibody (1:10000) dissolved in TBS-T. The detection was performed using the femtomolar enhanced chemiluminessence kit from Pharmacia. The primary antibodies were used at the following dilutions: anti-FLAG (M2) 1:2000 (Sigma F3165); anti-PARP [poly(ADP-ribose) polymerase] 1:3000 (Pharmingen 556362); anti-cleaved PARP 1:1000 (Cell Signaling Technology #9541); anti-cleaved caspase-3 1:10000 (Cell Signaling Technology #9661); anti-full-length caspase-3 1:5000 (BD Transduction Laboratories clone 19); anti-ICAD 1:6000 (Cell Sciences PX024 and PX023); anti-Hsp-90 1:5000, anti-XIAP 1:2000 (BD Transduction Laboratories); and anti-His 1:1000 (Qiagen 34660). For biotin detection, the blots were blocked in 2% (w/v) BSA (in TBS-T) overnight at 4 °C, then avidin–horseradish peroxidase (e-Bioscience; 18-4100-51) was added at 1:5000 dilution for 30 min at room temperature. Blots were washed three times for 10 min with TBS-T prior to ECL detection.
Crystallization and data collection
Crystals of caspase-3(V266E) were grown, and results were collected as described previously [22], except that the cryoprotectant consisted of 90% reservoir buffer and 10% 2-methyl-2,4-pentanediol (Hampton Research). Crystals were grown in the presence of the Ac-DEVD-CMK inhibitor and appeared within 4 days. A full data set was collected to a resolution of 1.63 Å (1 Å=0.1 nm) at 100 K at the SER-CAT beamline (Advanced Photon Source, Argonne, IL, U.S.A.). V266E crystallized with the symmetry of the orthorhombic space group I222 with one heterodimer in the asymmetric unit. The biological dimer of heterodimers is generated through a 2-fold symmetry axis. The structure of V266E was determined using molecular replacement with the previously determined structure of caspase-3 for initial phasing (PDB entry 2J30). The inhibitor and all water molecules were removed from the initial model and all B-factors for protein atoms were set to 20 A3. Inhibitor and water molecules were added in subsequent rounds of refinement performed with COOT [23] and CNS [24] and were positioned based on 2Fo−Fc and Fo−Fc electron density maps contoured at the 1σ and 3σ levels respectively. Crystallographic data collection and refinement statistics are shown in Supplementary Table S1 (http://www.BiochemJ.org/bj/424/bj4240335add.htm). Procaspase-3 homology models were generated as summarized in the Supplementary online data. The atomic co-ordinates and structure factors for caspase-3(V266E) have been deposited in the PDB under accession code 3ITN.
RESULTS
Caspase-3(V266E) interface mutants are active before processing
The enzyme activity of D3A,V266E was determined previously from the hydrolysis of a typical caspase fluorogenic substrate, Ac-DEVD-AFC [11]. In those studies, Western blot analysis showed that the high enzyme activity of D3A,V266E could not be explained by an alternately cleaved protein because the enzyme activity was only ~ 3–4-fold lower than that of the mature caspase-3. We examined this further by reacting the mutants with an active site probe developed for caspases, bEVD-AOMK, which labels the catalytic cysteine contained in a competent active site. As shown in Figure 1(C, top panel), the probe covalently labelled the large subunit of WT (positive control), but not the intact single-chain wild-type procaspase-3 (negative control). Importantly, single-chain (uncleaved) D3A,V266E was labelled nearly as well as the large subunit of cleaved V266E and WT. Because D3A,V266E was shown previously to have enzymatic activity equal to that of the mature V266E [11], these results clearly show that the V266E mutation allows for activity of single chain caspase-3 in the absence of cleavage in the IL, at least on small synthetic substrates. Furthermore, the V266E mutation does not change the oligomeric properties of caspase-3 from high micromolar to picomolar range of protein concentration (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/424/bj4240335add.htm) [11].
In order to determine whether the mutation changed the substrate specificity of the enzyme, we tested the P2–P4 sites in the V266E mutants using a positional scanning peptide library containing an aspartate residue fixed in the P1 position (Figure 1D). The results show that there are no substantial differences between the specificity of WT and the V266E mutants for the P2–P4 positions, where maximum activity was obtained for the tetrapeptide sequence DEVD. Overall, the results show that the substrate specificities of the V266E mutants are very similar to that of wild-type caspase-3, and suggest that the active site of the activated procaspase resembles that of the mature caspase.
V266E interface mutants kill mammalian cells more efficiently than wild-type caspase-3
One predicts that expression of a constitutively active procaspase-3 should kill cells efficiently; indeed this has been shown previously for a circular permutation of the caspase that mimics IL cleavage [25]. However, it was not clear whether the ~ 3–4-fold lower activity of the V266E mutants compared with fully active wild-type caspase-3 represented sufficient activity for cell death. To examine this, we transiently transfected HEK-293A cells with various caspase-3 mutants and monitored cell viability by Annexin V staining (Figure 2A). Interestingly, both V266E and D3A,V266E mutants resulted in robust cell death (~ 50%), which exceeded that produced by WT and D3A (~ 20%). The loss in cell viability produced by the interface mutants was as pronounced as that produced by Bax, a cytotoxic protein that initiates the intrinsic apoptotic pathway at the mitochondrial level (Figure 2A). In all cases, the levels of apoptosis were decreased in the presence of a caspase inhibitor, Z-VAD-FMK, suggesting that the increased levels of cell death were dependent on caspase activity. When protein production was monitored by Western blot analysis, only the WT and D3A species were detected by their reactivity to anti-FLAG antibodies (Figure 2B). Compared with the endogenous protein, the levels of WT were approx. 5–10-fold higher in transfected cells, as judged by the anti-caspase-3 immunoblot. The interface mutants could not be detected even after prolonged exposures of the FLAG immunoblots, after immunoblotting with anti-cleaved caspase-3 antibodies or when the proteasome inhibitor MG132 was added (results not shown). Thus the full-length procaspase-3 observed in the cells transfected with the interface mutants (Figure 2B, top panel) represents the endogenous protein. Immunoblots using cell lysates prepared at earlier time points post-transfection gave the same results (results not shown). However, RT–PCR (reverse transcription–PCR) reactions showed the presence of mRNA for the caspase variants (Supplementary Figure S2 at http://www.BiochemJ.org/bj/424/bj4240335add.htm), demonstrating that the genes were transcribed. The parsimonious explanation for the lack of immunostaining of transfected V266E variants is that cells expressing them die before sufficient protein can accumulate, suggesting a lethal nature of the interface mutation. In support of this assertion, we replaced the catalytic cysteine residue in the V266E variants with a serine residue to remove activity. We find that the inactive V266E variants no longer support apoptosis (Supplementary Figure S3A at http://www.BiochemJ.org/bj/424/bj4240335add.htm). In addition, the proteins are observed by anti-FLAG antibody staining, although the accumulation remains lower than that for WT. These data show that the toxicity of the V266E variants requires catalytic activity.
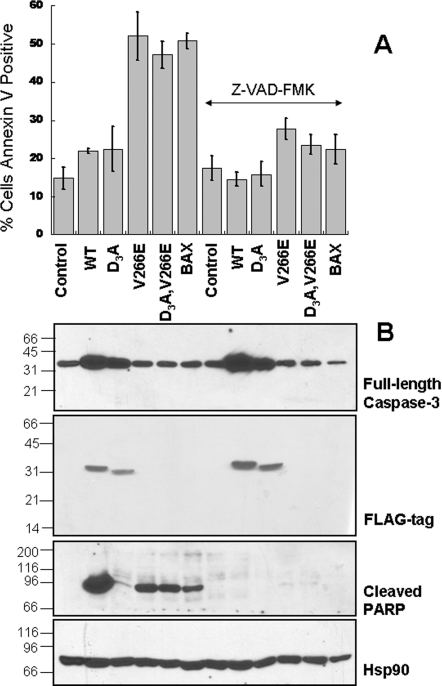
Figure 2. V266E mutants kill cells more efficiently than does the wild-type caspase-3.
(A) HEK-293A cells were transiently transfected with FLAG-tagged caspase-3 DNA (or 50 ng Bax/0.95 μg empty vector), and Annexin V-positive cells were quantified after 24 h. Z-VAD-FMK (100 μM) or DMSO was added to the cultures 2 h post-transfection. The values represent the means for three independent experiments±S.D. (B) Western blots of the cellular lysates from (A) against anti-full-length caspase-3 or anti-FLAG antibodies, for detection of the transfected constructs, or anti-cleaved PARP. The lower panel shows the loading control of Hsp90.
In order to show that the cell death mediated by the interface mutants involved typical caspase substrates, we examined the cleavage of the canonical substrate PARP (Figure 2B). Cleaved PARP exits the nucleus and resides in the cytosol, so we examined the presence of cleaved PARP in mRIPA (for composition see the Supplementary Experimental section)-soluble lysates. As expected, PARP was cleaved in all cells displaying high Annexin V staining, and its cleavage was suppressed by Z-VAD-FMK (Figure 2B). Cleaved PARP appeared to be in higher amounts in cells transfected with WT, which may indicate a higher degradation rate of cleaved PARP in cells transfected with the interface mutants or with Bax. We note that this experiment does not intend to quantify the amount of apoptosis, because the lysates were from cells collapsed in very late apoptosis, during which the process of protein degradation was advanced. The results in Figure 2(B) show the amount of cytosolic-cleaved PARP that leaked out of the nucleus at the specified time and had not been degraded yet by the proteasome. Owing to the fact that cells transfected with WT undergo very incipient apoptosis with no cleaved PARP degradation, the amount of cleaved PARP may seem higher in these cells in comparison with the cells transfected with V266E variants. However, in reality most of the PARP in the nucleus remained intact in cells transfected with WT, but was completely cleaved and removed from the nucleus in the V266E lysates. In addition, the cytosolic levels of cleaved PARP were much lower in the catalytically inactive C163S,V266E variants (Supplementary Figure S3B).
Overall, the results show that the constitutively active caspase-3 interface mutants were more efficient in killing transfected cells in culture than was wild-type caspase-3. Furthermore, the apoptotic activity of the V266E variants did not depend on the endogenous pool of procaspase-3. We transfected MCF7 cells, which lack endogenous caspase-3 [26], with plasmids containing the interface mutants, and we measured apoptosis by Annexin V staining (Supplementary Figure S4A at http://www.BiochemJ.org/bj/424/bj4240335add.htm). Although the transfection efficiency was lower than in the case of HEK-293A cells (~ 35% compared with ~ 60% respectively), the pattern of cell death observed in the HEK-293A cells (Figure 2A) was reproduced in the MCF7 cells. One should note that Bax, the positive control, was less toxic in MCF7 cells than were the interface mutants due to the lack of endogenous caspase-3, the main effector caspase. In all cases, the number of apoptotic cells diminished in the presence of the caspase inhibitor Z-VAD-FMK.
Similarly, we could show that recombinant V266E cleaved caspase substrates in the absence of caspase-3 when added to crude cellular extracts. We examined hypotonic lysates from Jurkat cells that were immunodepleted of the endogenous caspase-3 and reconstituted with recombinant caspase-3 proteins. The lysates were analysed by Western blotting for cleavage of ICAD (Supplementary Figure S4B). We observed that ICAD was cleaved efficiently by the active WT and V266E proteins, and it remained unprocessed upon addition of the less active procaspase-3. The cleavage of ICAD was judged by the disappearance of the full-length protein for which the antibodies were developed.
The intrinsic activity of interface mutants in transfected cells is lower than that of WT
We examined the threshold of caspase activity necessary to produce cell death by correlating the amount of death in cell culture with the total caspase activity in the corresponding lysates. We used the tetrapeptide substrate Ac-DEVD-AFC, which is cleaved primarily by effector caspases, to measure caspase activity in HEK-293A lysates that had been prepared from cells transfected with the interface mutants (same lysates as in Figure 2). Surprisingly, the lysates containing WT showed the highest DEVD-ase activity (Figure 3A). We expected to correlate this high activity to a high percentage of cell death in culture (see Figure 2A), which was not the case. In contrast, the lysates containing transfected interface mutants or Bax displayed less than 15% of the DEVD-ase activity of the WT lysates (Figure 3A), despite the high amounts of Annexin V-positive cells present in the cell culture (Figure 2A). Lysates prepared from cells transfected with WT displayed high DEVD-ase activity as early as 12 h post-transfection, and the activity increased over time (Supplementary Figure S5A at http://www.BiochemJ.org/bj/424/bj4240335add.htm). However, lysates prepared from cells transfected with V266E showed modest DEVD-ase activity compared with WT for each time considered, and the activity did not peak earlier than 24 h (Supplementary Figure S5A). Because the rate of cell death peaked as early as 16 h after transfection with V266E (results not shown), the results suggest that the apoptotic cells may have leaked the cytosolic caspase-3 [27]. In that case, DEVD-ase activity should be observed in the cell culture medium. However, when we measured caspase activity in the supernatant after removing the cells by centrifugation, we found that indeed there was DEVD-ase activity in the cellular medium (Supplementary Figure S5B), but the ratio between the activity of V266E mutants and the activity of WT reflected the ratio found in the lysates (Figure 3A). Therefore, the ‘missing’ DEVD-ase activity was not in the cellular medium.
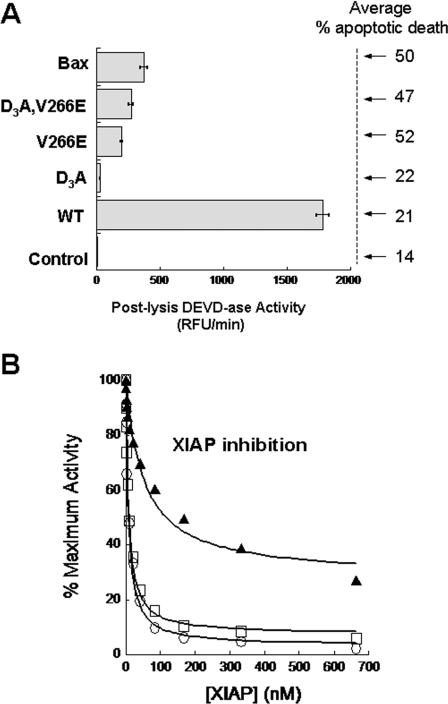
Figure 3. Wild-type caspase-3 lysates display higher DEVD-ase activity than those of V266E interface mutants.
(A) Lysates from cells transfected for 24 h with the indicated plasmids were assayed for enzymatic activity. The values represent the means ± S.D. for three independent experiments. Values for the mean percentage apoptotic death are from Figure 2(A). RFU, relative fluorescence units. (B) Caspase-3 mutants (300 pM) were incubated with XIAP at the indicated concentration in XIAP assay buffer (see Supplementary Experimental section) for 30 min at 37 °C, and the remaining activity was tested against Ac-DEVD-AFC (100 μM). The activity rates were plotted as the percentage of the maximum velocity in the absence of the inhibitor. The data were fitted to an equation describing the enzymatic activity in the presence of a reversible competitive inhibitor (see Supplementary Experimental section), and results of the fits are shown in Table 1.
XIAP is a poor inhibitor of D3A,V266E
The high DEVD-ase activity of caspase-3 in transfected cell lysates (Figure 3A) did not result in robust cell death (Figure 2A). It has been shown that cell death involves a XIAP-dependent threshold for activation of the effector caspases [28], where XIAP is an endogenous potent inhibitor of activated caspase-3 [29]. Our results suggest that caspase-3 could be inhibited in the cell by XIAP and that the mRIPA buffer (lysate preparation) and subsequent dilution into caspase assay buffer released XIAP from the active caspase-3, making measurement of the DEVD-ase activity possible. XIAP binds and inhibits cleaved caspase-3 via a two-step binding mechanism involving interactions with the active site and with the neo-epitope of the cleaved small subunit in L2′ [17]. Our data suggest that V266E becomes enzymatically active upon translation, without IL cleavage, so one predicts that XIAP will be a less potent inhibitor of the uncleaved procaspase D3A,V266E, since one binding site (L2′) may not be accessible to XIAP. In order to test this hypothesis, we performed inhibition studies using recombinant WT and V266E interface mutants (Figure 3B and Supplementary Figure S6 at http://www.BiochemJ.org/bj/424/bj4240335add.htm). The results show that XIAP efficiently inhibited WT and V266E, presumably due to the fact that both proteins are cleaved (see Figure 1B). The IC50 and the Ki values for the cleaved proteins were similar (Table 1) and close to the values reported previously [17]. In contrast, D3A,V266E was inhibited less efficiently by XIAP (Figure 3B), displaying a Ki value of ~ 8-fold higher in comparison with WT (Table 1). The results also showed that D3A,V266E could be inhibited by XIAP only up to ~ 70% of its maximum activity, compared with ~ 97% for WT (Table 1). This information, translated to the intracellular environment where the XIAP concentration does not exceed 50–70 nM [28], suggests that the uncleaved mutant will be mostly uninhibited in the cell. Because Asp175 in the IL of procaspase-3 is a poor substrate for cleavage by mature caspase-3 [30,31], we suggest that the V266E mutants remain unprocessed in the cell at low concentrations, effectively avoiding inhibition by XIAP. This explains the early and robust cell death upon transfection with caspase-3 interface mutants in comparison with WT. We have ensured that the active sites of the interface mutants were equally competent for binding other inhibitors that did not require interaction with the cleaved form of caspase-3. For example, we show that the interface mutants were inhibited by baculovirus p35 (Supplementary Figure S6C) similarly to wild-type caspase-3. Furthermore, we performed co-transfection experiments with plasmids encoding the interface mutants and XIAP (Supplementary Figure S7 at http://www.BiochemJ.org/bj/424/bj4240335add.htm). We observed that XIAP inhibited cell death only when present in large excess compared with the caspase plasmid (>5:1 XIAP/caspase). Our results are consistent with a low concentration of activated procaspase-3 carrying out apoptosis. This assertion is consistent with concentrations of active caspase-3 determined by molecular simulation (1 nM) in the absence of XIAP and proteasome degradation [32].
Table 1. XIAP inhibition constants for caspase-3 interface mutants.
| Caspase-3 protein | IC50 (nM) | Ki (nM) | Maximum inhibition (%) |
|---|---|---|---|
| WT | 7.4±0.8 | 1.75±0.25 | 97±2 |
| V266E | 7.6±0.5 | 1.88±0.11 | 93±1.2 |
| D3A,V266E | 51.4±11.5 | 13.4±2.6 | 72±4.6 |
The X-ray crystal structure of V266E
In order to examine structural changes caused by the replacement of Val266, we crystallized and determined the structure of cleaved V266E to 1.63 Å (Supplementary Table S1). The motivation for replacing Val266 with a glutamate residue was prompted by the hydrophilic dimer interface of caspase-1, which contains Glu390 (equivalent to Val266 in caspase-3) and Arg391. Glu390 forms a salt bridge with Arg286 (Arg164 in caspase-3), which is on L2, neutralizes the positive charge from that residue and appears to stabilize the active site.
Although the results show that the structure of V266E is very similar to that of WT (PDB code 2J30), with an overall RMSD (root mean square deviation) of 0.14 Å, two regions of the protein are different. Changes in one or both regions presumably result in the 3–4-fold lower enzyme activity. The first is derived from the IL, which in the uncleaved procaspase-3 is made of residues 169–190. Upon cleavage at Asp175 during maturation, residues 169–175 form L2 in the active site of one heterodimer and residues 176–190 reach across to the other heterodimer to form the active site L2′. L2′ is partially disordered in V266E, where we observe little or no electron density for amino acids 176–185. This is in contrast with the structure of WT, where the disorder is from residues 176–179. In WT, several residues in L2′ form a H-bonding network through water molecules with the P4 aspartate of the substrate (Figure 4A) [22], and the removal of these interactions was shown to decrease activity [33]. In V266E, we observe electron density starting with Lys186. Importantly, His185′ in WT forms two H-bonds with Thr245 at the C-terminus of helix 5. The contacts across the interface anchor the base of L4 with L2′, and these interactions are not observed in the mutant. Disruption of contacts at the C-terminus of helix 5 was shown previously to decrease the activity of procaspase-3 [34].
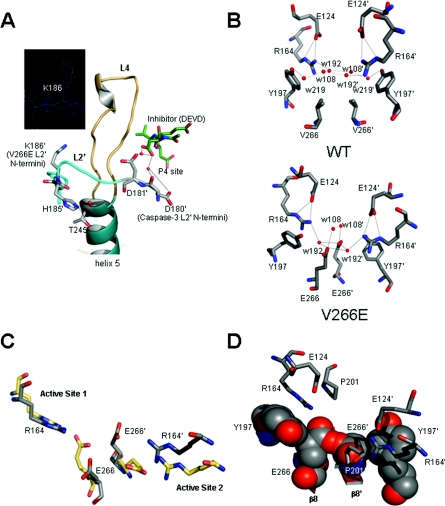
Figure 4. V266E changes the dimer interface.
(A) Interactions in L2′ (cyan) with L4 (brown) and the P4-binding site of WT. Inset: N-termini of L2′ for V266E begins at Lys186. (B) Dimer interface of WT (upper panel) demonstrating neutralization of the positive charge of Arg164 and six conserved water molecules (see Supplementary online data), and dimer interface of V266E (lower panel). Contacts between Glu124 and Arg164 are maintained in V266E, but two water molecules are removed and two are displaced by the carboxylate of Glu266. (C) Comparison of the dimer interface of V266E (grey) with that of caspase-1 (yellow). The rotamer at Glu390 in caspase-1 allows closer contact with the active site arginine. (D) V266E modelled using the rotamer found in caspase-1 demonstrates intra- and inter-subunit steric clashes with Tyr197 and Glu266′.
Secondly, the dimer interface of V266E has features in common with wild-type caspase-3 and with caspase-1, although Glu266 in the caspase-3 variant utilizes a different rotamer than does Glu390 in caspase-1. In this case, Glu266 interacts with Arg164 through a conserved water molecule (see Supplementary online data) rather than through direct interactions as observed in caspase-1 (Figures 4B and 4C). Interestingly, when Glu390 is replaced with an aspartate residue in caspase-1, the salt bridge with Arg286 is preserved but is mediated by a water molecule [9]. In WT, the positive charge of Arg164 is neutralized by Glu124, and these contacts are maintained in the mutant (Figure 4B). When the structure is superimposed with that of caspase-1, one finds that only one of the two active sites aligns well (Figure 4C). When we modelled V266E with the same rotamer as that of caspase-1, we found steric clashes between Glu266 and Glu266′, across the dimer interface, and between Glu266 and Tyr197, on the adjacent β-strand (Figure 4D). This issue is alleviated in caspase-1 because of the presence of an insertion in β-strand 8, Arg391, which allows Glu390 to adopt a rotamer that moves the carboxylate closer to Arg286 so that the two side chains are within H-bonding distance (2.8 Å), and because a cysteine residue (Cys331) is present at the position equivalent to Tyr197 in WT. So, the rotamer adopted for Glu266 in the caspase-3 mutant seems to be somewhat of a compromise between avoiding steric clashes with neighbouring amino acids and optimizing favourable interactions with neighbouring residues and nearby water molecules.
Models of (in)active procaspase-3
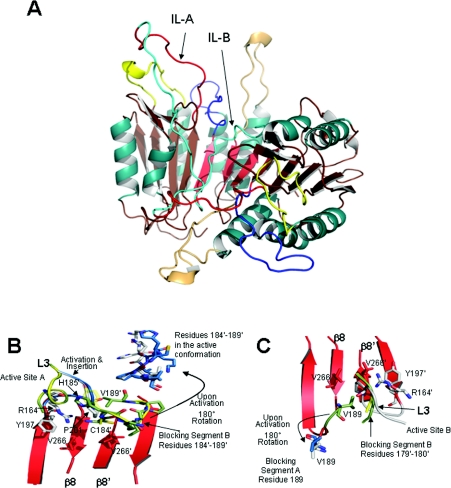
Our previous biochemical data for D3A,V266E [11], as well as that provided in the present study, show that the V266E mutation effectively shifts the zymogen to an active conformer. Currently there are no structures for procaspase-3, so we generated homology models of the putative active and inactive zymogen in order to examine the conformational switch. Owing to their high sequence identity, we modelled inactive WT after procaspase-7 [35,36], and energy minimized the structures as described in the Experimental section (and the Supplementary online data) to assure that our final model is energetically feasible. We find that, similar to procaspase-7, the active sites in our model of the inactive zymogen are disorganized and the IL occupies the central cavity. There are two monomers, represented by chains A and B, forming a complete dimer in the asymmetric unit of the procaspase-7 X-ray crystal structure. Our model of the inactive procaspase-3 is similar to the entire dimer. Although the IL from chain A (IL-A) in the procaspase-7 template mostly remains exposed to solvent, a large portion of IL-B is buried in the interface, making contacts with several interface residues and shielding the hydrophobic core from water (Figure 5A).
Figure 5. Homology models of (in)active procaspase-3.
(A) Model of inactive procaspase-3 demonstrating binding of the IL (cyan) in the dimer interface, preventing organization of the active site loops. Colour code is the same as that used in Figure 1(B). Note that L2 and L2′ are covalently connected in the IL. (B) Superposition of inactive procaspase-3 (green residues) and of procaspase-7 (yellow) shows the blocking segment of IL-B, residues 184′–189′ (procaspase-3 numbering), prevents insertion of active site loop 3 from monomer A. For clarity, only one residue from the blocking segment of procaspase-7 is shown (semi-transparent sticks, Tyr211), whereas all of the residues of the blocking segment of procaspase-3 are highlighted (green). Upon cleavage of the IL, L2′, where the blocking segment resides, rotates ~ 180 ° and vacates the interface. Subsequently, a portion of the substrate-binding loop is permitted to insert in the interface. Arg164, Tyr197 and Pro201 engage in a stacking interaction (shown as the white residues) once L3 inserts in the interface. Insertion of the substrate-binding loop is permitted in the active procaspase-3 (blue ribbon) as the blocking segment lifts out of the interface upon activation (blue residues). (C) Superposition of procaspase-7 (yellow) and of inactive procaspase-3 (green) reveals a blocking segment involving residues 179′–180′ (caspase-3 numbering) of IL-B and Val189 of IL-A, preventing insertion of L3 in the active site of monomer B. L3 (white ribbon, WT) cannot insert into the interface until the blocking segment (green, inactive procaspase-3; yellow, procaspase-7) vacates the interface.
An examination of the model for inactive procaspase-3 shows two features of the protein that may be involved in maintaining the inactive conformer. First, the dimer is asymmetric due to the binding of residues Val178–Glu190 from IL-B in the interface. As a result of the binding of IL-B, only residues His185–Glu190 from IL-A are observed near the interface, and these residues mostly interact with active site B. The asymmetric binding of the two ILs leads to inhibition of the two active sites (from chains A and B) in two complementary manners. For active site A, residues Cys184–Val189 from IL-B prevent insertion of active site loop 3 (chain A) into the dimer interface (Figure 5B). Cys184 and Val189 (IL-B) in the inactive procaspase-3 reside in the region occupied by Pro201 (chain A) in the active caspase-3. A similar sequence in caspase-7 was referred to as the ‘blocking segment’ because the residues prevent formation of the substrate-binding pocket [35]. In addition, steric clashes between the side chain of His185 (IL-B) and the backbone atoms of Gly202 (L3) and Arg164 (L2) (chain A) probably prevent rotation of L2 from chain A into the active conformation, where the side chain of Arg164 rotates into the dimer interface to intercalate between Pro201 and Tyr197, and the catalytic Cys163 rotates into the P1 site (see Figure 5B, grey model). Similar steric constraints are observed in procaspase-7 (Figure 5B, yellow model). In the active procaspase-3, as well as upon cleavage of the IL to yield caspase-3, removal of the blocking segment of IL-B from the interface allows Pro201 from chain A to move ~ 6 Å into the active conformation, thus forming the substrate-binding pocket for active site A. For active site B, a blocking segment also prevents insertion of Pro201 and the substrate-binding pocket, but the interactions differ due to the asymmetric nature of the inactive procaspase-3 dimer (Figure 5C). In this case, Val189 (IL-A) resides in the region occupied by Pro201 (chain B) in the active caspase-3, so Val189 behaves similarly for both ILs. However, the remainder of the blocking segment is comprised of the backbone atoms of residues 179–182 from IL-B, which also reside in the region occupied by Pro201-Tyr203 of the active caspase-3. Thus, although the mechanisms differ somewhat for each active site, the substrate-binding loops of both active sites are prevented from inserting into the interface because the interface is occupied by segments of IL-A and IL-B.
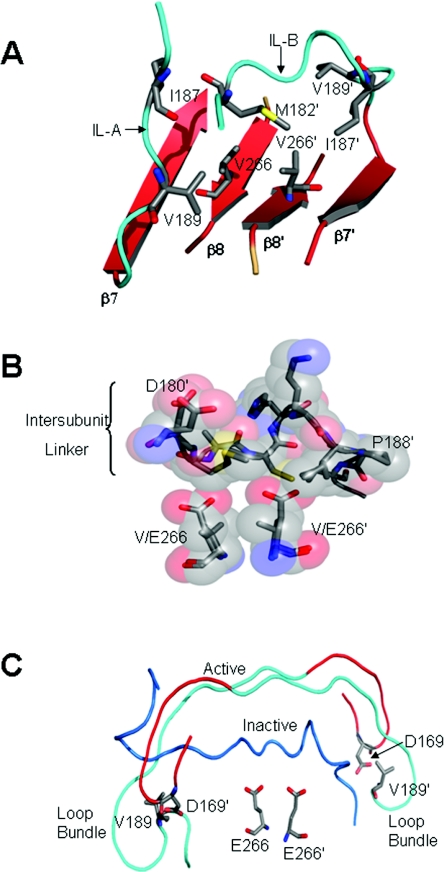
In addition to the blocking segments, a cluster of hydrophobic amino acid residues centred about Val266 appears to stabilize the inactive conformer (Figure 6A), and a substitution of a glutamate residue for Val266 in the model of the inactive procaspase-3 suggests a mechanism for activation resulting from the mutation. The hydrophobic cluster is comprised of seven residues: Met182′, from IL-B, and Ile187, Ile187′, Val189, Val189′, Val266, and Val266′ from both monomers (Figure 6A). On the basis of this analysis, we suggest that V266E activates the procaspase by disrupting the hydrophobic cluster and introducing steric clashes with Met182′ and Ile187′ (Figure 6B), essentially expelling the IL from the dimer interface: the first and most important step in activation. Because the glutamate side chain is approx. 2.5 Å longer than that of valine, the IL is prevented from rebinding in the interface after its release, and the protein conformation shifts to the active state. It is important to note that the conformational shift occurs without cleavage of the polypeptide chain at Asp175. In the active conformer, both ILs occupy a region above the central cavity, but have very few stabilizing contacts with the protease domain (Figure 6C). Ramachandran plots show that the backbone torsion angles are satisfactory (Supplementary Table S1), and energy minimization was performed in CHARMm [37] to alleviate energetically unfavourable conformations within the protein (see Supplementary Experimental section at http://www.BiochemJ.org/bj/424/bj4240335add.htm). Whether or not the conformational details are correct, the most important revelation obtained from our model of the active procaspase-3 (Figure 6C) is that the IL is indeed long enough to allow formation of the loop bundle at both active sites (contacts between Asp169 and Val189′). Similar to caspase-7, removal of the blocking segment from the central cavity allows L3 to move into the dimer interface, Arg164 to rotate out of the substrate-binding pocket and into the interface, and the catalytic groups to move into their active positions so that the active site is organized similarly to the mature caspase-3. Because L2 and L2′ remain covalently connected in the IL, however, putative stabilizing interactions provided by L2′ with the P4 aspartate of the substrate, described above, are not possible in the procaspase. Supplementary Movie S2 (at http://www.BiochemJ.org/bj/424/0335/bj4240335add.htm) shows the conformational switch between inactive and active procaspase-3.
Figure 6. V266E expels the IL from the interface.
(A) Hydrophobic cluster in the inactive procaspase-3 centred about Val266. Residues in the IL (cyan) of both monomers contribute to the cluster. (B) Because the glutamate side chain is longer than that of valine, the V266E mutation probably disrupts the hydrophobic cluster, preventing the IL from binding in the interface. (C) Comparison of the IL in the inactive and active procaspase-3. H-bonds contributed by the loop bundle, and centred about Asp169, are critical to active site stabilization. These contacts form only in the active conformer.
DISCUSSION
We show that the variant (pro)caspase-3 kills mammalian cells rapidly compared with wild-type caspase-3. D3A,V266E is active in the cell upon translation, and before processing at Asp175, and cleaves substrates similarly to the mature caspase. These features are important for two reasons. First, endogenous procaspase-3 is a poor substrate for D3A,V266E, so cell death occurs due to the activity of the procaspase variant on cellular substrates rather than its activation of endogenous procaspase-3. In support of this conclusion, we showed that MCF-7 cells, which lack procaspase-3, undergo apoptosis when transfected with D3A,V266E. Secondly, inhibition of the mutant by XIAP is weak because the procaspase is not processed. In this case, the neo-epitope at the N-terminus of L2′, which serves as a second binding region for XIAP, remains sequestered in the procaspase.
The low threshold of activated procaspase-3 is much lower than the total cellular concentration of caspase-3, which is ~ 100 nM in apoptotic Jurkat [4] and HEK-293 cells [30]. The concentration of the zymogen appears to be higher than that of mature caspase-3 [38], although HeLa cells are estimated to contain approx. 120 nM procaspase-3 [28]. Under normal cellular conditions, XIAP concentrations of 50–60 nM [28] are sufficient to inhibit the small amount of activated caspase-3 and to target the protein for degradation by the proteasome [39]. The induction of apoptosis leads to massive activation of endogenous procaspase-3, which overwhelms the control system and results in rapid cell death [32]. Because XIAP does not efficiently inhibit D3A,V266E, a low concentration of the pseudo-activated procaspase is sufficient for rapid cell death.
The interface mutation has little effect on the structure of the mature caspase, but it appears to have a larger effect on that of the procaspase. Our structural data show that L2′ is disordered in V266E, and conserved water molecules are displaced at the interface. At present, it is not clear how a mutation in the interface affects ordering of L2′, since the regions that are affected are >25 Å from the site of the mutation. Although residues in L2′ H-bond with the P4 carboxylate of the substrate, removal of those interactions do not significantly affect the enzymatic reaction. Val266 resides at the centre of β-strand 8 in a water-filled cavity, with the closest water molecules approx. 3.5 Å from the hydrophobic side chains, well within van der Waals contacts. As described by Wells and co-workers, this region of the interface encompasses a site for binding of allosteric inhibitors, which prevent active site formation, in general, by preventing the insertion of the substrate-binding loop (L3) into the active site cavity [10]. This is important because part of L3 extends into the dimer interface and forms new contacts with residues in L2 in addition to forming the base of the substrate-binding pocket. The allosteric binding pocket is occluded in the procaspase due to the positioning of the IL in the interface, and the ‘blocking segment’ of the IL prevents insertion of L3 into the active site. Our results from the present study suggest that the inhibition is alleviated in D3A,V266E so that the IL is removed from the allosteric site, allowing the active site loops to organize. Thus the allosteric site linked to inhibition of the active caspase also stabilizes the inactive procaspase.
The inactive conformer appears to be stabilized by hydrophobic and charge–charge interactions across the dimer interface, and these stabilizing contacts are disrupted in the active conformer. Upon removal from the interface-binding site, the IL moves across the surface of the protein to form the critical contacts contributed by Asp169, and the active conformer is stabilized primarily by backbone H-bonds between the two ILs and those that form at Asp169. Stabilizing interactions in the mature caspase, contributed by L2, L2′, L3 and L4, are unable to form in the active procaspase. The lack of these stabilizing interactions may explain, in part, why the inactive procaspase-3 is favoured.
Learning to selectively manipulate the level of apoptosis is expected to lead to therapeutic strategies for a number of diseases in which the dysregulation of apoptosis is a common factor. Cancer cells, for example, typically have gained the ability to circumvent apoptosis, so the proteins in the apoptotic cascades have become ideal targets for cancer therapy [40–42]. We suggest that directly targeting procaspase-3 could lead to more effective therapy and lower drug resistance, since effector caspases are the terminal proteases in the cell death programme. Our results suggest that small molecules could bind to the dimer interface of the zymogen and effectively mimic the effects of the V266E mutation by releasing the IL from the allosteric site in the interface. One advantage of this strategy, as shown in the present study, is that the activation of endogenous procaspase-3 could occur without cleavage at Asp175 and may circumvent inhibition by XIAP. Thus, compounds that induce a conformational change in the zymogen, as does the V266E mutation, may show great potential in drug design strategies. Modulation of caspase inactivation from the dimer interface has been proposed before [10], but we are the first to show that the reverse mechanism, activation, is possible by manipulating the dimer interface.
Online data
AUTHOR CONTRIBUTION
A. Clay Clark and Guy Salvesen designed the study. Jad Walters, Paul Swartz, Carla Mattos and A. Clay Clark carried out crystallographic and modelling studies. Cristina Pop, Fiona Scott and Guy Salvesen carried out cell biological studies. Marcin Drag synthesized the peptide library. Jad Walters, Cristina Pop, Carla Mattos, Guy Salvesen and A. Clay Clark wrote the manuscript.
ACKNOWLEDGEMENTS
We thank the research agencies of North Carolina State University and the North Carolina Agricultural Research Service for continued technical support.
FUNDING
This work was supported by a grant from the NIH (National Institutes of Health) [grant number GM065970 (to A.C.C.)]. The crystallography and modelling work were supported by an NIH grant [grant number CA096867 (to C.M.)]. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract number W-31-109-ENG-38.
References
- 1.Boatright K. M., Salvesen G. S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pop C., Fitzgerald P., Green D. R., Salvesen G. S. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry. 2007;46:4398–4407. doi: 10.1021/bi602623b. [DOI] [PubMed] [Google Scholar]
- 4.Pop C., Chen Y.-R., Smith B., Bose K., Bobay B., Tripathy A., Franzen S., Clark A. C. Removal of the pro-domain does not affect the conformation of the procaspase-3 dimer. Biochemistry. 2001;40:14224–14235. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- 5.Bose K., Pop C., Feeney B., Clark A. C. An uncleavable procaspase-3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase-3. Biochemistry. 2003;42:12298–12310. doi: 10.1021/bi034998x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacKenzie S. H., Clark A. C. Targeting cell death in tumors by activating caspases. Curr. Cancer Drug Targets. 2008;8:98–109. doi: 10.2174/156800908783769391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuentes-Prior P., Salvesen G. S. The protein structures that shape caspase activity, specificity activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y. Caspase activation, inhibition, and reactivation: a mechanistic view. Protein Sci. 2004;13:1979–1987. doi: 10.1110/ps.04789804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta D., Scheer J. M., Romanowski M. J., Wells J. A. An allosteric circuit in caspase-1. J. Mol. Biol. 2008;381:1157–1167. doi: 10.1016/j.jmb.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy J. A., Lam J., Nguyen J. T., O'Brien T., Wells J. A. Discovery of an allosteric site in the caspases. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pop C., Feeney B., Tripathy A., Clark A. C. Mutations in the procaspase-3 dimer interface affect the activity of the zymogen. Biochemistry. 2003;42:12311–12320. doi: 10.1021/bi034999p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed J. C. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 13.Diehl V., Stein H., Hummel M., Zollinger R., Connors J. M. Hodgkin's lymphoma: biology and treatment strategies for primary, refractory, and relapsed disease. Hematology. 2003;2003:225–247. doi: 10.1182/asheducation-2003.1.225. [DOI] [PubMed] [Google Scholar]
- 14.Estrov Z., Thall P. F., Talpaz M., Estey E. H., Kantarjian H. M., Andreeff M., Harris D., Van Q., Walterscheid M., Kornblau S. M. Caspase 2 and caspase 3 protein levels as predictors of survival in acute myelogenous leukemia. Blood. 1998;92:3090–3097. [PubMed] [Google Scholar]
- 15.Krepela E., Prochazka J., Fiala P., Zatloukal P., Selinger P. Expression of apoptosome pathway-related transcripts in non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2006;132:57–68. doi: 10.1007/s00432-005-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigoriev M. Y., Pozharissky K. M., Hanson K. P., Imyanitov E. N., Zhivotovsky B. Expression of caspase-3 and -7 does not correlate with the extent of apoptosis in primary breast carcinomas. Cell Cycle. 2002;1:337–342. [PubMed] [Google Scholar]
- 17.Scott F. L., Denault J.-B., Riedl S. J., Shin H., Renatus M., Salvesen G. S. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riedl S. J., Renatus M., Snipas S. J., Salvesen G. S. Mechanism-based inactivation of caspases by the apoptotic suppressor p35. Biochemistry. 2001;40:13274–13280. doi: 10.1021/bi010574w. [DOI] [PubMed] [Google Scholar]
- 19.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Carcia-Calvo M., Hotzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 20.Harris J. L., Backes B. J., Leonetti F., Mahrus S., Ellman J. A., Craik C. S. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drag M., Mikolajczyk J., Krishnakumar I. M., Huang Z., Salvesen G. S. Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem. J. 2008;409:461–469. doi: 10.1042/BJ20070940. [DOI] [PubMed] [Google Scholar]
- 22.Feeney B., Pop C., Swartz P., Mattos C., Clark A. C. Role of loop bundle hydrogen bonds in the maturation and activity of (pro)caspase-3. Biochemistry. 2006;45:13249–13263. doi: 10.1021/bi0611964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Panni N. S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasula S. M., Ahmad M., MacFarlane M., Luo Z., Huang Z., Fernandes-Alnemri T., Alnemri E. S. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J. Biol. Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 26.Janicke R. U., Sprengart M. L., Wati M. R., Porter A. G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 27.Krysko D. V., Vanden Berghe T., D'Herde K., Vandenabeele P. Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods. 2008;44:205–221. doi: 10.1016/j.ymeth.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Rehm M., Huber H. J., Dussmann H., Prehn J. H. M. Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J. 2006;25:4338–4349. doi: 10.1038/sj.emboj.7601295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckelman B. P., Salvesen G. S., Scott F. L. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stennicke H. R., Jurgensmeier J. M., Shin H., Deveraux Q., Wolf B. B., Yang X., Zhou Q., Ellerby M., Ellerby L. M., Bredesen D., et al. Pro-caspase-3 is a major physiologic target of caspase-8. J. Biol. Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 31.Liu H., Chang D. W., Yang X. Interdimer processing and linearity of procaspase-3 activation. J. Biol. Chem. 2005;280:11578–11582. doi: 10.1074/jbc.M414385200. [DOI] [PubMed] [Google Scholar]
- 32.Albeck J. G., Burke J. M., Aldridge B. B., Zhang M., Lauffenburger D. A., Sorger P. K. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol. Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesan R., Mittl P. R. E., Jelakovic S., Grutter M. G. Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis. J. Mol. Biol. 2006;359:1378–1388. doi: 10.1016/j.jmb.2006.04.051. [DOI] [PubMed] [Google Scholar]
- 34.Feeney B., Pop C., Tripathy A., Clark A. C. Ionic interactions near loop L4 are important for maintaining the active site environment and the dimer stability of (pro)caspase-3. Biochem. J. 2004;384:515–525. doi: 10.1042/BJ20040693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riedl S. J., Fuentes-Prior P., Renatus M., Kairies N., Krapp S., Huber R., Salvesen G. S., Bode W. Structural basis for the activation of human procaspase-7. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai J., Wu Q., Shiozaki E., Srinivasula S. M., Alnemri E. S., Shi Y. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 37.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 38.Saunders P. A., Cooper J. A., Roodell M. M., Schroeder D. A., Borchert C. J., Isaacson A. L., Schendel M. J., Godfrey K. G., Cahill D. R., Walz A. M., et al. Quantification of active caspase 3 in apoptotic cells. Anal. Biochem. 2000;284:114–124. doi: 10.1006/abio.2000.4690. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki Y., Nakabayashi Y., Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogelstein B., Kinzler K. W. Achilles' heel of cancer? Nature. 2001;412:865–866. doi: 10.1038/35091170. [DOI] [PubMed] [Google Scholar]
- 41.Kirkin V., Joos S., Zornig M. The role of Bcl-2 family members in tumorigenesis. Biochem. Biophys. Acta. 2004;1644:229–249. doi: 10.1016/j.bbamcr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Soengas M. S., Capodleci P., Polsky D., Mora J., Esteller M., Opitz-Araya X., McComble R., Herman J. G., Gerald W. L., Lazebnik Y. A., et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.