Abstract
Epithelial–mesenchymal interactions play a key role in the development of tissues such as tooth, lungs, and kidneys. To successfully engineer or repair such living tissues it is necessary to first understand the complex cell–cell and cell–matrix interactions underlying organogenesis. To mimic an in vivo setting it is necessary to assemble a three-dimensional matrix that would facilitate cell–cell interaction leading to proliferation and cellular differentiation. In this study, we have developed an in vitro three-dimensional multilayered coculture system using type I collagen and chitosan blends as matrices, to study epithelial–mesenchymal interactions that occur during tooth morphogenesis. Results from this study showed that the matrix composition influenced the migration, proliferation, and differentiation properties of the epithelial and mesenchymal cells. Specifically, the system supported the migration and differentiation of the HAT-7 epithelial cells and mesenchymal-derived dental pulp stem cells. Results from the in vivo implantation study of the coculture system in mice demonstrated a similar cellular migration and differentiation pattern that corroborates well with the in vitro model. Interestingly, the biopolymer matrix also permitted neovascularization in vivo.
Introduction
From the perspective of tissue engineering and regenerative medicine, it is of utmost importance to understand and emulate the complex interactions between multiple cell types that make up a tissue or organ. These interactions are carried out in a three-dimensional (3D) setting in vivo. To achieve such architecture in an in vitro model, a complex matrix system comprising appropriate bio-polymers and cell types should be employed. Such a scaffold should be fine-tuned to suit the intrinsic properties of various cell types to be used in specific tissue engineering applications.
Knockout mouse models have commonly been used to dissect complex interactions and they can be quite useful as defects and loss of development can be traced easily.1 However, knocking down certain genes may prove to be embryonically lethal, or conditional knockout may abrogate organogenesis. In the former case, no functional information can be obtained except for the fact that the gene is of extreme importance and in the latter case a similar problem arises wherein the importance of the knocked out gene at different stages of morphogenesis remains unknown. An alternative methodology to combat these limitations is to develop an in vitro coculture model to study the interactions between the involved cell types. The need for development of coculture models to study the interactions between different cell types and their importance have been extensively discussed in literature.2,3
In this study, we have specifically developed a coculture system to study epithelial–mesenchymal interactions during tooth formation. This interaction is one example of many complex and specific interactions occurring during organogenesis. Odontogenesis is a complex process that has been characterized as a series of inductive and reciprocal epithelial–mesenchymal interactions leading to proliferation, polarization, and differentiation of these cells, culminating with the formation of mineralized dentin and enamel.4
During tooth morphogenesis, epithelial–mesenchymal interactions are facilitated by several key molecules belonging to multiple conserved families. The primary players include growth factors such as transforming growth factor β, bone morphogenetic proteins (BMPs), fibroblast growth factor, transcription factors such as Cbfa1, Lhx6,7,4–8 and signaling molecules such as members of the hedgehog and Wnt family, dentin matrix protein 1 (DMP1), amelogenin, and dentin sialophosphoprotein.9–13 The functions of these signaling molecules, transcription factors, and extra cellular matrix (ECM) proteins have been studied in cell culture systems and knockout mouse models.
Recently, tissue recombination experiments8,14,15 have shown that recombined epithelia and mesenchymal tissues from developing embryonic tooth germ can form tooth-like structures in vitro and in vivo. Although these represent engineered tooth-like structures, they only serve to demonstrate the potential of these embryonic cells rather than their functionality in regenerative medicine. Development of an adult stem cell–based system that more closely resembles the in vivo scenario would be beneficial to study cellular interactions and to identify key players that are involved during epithelial and mesenchymal cell differentiation. Engineering the mammalian tooth has been a challenge for tissue engineers due to the multitude of interactions between the cell types involved and the complexity of the structure. Therefore, in this study we describe the development of a coculture model using HAT-7 dental epithelial cells and mesenchymal-derived dental pulp stem cells (DPSCs) embedded in a biomimetic collagen and chitosan copolymer matrix, to study the interactions between the two predominant cell types involved in enamel and dentin formation. HAT-7 and DPSCs are established dental epithelial and mesenchymal precursor cells. They can be cocultured with the same culture medium (Dulbecco's modified Eagle's medium/F12 with 10% fetal bovine serum), and hence were selected as representative cells for studying their differentiation process in the biopolymeric matrix. Collagen and chitosan are both naturally occurring biopolymers that have been shown to support 3D cell culture.
Materials and Methods
Cell culture
The cell types used in this study are human DPSCs (a gift from Dr. Songtao Shi, University of Southern California, Los Angeles, CA) and HAT-7 dental epithelial cells (a gift from Dr. Hidemitsu Harada, Osaka University, Osaka, Japan). DPSCs have been well characterized and have been shown to differentiate both in vitro and in vivo.16,17 HAT-7 cells have been shown to differentiate into ameloblasts and have been characterized as dental epithelial cells.18,19 Both cell types require the same culture conditions and were cultured in Dulbecco's modified Eagle's medium/F12 with 10% fetal bovine serum and 1% antibiotics.
Biomaterials
Two naturally occurring biopolymers, namely, type I collagen (BD Biosciences, San Jose, CA) (coll) and chitosan (Sigma, St. Louis, MD), were used in this study for the construction of the scaffold.
Preparation of collagen and chitosan hydrogel blends
Type I collagen and chitosan-blended hydrogels were prepared as described by Tan et al.20 Briefly, the hydrogels contained either pure collagen at 1 mg/mL or blends of collagen and chitosan with 1 mg/mL of type I collagen and 1–3 mg/mL chitosan in the growth medium with 1 × Hank's balanced salt solution and 5% (w/v) NaHCO3. Both collagen and chitosan are monomers in 0.1 M acetic acid. To aid polymerization of the monomeric mixture, the pH of the mixture was raised by the addition of 1 M NaOH solution and the mixture was incubated at 37°C and 5% CO2. The volume of NaOH to be added was estimated by titration for each of the different blends of collagen and chitosan. The cells were added to the monomer solutions before the addition of NaOH. Imaging of the actin fibrils of the embedded cells within the 3D construct was performed by fixing the construct in formalin and staining with phalloidin conjugated to rhodamine. A Zeiss LSM 510 laser scanning confocal microscope equipped with the appropriate filter sets was used.
Determination of the mechanical property of the scaffold using atomic force microscopy microindentation
Microindentation by atomic force microscopy (AFM) was performed as described by Titushkin et al.21 Force curves were obtained from at least 75 spots on the hydrogels. A bidomain polynomial was fit to the force curves using a standard least squares minimization algorithm. The polynomial was linear for the precontact period and was fit to Hertz equation for postcontact with the contact point adjustable. The hydrogels were immersed in a solution of phosphate-buffered saline at all times and were never allowed to dry before or during the experiment.
Cell proliferation experiment
DPSCs and HAT-7 cells were seeded at a density of 20,000 cells/well containing 50 μL of hydrogel in 96-well tissue culture plates. Four different hydrogels containing increasing concentrations of chitosan were used having 1 mg/mL of coll as constant, namely, 1 mg/mL type I collagen (Coll), Coll with 1 mg/mL chitosan (1:1 Coll:Chi), Coll with 2 mg/mL chitosan (1:2 Coll:Chi), and Coll with 3 mg/mL chitosan (1:3 Coll:Chi). The cells were allowed to attach overnight and were incubated with cell titer solution (Aqueous One Solution Cell Proliferation Assay, G3580; Promega, Madison, WI) as per the manufacturer's protocol. Basal absorbance at 450 nm was then measured. Absorbance was also measured after 6 and 10 days in culture to monitor increase in cell number.
Construction of the coculture system
To mimic the in vivo setup, the coculture construct consisted of three layers. Cells were premixed with the collagen and chitosan monomer solutions at a concentration of 1 × 106 cells/mL.
The bottom layer represented the mesenchymal layer and consisted of DPSCs embedded in a 1:1 Coll:Chi matrix. Specifically, the matrix consisted of type I collagen (1 mg/mL), chitosan (1 mg/mL), 1 × Hank's balanced salt solution, 5% (w/v) NaHCO3, and DPSCs made up to the required volume using the growth medium. The cells were added before the addition of NaOH solution to enable a homogeneous distribution. After addition of NaOH, 250 μL of the mixture was placed in a four-chambered cover glass (Labtek, Rochester, NY) and incubated at 37°C and 5% CO2 for 30 min.
Matrigel is a good substitute mimicking the composition of the in vivo basement membrane. Therefore, the middle layer contained growth factor–reduced Matrigel (BD Biosciences). Growth factor–reduced Matrigel was used to minimize the effect of Matrigel alone on the cell types. After 30 min of incubation with the mesenchymal matrix, 100 μL of Matrigel was added on top. The setup was incubated for an additional 30 min at 37°C and 5% CO2.
The upper layer represented the epithelial layer containing HAT-7 cells in a 1:2 Coll:Chi hydrogel. The hydrogel was prepared in a similar manner as described previously. The final layer was added on top of the Matrigel and incubated for 30 min at 37°C and 5% CO2. All the monomer solutions were placed on ice at all times during the process to avoid premature polymerization.
After the final incubation, the growth medium was added to the cells and cocultured at 37°C and 5% CO2 for the desired time period. The samples that were to be cultured in the differentiation medium (100 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10 mM dexamethasone) were placed in the same after incubating the coculture setup in the growth medium overnight. A fresh medium was added every other day; however, 250 μL of the medium was left behind in the wells in an attempt to prevent complete removal of growth factors and other signaling molecules that might have been secreted by the cells in the coculture. To study behavior of the two cell types in coculture, the assembly was cultured for 1, 8, 16, and 24 days. At the end of these time points the scaffolds were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned along the y–z direction (25 or 5 μm) so that each section contained all three layers.
To observe the homogenous distribution of the cells within the scaffold, cells were labeled with CellTrackerTM probes (Molecular Probes, Carlsbad, CA) red (DPSCs) and green (HAT-7) as per the manufacturer's instructions. For confocal microscopy, the scaffold containing the coculture was fixed with 4% paraformaldehyde and imaged. The mesenchymal layer was imaged first to obtain z-stack images. The matrix was then flipped and the epithelial layer was then subjected to similar imaging procedure.
Isolation of RNA and real-time PCR analysis
Three-dimensional coculture and monoculture constructs cultured in the growth medium were removed at 1-, 5-, and 10-day intervals. The layers were separated by tweezers for the coculture constructs and the gel was immersed in trizol (Invitrogen, Carlsbad, CA) solution. For monoculture constructs, the gels were directly immersed in trizol solution. Gels from a triplicate set were pooled for each time point. RNA was isolated and first-strand synthesis was carried out after DNAse (Gibco, Carlsbad, CA) treatment of the RNA using Superscript III (Invitrogen) according to manufacturer's protocol. Real-time PCR was then performed using the SYBR green method with forward and reverse primers for representative mesenchymal and epithelial genes. The following genes were tested for DPSCs: Runt-related transcription factor 2 (forward, 5′ ATGCTTCATTCGCCTCACAAAC3′; reverse, 5′ CCAAAAGAAGTTTTGCTGACAT GG 3′), coll (coll; forward, 5′ TGACGAGACCAAGAACTG 3′; reverse, 5′ CCATCCAAACCACTGAAACC 3′), and alkaline phosphatase (ALP; forward, 5′ ATCGCCTACCAGCTCATGCAT 3′) reverse, 5′ GTTCAGCTCGTACTGCATGTC 3′. The epithelial genes that were analyzed were amelogenin (forward, 5′ TGAG GTGCTTACCCCTTTGAAGTG 3′; reverse, 5′ GGAACTGGCATCATTGGTTGC 3′), dentin sialoprotein (DSP; forward, 5′ GGAGACGCCACCCTTGTC 3′; reverse, 5′ CTGATTTTGGCTC TGCCC 3′), and BMP2 (forward, 5′ ACAAATGCAGGAAGCTTTGG 3′; reverse, 5′ TTAAGACGCTTCCGCTGTTT 3′). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as internal control, and data from both coculture and monoculture were normalized to their respective GAPDH expression. The fold change in expression of the genes as a result of coculture was then calculated (2−ΔΔCT).
Implantation in mice
All experiments with animals were carried out as per the protocols established by the University of Illinois Office of Animal Care (assurance no. A3460.01). The coculture containing scaffolds were set up as described above and cultured in vitro for 48 h. They were implanted subcutaneously in immunocompromised CD-1 mice for 4 weeks. Two constructs per animal was used. Briefly, mice were anesthetized with ketamine/xylazine mixture (135 mg/kg and 15 mg/kg). The implants were placed subcutaneously on either side of the spine. At the end of 4 weeks the animals were euthanized and the implants were excised out and fixed in 4% paraformaldehyde, paraffin embedded, and sectioned into 5-μm sections. The implants were sectioned in a manner to reveal all the three layers.
Histological analysis
Apart from the in vitro and in vivo coculture paraffin-embedded sections, 20-day rat embryos and 5-day postnatal mouse heads embedded in paraffin were sectioned longitudinally to obtain 5-μm-thick slices. The sections from all the samples were deparaffinized in xylene and rehydrated by incubating in graded ethanol solutions. The endogenous peroxidases were quenched by incubation with 3% hydrogen peroxide for 30 min. Serial sections were then stained with hematoxylin and eosin to observe the cellular architecture; Masson's trichrome for identification of connective tissues and the cellular architecture can easily be observed, as they fluoresce in red; and von Kossa staining to observe the mineral deposits. Immunohistochemical analyses were performed using anti-DMP1 (produced in our laboratory), anti-DSP (produced in our laboratory), proliferating cell nuclear antigen (PCNA; Santa Cruz, Santa Cruz, CA), and anti-ameloblastin (a kind gift from Dr. Diekwisch, University of Illinois at Chicago) polyclonal antibodies. The sections were then incubated with a biotinylated anti-rabbit secondary antibody (Vectastain ABC peroxidase kit) and developed using the DAB kit (Vector Labs, Burlingame, CA). The sections were then imaged using a Zeiss Axio Observer D1 microscope along with Axiovision imaging software. Fluorescence micrographs were obtained using the same microscope equipped with the appropriate filter sets.
Electron microscopy
Paraffin-embedded sections were placed onto circular cover glass (Fisher, Pittsburgh, PA) and then deparaffinized in xylene and rehydrated in graded ethanol solutions. The sections were washed three times in cacodylate buffer and dehydrated with a series of graded alcohol solutions. The sections were then air-dried and mounted on to electron microscopy grids, sputter-coated, and examined using a field emission scanning electron microscope (JSM-6320F). For energy dispersive X-ray (EDX) analysis, samples were not sputter-coated and were imaged and analyzed using Hitachi S-3000N variable pressure scanning electron microscopy (SEM).
Results
Properties of collagen and chitosan blends
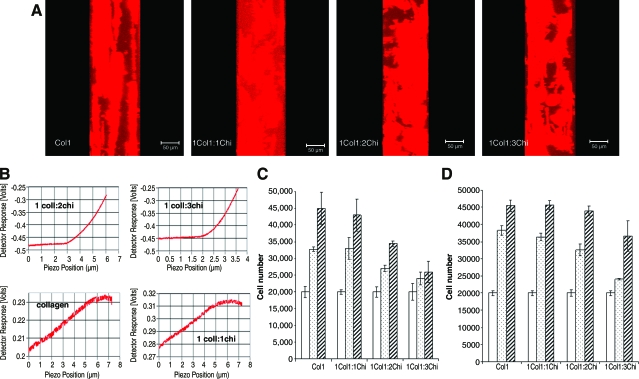
Blends of type I collagen and chitosan with the corresponding ratios 1:0, 1:1, 1:2, and 1:3 (type I collagen to chitosan) containing DPSCs at a concentration of 1 × 106 cells/mL were constructed and allowed to attach and proliferate in the regular growth medium for a period of 48 h. Figure 1A shows a y–z axis projection of representative z-stack confocal images spanning 100 μm in the z-axis for the cells in the various blends of type I collagen and chitosan. As can be observed from the figure, DPSCs in type I collagen gel by itself did not exhibit cell processes penetrating the hydrogel in the z-axis. This is evident from the cell thickness and the presence of cells in layers throughout the z-axis. However, upon addition of chitosan, the cells were observed to be extending their processes in the z-axis facilitating cell–cell contact. The respective panels in Figure 1A clearly illustrate this phenomenon. We believe that the increased density of the matrix by incorporating chitosan triggers the cells to put out processes in all three dimensions. We next proceeded to analyze if the mechanical strength of the hydrogels was affected by the addition of chitosan. The Young's modulus of the blends of type I collagen and chitosan were determined by AFM. For these experiments, we used scaffolds containing type I collagen at a concentration of 1 mg/mL and chitosan at a concentration ranging from 0 to 3 mg/mL. Figure 1B shows representative force curves obtained for the different matrix compositions. It can be seen from this figure that with an increase in chitosan concentration, the mechanical strength of the hydrogel increased proportionately. Pure type I collagen scaffold and scaffold comprising of 1:1 ratio of type I collagen to chitosan fell below the detection capacity of the AFM cantilever. However, 1:2 and 1:3 Col1:Chi ratio hydrogels yielded a Young's modulus of 1045 Pa (standard error of the mean of 45 measured over 90 spots) and 1652 Pa (standard error of the mean of 98 measured over 75 spots), respectively. Also, the force curves demonstrated that 1:1 ratio hydrogel was stiffer than the pure type I collagen by reaching a plateau at a lower piezo position, indicating that the tip obtained higher resistance from the hydrogel. These results suggest that the mechanical properties of the 3D scaffold can be varied by using different ratios of chitosan.
FIG. 1.
Characterization of the collagen chitosan matrix. (A) y–z axis projections of mesenchymal cells embedded in 3D matrices containing different ratios of collagen and chitosan. Scale bar represents 50 μm. (B) Representative experimental force curves obtained for the different blends of collagen and chitosan showing increase in strength with increase in ratio of chitosan. (C) Proliferation of DPSCs embedded in different ratios of type I collagen and chitosan-blended scaffolds. Empty bars represent number of cells at day 1, dotted bars at day 6, and striped bars at day 10. (D) Proliferation of HAT-7 cells in similar scaffolds containing type I collagen and chitosan. The bars represent the same time periods as in (C). Both (C) and (D) are data representing mean ± standard error of the mean of at least six replicates. Color images available online at www.liebertonline.com/ten.
Selection of optimum matrix composition to support proliferation of DPSCs and HAT-7 cells
Various compositions of the collagen–chitosan blends were used to determine the optimum concentration necessary for supporting the viability and proliferative capacity of DPSCs (Fig. 1C) and HAT-7 (Fig. 1D) when cocultured together. Identifying the optimum matrix concentration is important to ensure against overpopulation of one of the cell types that might have effects on the experimental outcome. Therefore, the results might not represent true interaction of the two cell types, but rather an artifact. Results presented in Figure 1C and D show that there is a decrease in rate of proliferation of the two cell types with increase in chitosan concentration. We ruled out the possibility of a 1:3 Coll:Chi matrix for DPSCs, as the proliferation rate decreased dramatically at this concentration. The two sets of data also reveal that the comparable proliferative capacity of DPSCs and HAT-7 cells were obtained in a 1:1 Coll:Chi and 1:2 Coll:Chi composition, respectively. Table 1 lists the rate of increase in cell number for both cell types under different ratios of collagen and chitosan. Considering the fact that the coculture systems would be in culture for close to 30 days, we selected the scaffold compositions that contained similar rates of proliferation for the two cell types. The cell titer solution was used to monitor living cells as the dye gets incorporated into the mitochondria to produce a color change. Taken together, these data show that the DPSC and HAT-7 cells are viable with comparable proliferation rates in 1:1 Coll:Chi mixture and 1:2 Coll:Chi mixture, respectively. It is to be noted that although type I collagen by itself showed good proliferation rates for both cell types, the integrity of the scaffold to maintain its shape with time under cell culture conditions was lost easily and the matrix showed considerable shrinkage (data not shown).
Table 1.
Rate of Increase in Cell Number
| |
Increase in cell number (no. of cells/day) |
|
|---|---|---|
| Type of matrix | Mesenchymal cells | Epithelial cells |
| Type I collagen | 2500 | 2550 |
| 1:1 Coll:Chi | 2300 | 2550 |
| 1:2 Coll:Chi | 1450 | 2330 |
| 1:3 Coll:Chi | 600 | 1660 |
This table indicates the average increase in cell number per day for both mesenchymal and epithelial cells cultured in three-dimensional matrices of different compositions of collagen and chitosan.
In vivo assembly of epithelial and mesenchymal cells
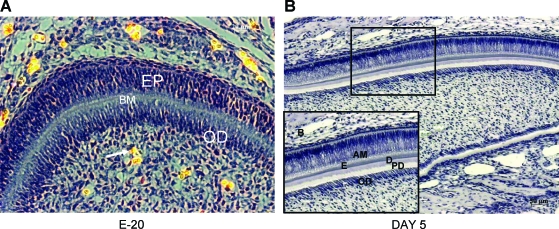
The scaffold containing the coculture was designed to mimic the in vivo assembly during odontogenesis. The in vivo assembly in a developing tooth is shown in Figure 2A and B, which depicts the spatial orientation of the ameloblasts (epithelial cells) and odontoblasts (mesenchymal cells) in a hematoxylin and eosin–stained section of a developing rat incisor at embryo day 20. The two cell types are separated by the basement membrane as indicated (Figure 2A). On day 5 the basement membrane is dismantled and the interaction between the synthesized products of the two cell types is responsible for the polarization and differentiation of ameloblasts and odontoblasts. Figure 2B is a picture of a hematoxylin-stained 5-day postnatal mouse incisor showing polarized odontoblasts and ameloblasts.
FIG. 2.
In vivo architecture of dental mesenchymal and epithelial cells. (A) Image showing a hematoxylin and eosin–stained section of a developing tooth germ from a 20-day rat embryo. The arrow points to a vascularized area in the dental pulp. Scale bar represents 20 μm. (B) A hematoxylin-stained section of a 5-day postnatal mouse incisor. Scale bar represents 50 μm. Note the arrangement of the epithelial and mesenchymal cells. AM, ameloblasts; EP, epithelial cells; BM, basement membrane; OD, odontoblasts; E, enamel matrix; D, dentin; PD, predentin. Color images available online at www.liebertonline.com/ten.
Histological characterization of the coculture setup
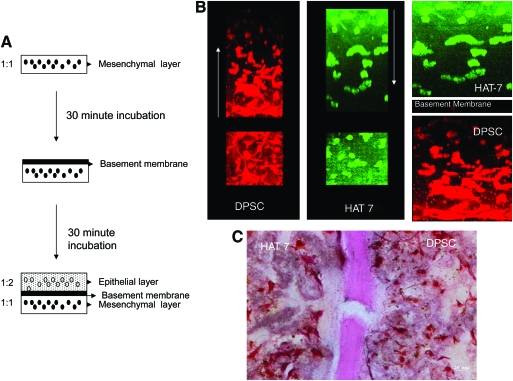
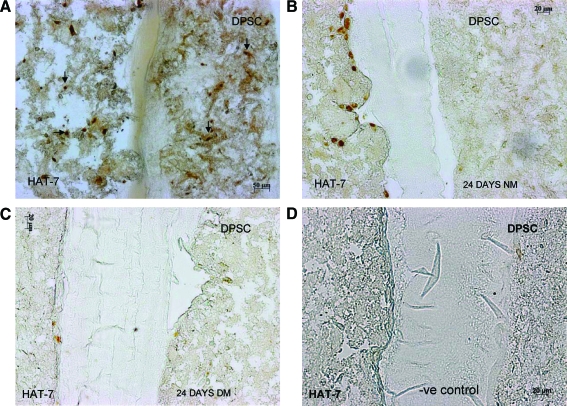
Our goal is to establish a coculture system similar to an in vivo setting as shown in Figure 2A. Figure 3A shows a schematic diagram of the scaffold setup. Figure 3B represents 3D confocal images of the coculture setup. DPSCs were prestained with Celltracker red and HAT-7 cells with Celltracker green as per the manufacturer's protocol. The 3D coculture matrix was then assembled as described in the Materials and Methods section. The cells were allowed to attach overnight in growth medium and were fixed and imaged. The merged image in Figure 3B is a pseudo-experimental (not to scale) representation of the setup showing the orientation of DPSCs and HAT-7 cells. The arrows in the individual images point to the scanning direction. It should be noted that the reduction in fluorescence with depth is due to decreased fluorescence intensity with increasing distance from the objective rather than decrease in cell number. Figure 3C is a von Kossa–stained section of the coculture matrix after 24 h of coculture in the growth medium showing all three layers. As expected, there are no mineral deposits after 1 day in culture. Figure 3C will serve as a control for basal von Kossa staining. Panels in Figure 3B and C illustrate the homogeneous distribution of the two cell types in their respective matrices. Another distinctly visible feature is the morphological difference between the two cell types (Fig. 3B). DPSCs displayed a tendency to put out extremely long processes in all three dimensions. On the other hand, HAT-7 cells displayed a more rounded morphology. Such morphological differences might facilitate increased proliferation of HAT-7 cells in denser matrices compared with DPSCs.
FIG. 3.
Coculture setup and tracking the cells in the scaffold. (A) A step-by-step schematic representation of the setup of the coculture model. (B) 3D confocal images of DPSCs (red) and HAT-7 cells (green), and the pseudo-experimental representation of the coculture setup. Arrows point to the direction of imaging. (C) A von Kossa–stained section of the coculture after 1 day of coculture in the normal growth medium. Scale bar represents 20 μm. Note the absence of mineral deposits. Color images available online at www.liebertonline.com/ten.
Migration of HAT-7 epithelial cells
The biomimetic scaffold containing the coculture was subjected to histological evaluation to determine the migratory patterns of the two cell types at 1, 8, 16, and 24 days in culture. One of the striking features exhibited by the HAT-7 cells was their migration toward the inner boundary (toward the Matrigel) with time. Panels in Figure 4A–D represent fluorescent micrographs taken from sections of cocultures after 1, 8, 16, and 24 days in culture, respectively, and collectively elucidate this process. The sections were stained with Masson's trichrome enabling the cells to fluoresce in red. As can be seen from the figures, the HAT-7 cells were homogeneously distributed within the matrix on day 1 (Fig. 4A); however, on day 8 (Fig. 4B) a migratory pattern is observed with a few cells lining the inner boundary (white arrows). By 16 days, most of the HAT-7 cells had accumulated at the inner boundary (Fig. 4C) and continued to remain at the interface at 24 days (Fig. 4D). We observed that once a monolayer of HAT-7 cells assembles close to the Matrigel, they stopped migrating. It is to be noted that the images shown are from 25-μm sections of the 3D matrix; hence, this phenomenon would have resulted in an entire plane of cells at the inner boundary. Overall, the integrity of the setup remained unchanged and no degradation was observed throughout the entire period of study.
FIG. 4.
Migration of HAT-7 cells in the matrix toward the Matrigel interface. (A), (B), (C), and (D) represent fluorescent micrographs of Masson's trichrome–stained sections of the coculture setup in the growth medium after 1, 8, 16, and 24 days in culture, respectively. Scale bar represents 10 μm for all images. Arrows point to migrating HAT-7 cells toward the Matrigel interface. Color images available online at www.liebertonline.com/ten.
Evidence of interaction between DPSCs and HAT-7 cells
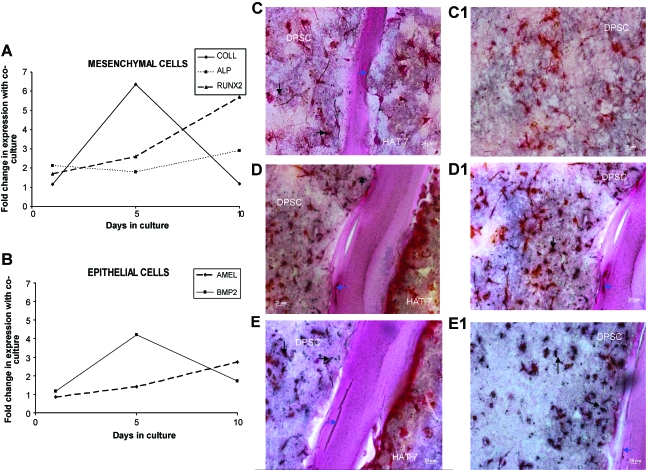
Gene expression analysis of various early and late markers expressed by epithelial and mesenchymal cells during tooth formation was performed after 1, 5, and 10 days in coculture in the presence of the growth medium and compared with the expression of the same genes in monocultures for identical time points. Figure 5A shows the change in expression of runx2, type I collagen (coll), and ALP in DPSCs. Figure 5B shows the change in expression of BMP2 and amelogenin in HAT-7 cells. DSP was only expressed in epithelial cells subjected to coculture for 10 days (data not shown in graph). All of the above-mentioned markers have been reported to be involved in the differentiation of mesenchymal and epithelial cells in vivo; further, their expression levels vary during various stages of differentiation. The varying gene expression profile is a clear indication of interactions between DPSC and HAT-7 cells in coculture.
FIG. 5.
Presence of calcified deposits in the matrix and gene expression in the presence of the growth medium. Graphs (A) and (B) represent fold change in expression profiles of different genes in the mesenchymal and epithelial cells, respectively, after 1, 5, and 10 days in coculture with respect to their expression in monoculture. (C) and (C1) are histological images of von Kossa–stained sections after 8 days of coculture in the growth medium. (D) and (D1) are images of von Kossa–stained sections after 16 days of coculture in the growth medium. (E) and (E1) are images of von Kossa–stained sections after 24 days of coculture in the growth medium. Arrows point to calcium deposition in and around the cells, and in the matrix. (C1), (D1), and (E1) are images of the mesenchymal areas alone. Scale bar represents 20 μm for all images. Color images available online at www.liebertonline.com/ten.
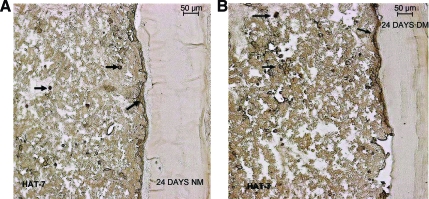
Assessment of differentiation of DPSCs and HAT-7 cells
Under differentiation conditions, DPSCs have been shown to undergo terminal differentiation into odontoblast-like cells producing a mineralized matrix in vitro.16,17 Similarly, HAT-7 cells have also been characterized to differentiate into ameloblast-like cells in vitro.18,19 To test if the coculture of the two cell types triggered differentiation, they were placed in the presence and absence of differentiation medium. The experiments were conducted as described in the Materials and Methods section, and samples were analyzed after 1, 8, 16, and 24 days in culture. Figure 5 shows the results from the coculture setup in the presence of the growth medium. Panels in Figure 5C–E are von Kossa–stained sections showing the calcified deposits at the end of 8, 16, and 24 days, respectively. Panels in Figure 5C1–E1 show images of the mesenchymal layer, wherein a progressive increase in calcified deposits is seen. Formation of such deposits suggests that the DPSCs might be differentiating. The basal staining for calcium is shown in Figure 3C. The panel in Figure 6 shows differentiation of the DPSCs in the coculture in the presence of the differentiation medium. von Kossa results show similar staining characteristics along the Matrigel layer for mineralized deposits as compared to Figure 5 (time points and arrangement of figures are similar to the representation in Fig. 5). This result provides evidence that the DPSCs cocultured with HAT-7 cells were able to initiate the differentiation cascade without any external additives. Also, migration of epithelial cells was observed. Although the epithelial cell layer, in the presence of the growth medium, did have areas that stained positive with von Kossa stain, the staining of epithelial cells in the presence of the growth medium was neither as pronounced as the DPSCs nor as much the staining in the presence of the differentiation medium. For the duration of the experiment, both cell types remained in their respective matrices without crossing over.
FIG. 6.
Presence of calcium deposits in the matrix in the presence of the differentiation medium. (A) and (A1) are light microscopic images of von Kossa–stained sections after 8 days of coculture in the differentiation medium. (B) and (B1) are images of von Kossa–stained sections after 16 days of coculture in the differentiation medium. (C) and (C1) are images of von Kossa–stained sections after 24 days of coculture in the differentiation medium. Arrows point to calcium deposition in and around the cells, and in the matrix. (A1), (B1), and (C1) are images of the mesenchymal areas alone. Scale bar represents 20 μm for all images. Color images available online at www.liebertonline.com/ten.
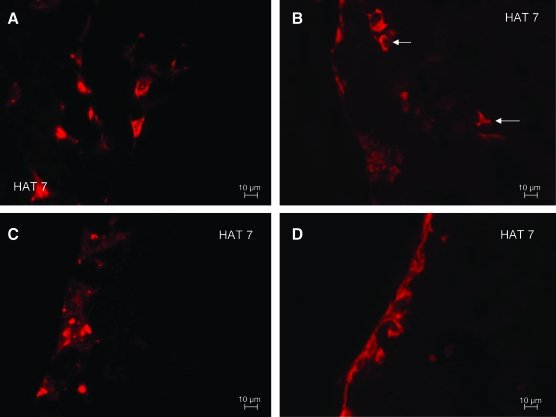
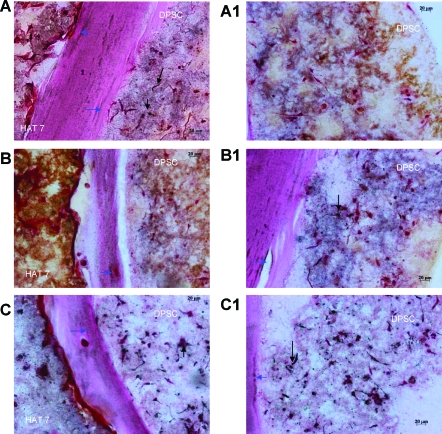
To further confirm that the DPSCs were undergoing differentiation, representative sections obtained from cocultures with the two experimental conditions were immunostained with PCNA antibody. This antibody staining is specific for cells in the proliferative stage. Results in Figure 7B and C show the absence of PCNA staining in the DPSC layer in the presence and absence of the differentiation medium, indicating the differentiated state of the DPSCs. However, positive staining was observed in a few HAT-7 cells (Fig. 7B) in the presence of the normal growth medium. In the presence of the differentiation medium, both DPSCs and HAT-7 cells did not show positive staining (Fig. 7C). The presence of only a few PCNA-stained HAT-7 cells demonstrates that most of the cells have ceased proliferation and have started their differentiation phase. However, they are yet to synthesize a mineralized matrix. This explains the lack of von Kossa staining in the matrix containing HAT-7 cells (Fig. 5). On the other hand, the control cells 24 h after the start of the coculture (Fig. 7A) showed primarily a proliferating population. From this experiment, we could conclude that the coculture environment alone might initiate the differentiation of almost all of the mesenchymal cells and a partial differentiation of the epithelial cells. Figure 7D is a representative image of immunohistochemical staining with secondary antibody alone and serves as a negative control for all the immunostained sections in this article.
FIG. 7.
Detection of proliferating cells by proliferating cell nuclear antigen immunostaining. PCNA immunostained sections from 1 day, 24 days co-culture. (A) represents control section immunostained after 1 day of co-culture and shows a mixed population of proliferating and nonproliferating cells. Scale bar represents 50 μm. Note that this section was imaged at a lower magnification, as the cells were homogeneously spread out in the matrix. Arrows point to positively stained cells. (B) and (C) represent images of immunostained sections after 24 days of coculture in normal (B) and the differentiation (C) medium. Scale bar represents 20 μm for (B) and (C). Presence of brown nuclear stain indicates proliferating cells. Absence of staining denotes nonproliferating and possibly differentiating cells. NM represents the normal culture medium (growth medium) and DM represents the differentiation culture medium. (D) represents secondary antibody negative control. Scale bar represents 20 μm. Color images available online at www.liebertonline.com/ten.
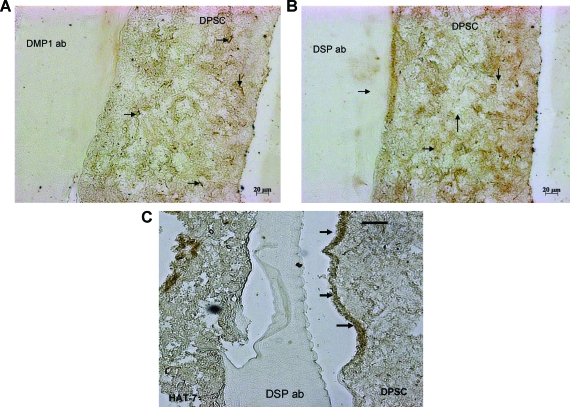
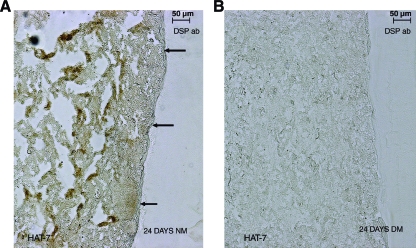
To confirm that the DPSCs were differentiating toward an odontoblast phenotype at the end of 24 days, immunohistochemical analysis was performed using anti-DMP1 and anti-DSP antibodies. Positive staining of the mesenchymal cells (Fig. 8A and B, respectively) indicated that the mesenchymal cells were differentiating toward matrix-secreting functional odontoblasts. Negative controls with secondary antibody alone did not show any staining (Fig. 7D). Migration of DPSCs was not as pronounced as the epithelial cells. However, few migratory cells that stained positive for DSP could be observed along the interface (marked by arrows in Fig. 8B, C). An interesting observation was the presence of positive DSP staining in the epithelial layer of 24-day coculture setup cultured in the growth medium. However, these cells were limited to the middle section of the matrix and the cells that had migrated to the interface did not show positive staining (Fig. 9A). No positive staining was observed in similar sections from coculture in the presence of the differentiation medium (Fig. 9B). Published reports22 indicate that dental epithelial cells produce DSP in their presecretory phase and not during the secretory phase. Thus, the epithelial cells in the middle of the matrix might contain HAT-7 cells that have differentiated to the presecretory phase and the cells closer to the Matrigel could be in a more differentiated state.
FIG. 8.
Immunostaining with anti-dentin matrix protein 1 and anti-dentin sialoprotein (DSP) antibodies. Anti-dentin matrix protein 1 antibody (A) and anti-DSP antibody (B) and (C) were used to detect differentiation of DPSCs toward odontoblast lineage in 24-day coculture sections. Arrows point to positive staining in both the images. Scale bar represents 20 μm for A and B images and 50 μm for C. Color images available online at www.liebertonline.com/ten.
FIG. 9.
DSP immunostaining in HAT-7 cells. (A) and (B) represent DSP-immunostained HAT-7 cells from sections of 24-day coculture setups in the growth medium and differentiation medium, respectively. Arrows point to negatively stained HAT-7 cells at the interface. Scale bar represents 50 μm for both images. Color images available online at www.liebertonline.com/ten.
Additionally, HAT-7 cells from 24-day coculture sections showed positive staining for ameloblastin both in the absence and presence of the differentiation medium (Fig. 10A, B). Ameloblastin is a marker for dental epithelial cells, and positive staining indicates that the epithelial cells are differentiating toward the ameloblastic lineage.
FIG. 10.
Ameloblastin immunohistochemistry. (A) and (B) represent ameloblastin-immunostained HAT-7 cells from sections of 24-day coculture setups in the growth medium and differentiation medium, respectively. Arrows point to positively stained cells. Scale bar represents 50 μm for both images. Color images available online at www.liebertonline.com/ten.
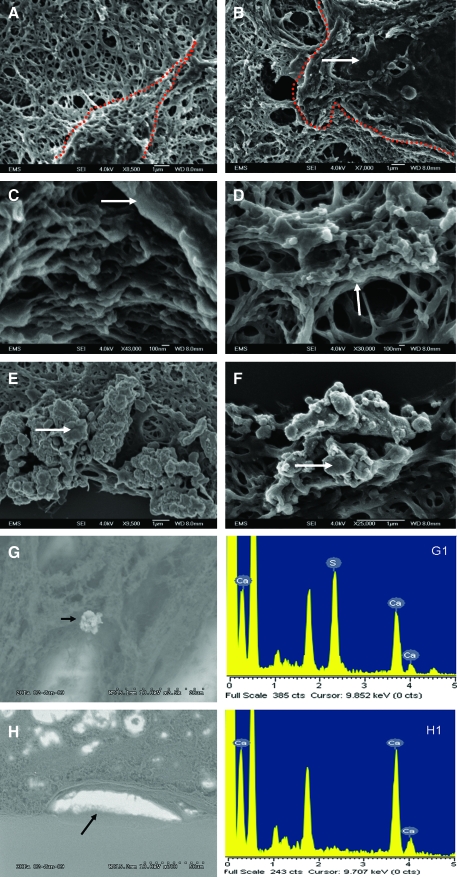
Analysis of the synthesized matrix and the cellular architecture by SEM
Panels in Figure 11A and B show a DPSC and a HAT-7 cell embedded in their respective matrix from a 24-day coculture setup. The dotted line marks the outline of the two cell types. Panels in Figure 11C and D are higher magnification images showing attachment of the cellular processes of DPSCs and HAT-7 cells to the matrix. These SEM images show the integration of the cells within the matrix. Panels in Figure 11E and F are SEM images of calcium deposits seen in the mesenchymal matrix after 24 days of coculture in normal (Fig. 11E) and differentiation media (Fig. 11F). EDX analysis was performed on the sections to show the presence of calcium. Figure 11G is a representative SEM micrograph of a section from 24-day growth medium coculture, and Figure 11G1 is the EDX analysis of the deposit marked by a black arrow in Figure 11G. Panels in Figure 11H and H1 show similar data with a 24-day differentiation medium coculture section. Taken together, these data show that both DPSCs and HAT-7 cells were well integrated within the 3D matrix and were capable of producing a mineralized matrix.
FIG. 11.
Scanning electron microscopy. (A) and (B) are electron micrographs of a DPSC and a HAT-7 cell in their respective matrices after 1 day of coculture. The red dotted lines indicate boundaries of the cells. Arrow points to a cell. (C) and (D) are electron micrographs of processes of DPSCs and HAT-7 cells adhering to the ECM. Arrows point to collagen fibers with cell processes. (E) and (F) are electron micrographs of the calcium deposits observed in the mesenchymal matrix after 24 days of coculture in the normal medium (E) and the differentiation medium (F). Arrows point to the deposits. Magnification is specified under each image. (G) and (H) are electron micrographs of coculture sections in the growth medium and differentiation medium, respectively, showing calcium deposits (black arrows). (G1) and (H1) are the respective energy dispersive X-ray analysis data for the deposits marked by arrows in figures (G) and (H). Color images available online at www.liebertonline.com/ten.
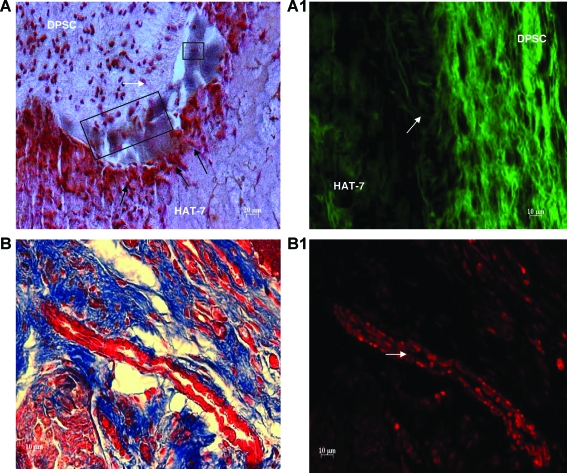
Integrity of the coculture system in an in vivo environment
To examine the integrity and behavior of the coculture setup in an in vivo setting, we implanted the coculture construct subcutaneously in immunocompromised mice. The implant was removed at the end of 4 weeks, embedded in paraffin, and sectioned. von Kossa staining shows the presence of calcium deposition at the interface (Fig. 12A) similar to the results obtained from in vitro analysis, and the polarizing nature of the DPSCs could be clearly observed (arrow and boxes in Fig. 12A). Migration of the epithelial cells to the interface was similar in nature to the in vitro setup (Fig. 12A), and the beginning of epithelial polarization could be observed (black arrows in Fig. 12A). Autofluorescence from the collagen fibers when viewed under the fluorescence microscope indicated that the differentiating DPSCs were actively synthesizing a collagenous matrix. This is evident from Figure 12A1, showing de novo synthesized collagen fibrils running perpendicularly to the matrix at the interface (white arrow in Fig. 12A1). It is to be noted that existing collagen fibers do not get rearranged in a perpendicular direction; therefore, the perpendicular collagen fibers should be de novo collagen synthesized by DPSCs. Deposition and assembly of collagen matrix is a requirement for calcified matrix formation.
FIG. 12.
Histological analysis of coculture constructs implanted in immunocompromised mice. (A) represents a von Kossa–stained section of the coculture construct excised after 4 weeks of implantation. Scale bar represents 20 μm. White arrow and black boxes indicate polarizing DPSCs. Black arrows point to polarizing HAT-7 cells. Note the presence of positive von Kossa staining in the interface between the two cell types. (A1) represents a fluorescent micrograph demonstrating collagen autofluorescence from a section of the coculture construct after 4 weeks of implantation. The arrow points to de novo–synthesized collagen fibrils arranged perpendicular to the matrix. (B) is an image of Masson's trichrome–stained section of the coculture construct excised after 4 weeks of implantation showing neovascularization. (B1) is a fluorescent micrograph of (B). Arrow points to blood vessel with red blood corpuscles. Scale bar represents 10 μm for all images except (A). Color images available online at www.liebertonline.com/ten.
Angiogenesis is observed during in vivo implantation of the construct
Masson's trichrome staining combined with fluorescence microscopy of the coculture implants showed the presence of vascularization in the mesenchymal matrix. Panels in Figure 12B and B1 show neovascularization in the mesenchymal matrix. Figure 12B1 is a fluorescent micrograph of the same section shown in Figure 12B. The arrow points to the newly formed blood vessels and also clearly shows the presence of red blood corpuscles. Support of angiogenic activity by the matrix is particularly important when the coculture construct would be used for various tissue engineering applications in the future.
Discussion and Conclusion
Fabrication of biomaterials using biopolymers such as type I collagen and chitosan would be highly valuable to study cellular activities and multiple cell interactions in a 3D environment. Several collagen-based scaffolds such as Collagraft are currently being used for mineralized tissue regeneration. Chitosan on the other hand is obtained from deacylation of chitin present in the exoskeleton of crustaceans and has a structural similarity to glycosaminoglycans.23 Chitosan is biocompatible and biodegradable and is currently used in a variety of tissue engineering applications.24–26 As an additional incentive, chitosan also possesses antimicrobial properties.27 The ability of type I collagen and chitosan blends to support cell culture has been reported earlier20,28 and proven to be an excellent biomaterial. In this report we show that an increase in chitosan concentration favors cell–cell interaction and increases the stiffness and integrity of the hydrogel. This system can be manipulated to obtain hydrogels of required mechanical strength by adjusting the collagen–chitosan ratio in the scaffold to suit the proliferation rates of the different cell types that could be used in various applications.
In this study we have employed type I collagen and chitosan blends to create a biomimetic environment that would support cell growth and differentiation. We show that the proliferation rate can be controlled by using different blends of collagen and chitosan and that these 3D coculture constructs can be used to study the interaction between the dental epithelial and mesenchymal cells.
Various coculture models have been investigated that involve 2D micropatterning, microfluidics.29–41 and coculture with inserts.16 In these studies the two cell types are spatially fixed in the same 2D substrate to study their interactions. The drawback of these systems are (1) they often require confluent monolayers of cells that will be difficult to maintain for extended periods of time, (2) they do not permit cell migration, and (3) they do not represent the cellular arrangement seen in vivo. Overall, interpretation of results obtained from such systems is not relevant, as the cell arrangement does not mimic the in vivo architecture. A published in vitro study involving random coculture of dental epithelial cells and mesenchymal cells in tissue culture dishes42 demonstrated an increase in ALP activity in the mesenchymal cells after coculture. However, the above-mentioned drawbacks apply to this system too.
The 3D coculture models described in literature involve the random coculture of two cell types in a scaffold43,44 or coculture of pelleted cells to form spheroids.45 Tan et al. have described a layer-by-layer microfluidic setup to mimic the blood vessel in microscale,46 but the study lacked description of the behavior of the cell types over time. A multitude of coculture models with most of them being 2D and a few being pseudo 3D (cells seeded on top of a 3D matrix) exist in literature.
In comparison, results from this study show the successful construction of a 3D biomimetic scaffold with collagen and chitosan forming the polymer matrix facilitating coculture of epithelial and mesenchymal cells. The composition of the matrix was optimized to match the proliferation rates of the two cell types. The novelty of this scaffold lies in the use of layered macroscale biomimetic setup with tunable mechanical properties that provides freedom of movement in all directions for the two cell types. The nature of this setup also facilitates easy isolation of protein and RNA individually from the two cell types for quantitative analysis. Moreover, the stability of the construct was remarkable. No disintegration of the scaffold was observed within the 24-day time period used in coculture experiments. This is evident from the histological images showing intact layers. The absence of PCNA staining, presence of calcified deposits in the matrix, and positive staining for DMP1 and DSP indicate probable differentiation of DPSCs toward and odontogenic lineage. On the other hand, HAT-7 cells showed expression of ameloblastin and spatial changes in expression of DSP. Also, differential expression patterns of various marker genes were observed with time in coculture for both epithelial and mesenchymal cells, providing evidence for interaction. This demonstrates that the 3D coculture setup provides the cells with an environment conducive for growth and differentiation rather than independent behavior that is observed when the cells are confined spatially in two dimensions. Studies are underway to systematically identify activation of different signaling pathways at different stages of coculture. This information could provide critical clues to the importance of different growth factors and signaling molecules that might be required at different stages of differentiation.
The migration of HAT-7 cells to the interface was an interesting observation. However, migration of the mesenchymal cells to the interface of the coculture setup in vitro was not as significant, although few migratory DPSCs positive for DSP were observed in the in vitro samples. Polarization of the DPSCs was observed to a greater extent in the in vivo implants. It is possible that additional factors might be required for complete migration and polarization of DPSCs in vitro.
Taken together, the in vitro results from this study show that this biomimetic scaffold can be further exploited to study epithelial–mesenchymal interactions. Another important conclusion that can be drawn from this study is that the interacting cells can initiate cellular differentiation in the absence of the differentiation medium. In fact, the presence of the differentiation medium might result in simultaneous differentiation of the two cell types negating any effects of interaction. This is evident to an extent from the immunohistochemical analysis with PCNA and DSP antibodies that show the epithelial cell layer differentiating at a slower pace compared to the mesenchymal layer in the presence of the growth medium. Also, spatially different expression patterns of DSP in the epithelial cells indicate that there might be reciprocal interactions between the two cell types. Changes in gene expression profiles add further support to this coculture model. We also show that the coculture setup had similar characteristics in an in vivo environment. Implantation of the scaffold subcutaneously in an immunocompromised mouse was performed to test the stability of the scaffold and to observe whether the two cell types exhibited similar differentiation cascade both in vitro and in vivo. Immunocompromised mice were used to avoid immune reactions to the scaffold. An interesting observation is that the scaffold, in an in vivo environment, could favor the formation of microcapillary-like structures. Collagen–chitosan blended scaffolds have been previously shown to induce angiogenesis when implanted in vivo.26 We believe that his property of the scaffold would be an important tenet during tissue regeneration process and ensure proper integration of the biomaterial with the surrounding tissue.
To our knowledge, this is the first successful 3D multilayered coculture system established that can be exploited to study the interactions between dental epithelial and mesenchymal cells. This system could be extended to other epithelial and mesenchymal interaction–based organogenesis as in the formation of lungs and kidneys. The study of angiogenesis in the presence of cancer cells, effect of cancer cells on their corresponding nonmalignant cells, and invasion of malignant cells into normal tissue beds are some examples of complex interactions that could be studied using such an approach by incorporating the relevant cell types in 3D matrices.
Acknowledgments
This work was supported by the National Institutes of Health Grant DE 16533. We thank Ms. Verna Brown for her help with the histological staining. We thank Dr. Igor Titushkin and Dr. Michael Cho of the Bioengineering Department at the University of Illinois at Chicago for their assistance in using their AFM.
Disclosure Statement
No competing financial interests exist.
References
- 1.Xu Y. Zhou Y.L. Gonzalez F.J. Snead M.L. C/EBP-delta maintains amelogenin expression in the absence of C/EBP-alpha in vivo. J Biol Chem. 2007;282:29882. doi: 10.1074/jbc.M702097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriks J. Riesle J. van Blitterswijk C.A. Co-culture in cartilage tissue engineering. J Tissue Eng Regen Med. 2007;1:170. doi: 10.1002/term.19. [DOI] [PubMed] [Google Scholar]
- 3.Kirkpatrick C.J. Fuchs S. Hermanns M.I. Peters K. Unger R.E. Cell culture methods of higher complexity in tissue engineering and regenerative medicine. Biomaterials. 2007;28:5193. doi: 10.1016/j.biomaterials.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003;116:1647. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- 5.Jernvall J. Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 6.D'Souza R.N. Åberg T. Gaikwad J. Cavender A. Owen M. Karsenty G. Thesleff I. Cbfa 1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- 7.Thesleff I. Mikkola M. The role of growth factors in tooth development. Int Rev Cytol. 2002;217:93. doi: 10.1016/s0074-7696(02)17013-6. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto H. Cho S.W. Song S.J. Hwang H.J. Lee M.J. Kim J.Y. Jung H.S. Characteristic tissue interaction of the diastema region in mice. Arch Oral Biol. 2005;50:189. doi: 10.1016/j.archoralbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Yamashiro T. Zheng L. Shitaku Y. Saito M. Tsubakimoto T. Takada K. Yamamoto T.T. Thesleff I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;75:452. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 10.Narayanan K. Gajjeraman S. Ramachandran A. Hao J. George A. Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J Biol Chem. 2006;281:19064. doi: 10.1074/jbc.M600714200. [DOI] [PubMed] [Google Scholar]
- 11.Tompkins K. George A. Veis A. Characterization of a mouse amelogenin [A-4]/M59 cell surface receptor. Bone. 2006;38:172. doi: 10.1016/j.bone.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Shapiro J.L. Wen X. Okamoto C.T. Wang H.J. Lyngstadaas S.P. Goldberg M. Snead M.L. Paine M.-L. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell Mol Life Sci. 2007;64:244. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan K. Ramachandran A. Hao J. He G. Park K.W. Cho M. George A. Dual functional roles of dentin matrix protein 1. Implications in biomineralization and gene transcription by activation of intracellular Ca2 + store. J Biol Chem. 2003;278:17500. doi: 10.1074/jbc.M212700200. [DOI] [PubMed] [Google Scholar]
- 14.Mitsiadis T.A. Drouin J. Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev Biol. 2008;313:887. doi: 10.1016/j.ydbio.2007.10.055. [DOI] [PubMed] [Google Scholar]
- 15.Nakao K. Morita R. Saji Y. Ishida K. Tomita Y. Ogawa M. Saitoh M. Tomooka Y. Tsuji T. The development of a bioengineered organ germ method. Nat Methods. 2007;4:227. doi: 10.1038/nmeth1012. [DOI] [PubMed] [Google Scholar]
- 16.Gronthos S. Mankani M. Brahim J. Robey P.G. Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batouli S. Miura M. Brahim J. Tsutsui T.W. Fisher L.W. Gronthos S. Robey P.G. Shi S. Comparison of stem-cell-mediated osteogenesis and dentinogenesis. J Dent Res. 2003;82:976. doi: 10.1177/154405910308201208. [DOI] [PubMed] [Google Scholar]
- 18.Harada H. Cell dynamics in the growth and differentiation of dental epithelium during tooth development: stratum intermedium cells originated from inner enamel epithelium. J Hard Tissue Biol. 2005;14:172. [Google Scholar]
- 19.Harada H. Ichimori Y. Tamaki T.Y. Ohshima H. Kawano S. Katsube K.I. Wakisaka S. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 2006;340:611. doi: 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 20.Tan W. Krishnaraj R. Desai T.A. Evaluation of nanostructured composite collagen-chitosan matrices for tissue engineering. Tissue Eng. 2001;7:203. doi: 10.1089/107632701300062831. [DOI] [PubMed] [Google Scholar]
- 21.Titushkin I. Cho M. Modulation of cellular mechanics during osteogenic differentiation of human mesenchymal stem cells. Biophys J. 2007;93:3693. doi: 10.1529/biophysj.107.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paine M.L. Luo W. Wang H.J. Bringas P., Jr. Ngan A.Y. Miklus V.G. Zhu D.H. MacDougall M. White S.N. Snead M.L. Dentin sialoprotein and dentin phosphoprotein overexpression during amelogenesis. J Biol Chem. 2005;280:31991. doi: 10.1074/jbc.M502991200. [DOI] [PubMed] [Google Scholar]
- 23.Chandy T. Sharma C. Chitosan as a Biomaterial. Biomater Art Cells Art Org. 1990;18:1. doi: 10.3109/10731199009117286. [DOI] [PubMed] [Google Scholar]
- 24.Xia W. Liu W. Cui L. Liu Y. Zhong W. Liu D. Wu J. Chua K. Cao Y. Tissue engineering of cartilage with the use of chitosan-gelatin complex scaffolds. J Biomed Mater Res B Appl Biomater. 2004;71:373. doi: 10.1002/jbm.b.30087. [DOI] [PubMed] [Google Scholar]
- 25.Chávez-Delgado M.E. Mora-Galindo J. Gómez-Pinedo U. Feria-Velasco A. Castro-Castañeda S. Lopez-Dellamary Toral F.A. Luquin-De Anda S. Garcia Segura L. Garcia-Estrada J. Facial nerve regeneration through progesterone-loaded chitosan prosthesis. A preliminary report. J Biomed Mater Res B Appl Biomater. 2003;67:702. doi: 10.1002/jbm.b.10059. [DOI] [PubMed] [Google Scholar]
- 26.Wu X. Black L. Santacana-Laffitte G. Patrick C.W. Preparation and assessment of glutaraldehyde-crosslinked collagen-chitosan hydrogels for adipose tissue engineering. J Biomed Mater Res A. 2007;81:59. doi: 10.1002/jbm.a.31003. [DOI] [PubMed] [Google Scholar]
- 27.Kim K.W. Thomas R.L. Lee C. Park H.J. Antimicrobial activity of native chitosan, degraded chitosan, and O-carboxymethylated chitosan. J Food Prot. 2003;66:1495. doi: 10.4315/0362-028x-66.8.1495. [DOI] [PubMed] [Google Scholar]
- 28.Tan W. Desai T.A. Microscale multilayer cocultures for biomimetic blood vessels. J Biomed Mater Res A. 2005;72:146. doi: 10.1002/jbm.a.30182. [DOI] [PubMed] [Google Scholar]
- 29.Das M. Rumsey J.W. Gregory C.A. Bhargava N. Kang J.F. Molnar P. Riedel L. Guo X. Hickman J.J. Embryonic motoneuron-skeletal muscle co-culture in a defined system. Neuroscience. 2007;146:481. doi: 10.1016/j.neuroscience.2007.01.068. [DOI] [PubMed] [Google Scholar]
- 30.Hoben G.M. Hu J.C. James R.A. Athanasiou K.A. Comparison of scaffolds and culture conditions for tissue engineering of the knee meniscus. Tissue Eng. 2007;13:939. doi: 10.1089/ten.2005.11.1095. [DOI] [PubMed] [Google Scholar]
- 31.Eves P.C. Beck A.J. Shard A.G. Mac Neil S. A chemically defined surface for the co-culture of melanocytes and keratinocytes. Biomaterials. 2005;26:7068. doi: 10.1016/j.biomaterials.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia S. Yarmush M. Toner M. Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts. J Biomed Mater Res. 1997;34:189. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Yamato M. Utsumi M. Kushida A. Konno C. Kikuchi A. Okano T. Thermo-responsive culture dishes allow the intact harvest of multilayered keratinocyte sheets without dispase by reducing temperature. Tissue Eng. 2001;7:473. doi: 10.1089/10763270152436517. [DOI] [PubMed] [Google Scholar]
- 34.Tsuda Y. Kikuchi A. Yamato M. Nakao A. Sakurai Y. Umezu M. Okano J. The use of patterned dual thermoresponsive surfaces for the collective recovery as co-cultured cell sheets. Biomaterials. 2005;26:1885. doi: 10.1016/j.biomaterials.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Co C.C. Wang Y.C. Ho C.C. Biocompatible micropatterning of two different cell types. J Am Chem Soc. 2005;127:1598. doi: 10.1021/ja044382a. [DOI] [PubMed] [Google Scholar]
- 36.Leclerc E. Furukawa K.S. Miyata F. Sakai Y. Ushida T. Fujii T. Fabrication of microstructures in photosensitive biodegradable polymers for tissue engineering applications. Biomaterials. 2004;25:4683. doi: 10.1016/j.biomaterials.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 37.Mooney D. Hansen L. Vacanti J. Lanter R. Stephen F. Ingber D. Switching from differentiation to growth in hepatocytes: control by extracellular matrix. J Cell Physiol. 1992;151:497. doi: 10.1002/jcp.1041510308. [DOI] [PubMed] [Google Scholar]
- 38.Chen C.S. Mrksich M. Huang S. Whitesides G.M. Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 39.Bhatia S. Balis U. Yarmush M. Toner M. Effect of cell–cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 40.Tsuda Y. Kikuchi A. Yamato M. Chen G. Okano T. Heterotypic cell interactions on a dually patterned surface. Biochem Biophys Res Commun. 2006;348:937. doi: 10.1016/j.bbrc.2006.07.138. [DOI] [PubMed] [Google Scholar]
- 41.Khatiwala C.B. Peyton S.R. Putnam A.J. Intrinsic mechanical properties of the extracellular matrix affect the behavior of pre-osteoblastic MC3T3-E1 cells. Am J Physiol Cell Physiol. 2006;290:1640. doi: 10.1152/ajpcell.00455.2005. [DOI] [PubMed] [Google Scholar]
- 42.Shiba H. Mouri Y. Komatsuzwa H. Mizuno N. Xu W. Noguchi T. Nakamura S. Sugai M. Kato Y. Kurihara H. Enhancement of alkaline phosphatase synthesis in pulp cells co-cultured with epithelial cells derived from lower rabbit incisors. Cell Biol Int. 2003;27:815. doi: 10.1016/s1065-6995(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 43.Fan H. Liu H. Toh S.L. Goh J.C.H. Enhanced differentiation of mesenchymal stem cells co-cultured with ligament fibroblasts on gelatin/silk fibroin hybrid scaffold. Biomaterials. 2008;29:1017. doi: 10.1016/j.biomaterials.2007.10.048. [DOI] [PubMed] [Google Scholar]
- 44.Hofmann A. Ritz U. Verrier S. Eglin D. Alini M. Fuchs S. Kirkpatrick C.-J. Rommens P.M. The effect of human osteoblasts on proliferation and neo-vessel formation of human umbilical vein endothelial cells in a long-term 3D co-culture on polyurethane scaffolds. Biomaterials. 2008;29:4217. doi: 10.1016/j.biomaterials.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 45.Rouwkema J. Boer J. Clemens A. Van Blitterswijk Endothelial cells assemble into a 3-dimensional prevascular network in a bone tissue engineering construct. Tissue Eng. 2006;12:2685. doi: 10.1089/ten.2006.12.2685. [DOI] [PubMed] [Google Scholar]
- 46.Tan W. Desai T.A. Layer-by-layer microfluidics for biomimetic three-dimensional structures. Biomaterials. 2004;25:1355. doi: 10.1016/j.biomaterials.2003.08.021. [DOI] [PubMed] [Google Scholar]