Abstract
Background
Adenovirus type 3 (HAdV3) is one of the most prevalent serotypes detected globally. Variants of HAdV3 have been associated with outbreaks of severe disease.
Objectives
To better understand genetic diversity of circulating HAdV3s and examine risk factors for severe disease.
Study Design
Restriction enzyme analysis for genomic characterization of clinical HAdV3 isolates detected by 15 collaborative US laboratories during the period July 2004 to May 2007. Multivariate modeling was employed for statistical analyses.
Results
The most common HAdV3 types of 516 isolates studied were HAdV3a2 (36.9%), HAdV3a50 (27.1%), HAdV3a51 (18.0%), and HAdV3a17 (4.6%). Non-HAdV3a genome types were rare (1.2%). HAdV3a50 and HAdV3a51 are newly described variants which became more prevalent in 2006 and 2007 and have been associated with at least one epidemic. Their uniqueness was determined by specific banding profiles generated by digests with endonucleases Bcl I, Bgl II, and Hind III. Multivariable risk factor modeling demonstrated that children under 2 years of age (OR=2.7; 95%CI 1.6 to 4.6), persons with chronic disease (OR=5.1; 95%CI 2.6–9.8), persons infected with HAdV3a2 (OR=3.0; 95%CI 1.5 to 6.0), with HAdV3a50 (OR=2.5; 95%CI 1.2 to 5.2), or with multiple or rare strains (OR=2.8; 95%CI 1.3–6.5) were at increased risk of severe HAdV3 clinical disease.
Conclusions
In the study period considerable genetic diversity was found among US clinical HAdV3 strains. Novel variants emerged and became prevalent. One such emergent strain may be associated with more severe clinical disease.
Key words (MESH): Adenoviridae, restriction mapping, infectious diseases, epidemiology, molecular
Background
Adenoviruses (HAdVs) are a frequent cause of acute respiratory illness in man. HAdVs also cause conjunctivitis, gastrointestinal and urinary tract infections, and occasionally encephalitis.1 HAdV infections may be particularly severe among young children and the immunocompromised. Traditionally, HAdVs have been characterized by hemagglutination and viral neutralization to identify species (e.g. species A–G) and by neutralization or partial genome sequencing to identify serotype (e.g. HAdV1-HAdV52).2 Restriction enzyme analysis (REA) has been used to determine specific genome types within a serotype (e.g. HAdV3a vs. HAdV3b). Further subgenome type analyses may be performed by using multiple endonucleases and comparing digest patterns. As HAdV serotypes, genome types, and subgenome types may differ in tissue tropism and virulence, their characterization can be clinically and epidemiologically important.3,4
We have adapted molecular typing methods to more rapidly identify HAdV serotypes.5 A total of 15 collaborating civilian laboratories contributed HAdV specimens in US national surveillance efforts during the period July 2004 through May 2007. Sequencing a portion of the hexon gene correlated well with classical HAdV serotyping. HAdV3 was revealed to be the most prevalent type, comprising approximately 917 (29.9%) of the 3065 clinical Ads detected.5 Surveillance study data suggest that recent HAdV3 prevalence was elevated as compared to survey data generated from 1967 to 1976.6
Objectives
The purpose of this study was to more fully characterize recent US clinical HAdV3 isolates with full-genome REA DNA digests. In an effort to better understand the recent epidemiology of HAdV3 infections multivariate risk factor modeling was employed.
Study Design
Population
In our previous 3-year study5 we evaluated more than 3000 clinical HAdV isolates with multiple methods of gene sequence typing.7,8 Any isolate that was sequence-typed as HAdV3 from the beginning of the study (July 2004) to April 2007 was further studied with REA techniques in this sub study. This sample of 516 isolates represented 56.3% of the 917 HAdV3s detected in the original study.
Viral Culture
HAdV3 specimens were preserved at −80°C until propagated in A549 cells. A549 cells in 6-well cluster plates were inoculated with 150 μl of HAdV positive specimen or isolate. Cultures were incubated and observed for 2 to 10 days for cytopathic effect (CPE). If no CPE was detected after 10 days, infected cell supernatants were passaged and the observations repeated. Once CPE was noticed the isolate was harvested for DNA extraction.
DNA Extraction
Following viral harvest and centrifugation at 1200 rpm for 10 minutes, infected cell pellets were resuspended in 200 μl of corresponding viral supernatant. DNA was extracted from the infected cell/supernatant mix using the QIAamp DNA Blood Mini Kit (QIAgen, Valencia CA) according to manufacturer’s instructions.
Restriction Enzyme Digests
Nucleic acid concentrations from eluted samples were analyzed using a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Standard sodium acetate/ethanol precipitations were performed to yield approximately 1–2 μg of DNA per enzymatic reaction.9
Enzymatic reaction conditions were determined for each endonuclease according to the manufacturer’s recommendations (New England Biolabs, Ipswich, MA, USA). Initially, endonucleases Bam HI, Bgl II, and Bcl I were used in early characterization followed by subsequent digests with Hpa I, Hind III, Sal I, and Sma I.10
Agarose electrophoresis of DNA restriction fragments
The DNA fragments were separated by electrophoresis on 1.0% agarose horizontal slab gels prepared and run in 40mM Tris-acetate-EDTA. Gels were stained in ethidium bromide (0.5 μg/ml). Gel banding patterns were analyzed with Quantity One version 4.3.1 software (Bio Rad, Hercules, CA) with Gel Doc transillumination. Resultant banding profiles were then compared to data from previously published HAdV3 characterizations.1,10,11
Statistical analyses
Questionnaire and laboratory data were linked by the unique specimen number. During studies of clinical outcomes, only the first adenovirus specimen from a specific patient’s illness was considered in studies of clinical events. Chi-square and Fisher’s exact tests were used to examine potential risk factor associations. The exact Cochran-Armitage trend test was used to examine adenovirus variant genome type prevalence across years. Logistic regression modeling was used to examine potential risk factors for outcomes for binary severity (death or intensive care hospitalization versus other clinical encounters). The proportional odds model was used to examine risk factors for severe adenovirus infection considering the outcomes of hospitalization, intensive care unit stay, and death in a mutually exclusive, ordinal severity scale. Analyses were performed using SAS software version 9.1 (SAS Institute, Inc., Cary, NC).
Results
Digests of the 516 clinical HAdV3 isolates with Bam HI (Figure 1) showed that most study isolates were of the HAdV3a genome cluster with the exception of 1 isolate of HAdV3p1 (Figure 1, Lane p1). Further digests with other enzymes were used in the identification of two new subgenome types, designated HAdV3a50 and HAdV3a51. These genotypes demonstrated unique REA profiles when compared to previously reported HAdV3 profiles. HAdV3a50 was distinguished by digest with endonuclease Bcl I (Figure 2 Lane a50), while HAdV3a51 was distinguished with endonuclease Bgl II (Figure 3, Panel A, Lane a51). Though the HAdV3a51 REA profile was consistent with that found for the HAdV3a1 genome type10 as depicted by an additional fragment of approximately 2.7 kilobases, verification by digest using endonuclease Hind III (Figure 3, Panel B, Lane a51) suggests a new variant within the HAdV3a genome. The two novel strains were unique compared to another recently characterized HAdV3 variant, identified as HAdV3a17 by digest with Bgl II (Figure 3, Panel A, Lane a17) and Sma I (Figure 4 Lane a17).3
Figure 1.
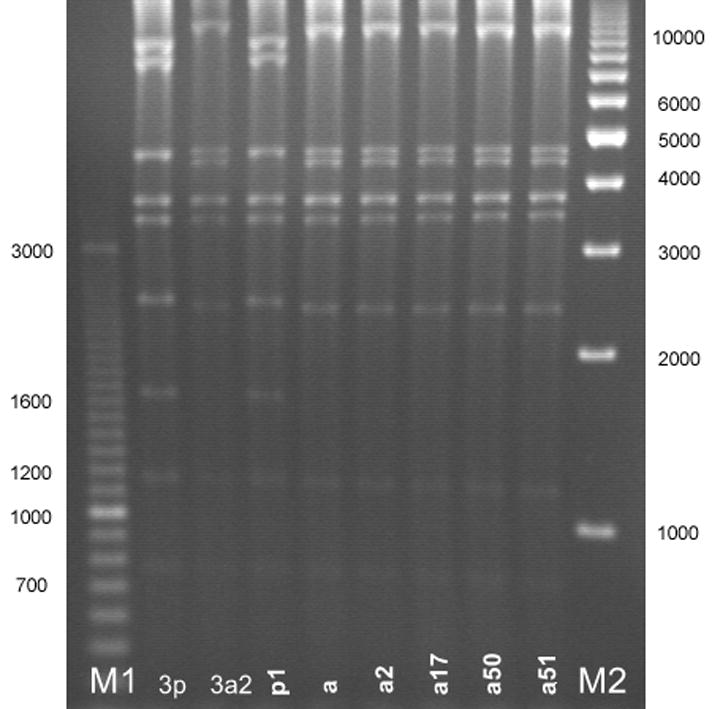
HAdV3 genome types as identified by restriction enzyme analysis with Bam HI. The gel patterns reveal that most of the HAdV3 genome types found were in the HAdV3a genome cluster save for HAdV3p1 (lane p1). Lane designations: M1 and M2 are Bio-Rad EZ Load 100 bp (3 kb range) and 1 kb molecular rulers, respectively (Hercules, CA). Lane 3p=prototype HAdV3p virus (strain GB, US, 1953); lane 3a2=reference HAdV3a2 virus (CDC, 2005). HAdV3 REA patterns from study specimens: p1=HAdV3p1; a=HAdV3a; a2=HAdV3a2; a17=HAdV3a17; a50=HAdV3a50; a51=HAdV3a51.
Figure 2.
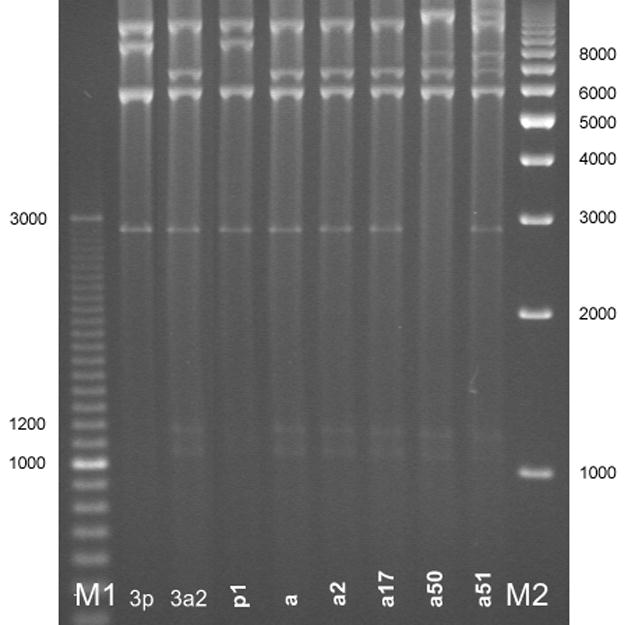
New variant HAdV3a50 (lane a50) as distinguished from other genome types by restriction enzyme analysis with Bcl I. Lane designations: M1 and M2 are Bio-Rad EZ Load 100 bp (3 kb range) and 1 kb molecular rulers, respectively (Hercules, CA). Lane 3p=prototype HAdV3p virus (strain GB, US, 1953); lane 3a2= reference HAdV3a2 virus (CDC, 2005). HAdV3 REA patterns from study specimens: p1=HAdV3p1; a=HAdV3a; a2=HAdV3a2; a17=HAdV3a17; a50=HAdV3a50; a51=HAdV3a51.
Figure 3.
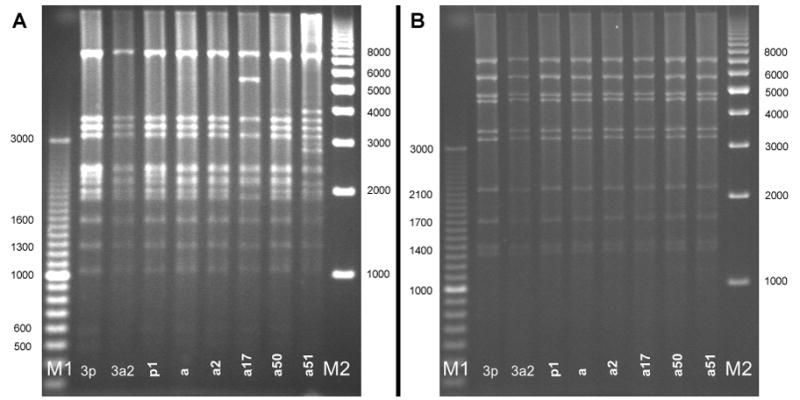
New variant HAdV3a51 (lane a51) as distinguished from other genome types by restriction enzyme analysis with Bgl II (panel A) and Hind III (panel B). Lane designations: M1 and M2 are Bio-Rad EZ Load 100 bp (3 kb range) and 1 kb molecular rulers, respectively (Hercules, CA). Lane 3p=prototype HAdV3p virus (strain GB, US, 1953); lane 3a2= reference HAdV3a2 virus (CDC, 2005). HAdV3 REA patterns from study specimens: p1=HAdV3 p1; a=HAdV3a; a2=HAdV3a2; a17=HAdV3a17; a50=HAdV3a50; a51=HAdV3a51.
Figure 4.
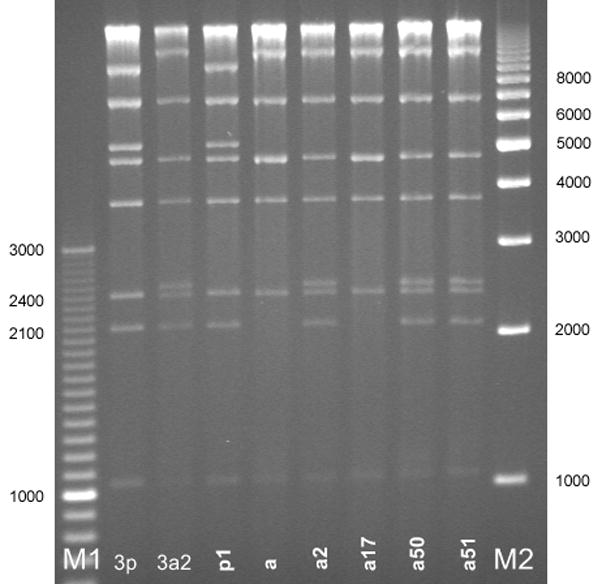
HAdV3 genome types identified by restriction enzyme analysis with Sma I. Lane designations: M1 and M2 are Bio-Rad EZ Load 100 bp (3 kb range) and 1 kb molecular rulers, respectively (Hercules, CA). Lane 3p=prototype HAdV3p virus (strain GB, US, 1953); lane 3a2= reference HAdV3a2 virus (CDC, 2005). HAdV3 REA patterns from study specimens: p1=HAdV3 p1; a=HAdV3a; a2=HAdV3a2; a17=HAdV3a17; a50=HAdV3a50; a51=HAdV3a51.
Among the 516 clinical HAdV3 isolates studied, the most common HAdV3 subgenome types were HAdV3a2 (36.9%), HAdV3a50 (27.1%), HAdV3a51 (18.0%), and HAdV3a17 (4.6%). Non-HAdV3a genome types were rare (1.2%). Among the 516 isolates, 505 were from civilian laboratories which provided some patient data. Through record review5 it was found that 484 of the 505 specimens had complete questionnaire data and were the first HAdV isolated from a patient’s unique clinical event (Table 1). Of these 484 civilian isolates, a high prevalence of newly described HAdV3a genomic variants was found: 131 isolates of HAdV3a50 and 87 isolates of HAdV3a51 (Table 1). Collection year data (Table 1) and a supplemental REA examination with endonucleases Bam HI, Bgl II, Bcl I, and Sma I of 24 archived HAdV3 isolates from the State of California, Department of Public Health laboratory revealed that the HAdV3a genome cluster has been the most prevalent genome type in California, consistent with what has been reported globally since 1963.10,12–16 However, within the HAdV3a genome type, subgenome types HAdV3a50 and HAdV3a51 have recently emerged in the United States. Analysis of these archived isolates has revealed that HAdV3a50 may be found as far back as 2002. Many of these novel HAdV3a50 and HAdV3a51 study strains were obtained from the collaborating laboratory in New Haven, CT (Table 1) which experienced an epidemic of HAdV3 detections during the later part of the study period.17 Most of the novel subgenome type detections were associated with upper respiratory tract infections among otherwise healthy children. Though sample size was small (n=11), the most prevalent HAdV3 genome type among military isolates studied was also identified as HAdV3a2 comprising 8 of the total 11 HAdV3 identified (data not shown).
Table 1.
Distribution of most prevalent adenovirus type 3 (HAdV3) genotypes among a national sample of patients with adenovirus positive clinical specimens.
| Adenovirus types with column percentages |
||||||
|---|---|---|---|---|---|---|
| Variables | total | HAdV3a2 (n=180) | HAdV3a17 (n=22) | HAdV3a50 (n=136) | HAdV3a51 (n=87) | Other HAdV3a (n=66) |
| Collection year** | ||||||
| 2004 | 93 | 58 (32.6) | 2 (9.1) | 18 (13.7) | 1 (1.1) | 14 (21.2) |
| 2005 | 183 | 72 (40.4) | 11 (50) | 62 (47.3) | 9 (10.3) | 29 (43.9) |
| 2006 | 137 | 43 (24.2) | 8 (36.4) | 32 (24.4) | 34 (39.1) | 20 (30.3) |
| 2007 | 71 | 5 (2.8) | 1 (4.5) | 19 (14.5) | 43 (49.4) | 3 (4.5) |
| Patient hospitalized when culture obtained** | ||||||
| No | 311 | 94 (52.8) | 13 (59.1) | 83 (63.4) | 82 (94.3) | 39 (59.1) |
| Yes | 143 | 66 (37.1) | 6 (27.3) | 40 (30.5) | 5 (5.7) | 26 (39.4) |
| Uncertain | 30 | 18 (10.1) | 3 (13.6) | 8 (6.1) | 0 (0) | 1 (1.5) |
| Specimen source** | ||||||
| Upper respiratory tract | 437 | 155 (86.1) | 21 (95.5) | 124 (91.2) | 85 (97.7) | 52 (78.8) |
| Lower respiratory tract | 8 | 3 (1.7) | 0 (0) | 2 (1.5) | 0 (0) | 3 (4.5) |
| Gastrointestinal | 17 | 9 (5) | 0 (0) | 2 (1.5) | 0 (0) | 6 (9.1) |
| Urine | 4 | 2 (1.1) | 0 (0) | 0 (0) | 0 (0) | 2 (3) |
| Blood | 3 | 2 (1.1) | 0 (0) | 1 (0.7) | 0 (0) | 0 (0) |
| Ocular | 6 | 2 (1.1) | 0 (0) | 1 (0.7) | 0 (0) | 3 (4.5) |
| Others | 16 | 7 (3.9) | 1 (4.5) | 6 (4.4) | 2 (2.3) | 0 (0) |
Due to missing values the sum may not add to the total.
“Other HAdV3” include 7 patients with a rare HAdV3 variant type (3a, 3a11, 3p1) and 59 subjects with coinfections. Coinfections are mixed infections with more than one adenovirus type based on REA banding profiles.
Statistically significant with chi-squared test at 5% significance level
Additional clinical data were available from 373 of the 484 patients who were <7 yrs of age or who had undergone a transplantation procedure up to six months prior to adenovirus specimen collection.5 Of these 373 patients, 99.5% were <7 yrs of age, 57.9% were males, 1.6% reported to have been a transplant patient, and 1.6% were diagnosed with cancer (Table 2). In general, relatively few had an underlying chronic disease condition and most suffered simply an upper respiratory tract illness, yet the clinical impact of HAdV3 infection was significant with 136 patients being hospitalized, and 17 receiving intensive care.
Table 2.
Demographic and clinical characteristics of a national sample of patients whose specimens yielded an adenovirus type 3 (HAdV3). Patients were either recent transplant recipients or <7yrs of age. Each patient contributed only 1 positive adenovirus specimen per unique clinical event.
| Adenovirus types with column percentages |
||||||
|---|---|---|---|---|---|---|
| Variables | Total | HAdV3a2 (n=134) | HAdV3a17 (n=17) | HAdV3a50 (n=102) | HAdV3a51 (n=74) | Other HAdV3a (n=46) |
| Age group** | ||||||
| < 7 years old | 371 | 134 (100) | 17 (100) | 102 (100) | 74 (100) | 44 (95.7) |
| ≥ 7 years old | 2 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (4.3) |
| Immunocompromised** | ||||||
| Cancer | 2 | 1 (0.7) | 0 (0) | 0 (0) | 0 (0) | 1 (2.2) |
| Cancer and transplant | 4 | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 3 (6.5) |
| Nothing reported | 353 | 123 (91.8) | 15 (88.2) | 100 (98) | 74 (100) | 41 (89.1) |
| Transplant | 14 | 10 (7.5) | 2 (11.8) | 1 (1) | 0 (0) | 1 (2.2) |
| Cancer types** | ||||||
| None reported | 367 | 133 (99.3) | 17 (100) | 101 (99) | 74 (100) | 42 (91.3) |
| Lymphatic and hematopoietic tissue | 5 | 1 (0.7) | 0 (0) | 1 (1) | 0 (0) | 3 (6.5) |
| Respiratory or intrathoracic organs | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.2) |
| Transplant types** | ||||||
| None reported | 367 | 133 (99.3) | 17 (100) | 101 (99) | 74 (100) | 42 (91.3) |
| Bone marrow | 4 | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 3 (6.5) |
| Heart | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.2) |
| Kidney | 1 | 1 (0.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Upper respiratory tract infection** | (0) | (0) | (0) | (0) | (0) | |
| Yes | 222 | 63 (47) | 9 (52.9) | 51 (50) | 71 (95.9) | 28 (60.9) |
| Not reported | 151 | 71 (53) | 8 (47.1) | 51 (50) | 3 (4.1) | 18 (39.1) |
| Conjunctivitis** | ||||||
| Yes | 51 | 13 (9.7) | 2 (11.8) | 10 (9.8) | 16 (21.6) | 10 (21.7) |
| Not reported | 322 | 121 (90.3) | 15 (88.2) | 92 (90.2) | 58 (78.4) | 36 (78.3) |
| Days in the hospital**† | ||||||
| Missing | 17 | 11 (8.2) | 3 (17.6) | 3 (2.9) | 0 (0) | 0 (0) |
| No | 220 | 68 (50.7) | 7 (41.2) | 59 (57.8) | 61 (82.4) | 25 (54.3) |
| Yes, 0–2 days | 67 | 29 (21.6) | 3 (17.6) | 24 (23.5) | 5 (6.8) | 6 (13) |
| Yes, 3–385 days | 68 | 26 (19.4) | 4 (23.5) | 15 (14.7) | 8 (10.8) | 15 (32.6) |
| Yes, number of days unknown | 1 | 0 (0) | 0 (0) | 1 (1) | 0 (0) | 0 (0) |
“Other HAdV3” includes two patients with a rare HAdV3 variant type (3a, 3a11) and 44 subjects with coinfections. Coinfections are mixed infections with more than one adenovirus type based on REA banding profiles.
Statistically significant with chi-squared test at 5% significance level
Missing and “Yes, number of days unknown” not included in the statistical test.
Multivariable risk factor modeling for adenovirus 3 disease severity (Table 3) demonstrated that children under 2 years of age (OR=2.7; 95%CI 1.6 to 4.6), persons with chronic disease (OR=5.1; 95%CI 2.6–9.8), persons infected with HAdV3a2 (OR=3.0; 95%CI 1.5 to 6.0), with HAdV3a50 (OR=2.5; 95%CI 1.2 to 5.2), or with multiple or rare HAdV strains (OR=2.8; 95%CI 1.3–6.5) were at increased risk of severe HAdV3 clinical disease.
Table 3.
Risk factors for having a severe adenovirus infection, by proportional odds model.
| Variables | n | Unadjusted OR (95% CI) | Adjusted OR (95% CI) |
|---|---|---|---|
| Gender | |||
| Male | 216 | 1.1 (0.7–1.7) | --- |
| Female | 157 | reference | --- |
| Age group (yrs) | |||
| 0–1 | 126 | 2.6 (1.6–4.3) | 2.7 (1.6–4.6) |
| 2–3 | 103 | 1.6 (0.9–2.8) | 1.7 (0.979–3.1) |
| >4 | 144 | reference | reference |
| Adenovirus 3 variant type | |||
| a2 | 134 | 3.2 (1.6–6.4) | 3 (1.5–6) |
| a17 | 17 | 3 (1–9.2) | 2.2 (0.7–7.1) |
| a50 | 102 | 2.8 (1.4–5.8) | 2.5 (1.2–5.2) |
| Other HAdV3a | 46 | 4.1 (1.8–9.2) | 2.8 (1.2–6.5) |
| a51 | 74 | reference | reference |
| Chronic disease | |||
| Yes | 43 | 5.6 (3–10.8) | 5.1 (2.6–9.8) |
| No/Unknown | 330 | reference | reference |
“Other HAdV3” includes two patients with a rare HAdV3 variant type (3a, 3a11) and 44 subjects with coinfections. Coinfections are mixed infections with more than one adenovirus type based on REA banding profiles.
Discussion
For nearly 25 years epidemiologists in a number of countries have noticed that certain types of adenovirus strains are often associated with clinical disease and epidemics.1–5,7,10,11,13–16,18–32 Antigenic and genetic variability clearly play a role in explaining epidemics and severe disease outcomes.1–5,7,13,14,16,19,21,24,28–30
In our previous 3-year study5 we evaluated more than 3000 clinical HAdV isolates with hexon gene sequence typing. HAdV3 was the most prevalent serotype found (34.6%).
The majority of HAdV3 genomic characterization work has been reported by investigators outside the United States, particularly in Asia and South America. Our study’s findings are consistent with limited historical data generated by the investigation of isolates collected in North America, either as studies designed to examine the global distribution of HAdV or as response to outbreak events. The most prevalent HAdV3 subgenome type identified by this study was HAdV3a2. This variant was first identified from a panel of HAdV positive specimens isolated in China between the years of 1962–198510 and has been considered the most prevalent HAdV3 genome type for decades. It has been suggested that HAdV3 has the capacity to remain somewhat genetically stable within certain geographic regions while exhibiting a great deal of variability as distributed globally.30
Having developed a typing algorithm based on previous studies,10,28 we identified two new HAdV3 subgenome type variants, HAdV3a50 and HAdV3a51, provided evidence suggesting their recent emergence, and reported an association of HAdV3a51 with more severe disease. The nomenclature for the two new virus strains is arbitrary as there is no central repository of REA defined subgenome types, no catalog of high quality REA images, and no library of supporting sequence data. One must rely upon the indexed medical literature and such literature is not without controversy. While a number of REA studies seem to agree in following established REA typing algorithms (i.e. 4- vs. 6- base restriction site enzymatic digests), there has been a lack of agreement in genome and subgenome type nomenclature, especially with suspected newly emergent variant types of HAdV3. When specific endonucleases have been left out of REA characterization efforts, resultant typing data may be inconsistent. A better nomenclature system and consensus libraries of REA image data are needed for classifying human adenoviruses through REA techniques. DNA sequence data should be used to validate these prototype virus gel profiles.
This study had a number of limitations. While the surveillance study was designed to capture risk information from children and patients with recent transplants, the medical record information may not have included additional risk factors for evaluation. Multiple laboratory staff and laboratory interns contributed to this effort and this variability could have introduced various biases. However, the first author developed well-documented laboratory protocols and was directly involved with all data collection methods. Performing REA with several endonucleases on more than 500 HAdV3s presented a variety of challenges, particularly obtaining adequate DNA for thorough REA characterization. While numerous REAs were repeated, the challenge was met in large part by standardization of viral culture techniques and innovations such as the use of tissue cluster plates and the harvest of and DNA extraction from infected cell pellets coupled with viral supernatant. The study was further limited by the quality of REA images in the available literature. Sometimes it was difficult to distinguish important bands from these images.
Due to its specialized and laborious nature, REA has been an infrequent method of adenovirus characterization. It appears that PCR-based identification methods7,33,34 are supplanting traditional serotyping and REA methods. While we embraced PCR and sequencing-based typing methods it is important to note that these methods focus upon limited areas of the adenovirus genome and have potential to overlook and mistype new HAdV variants which differ genetically in other gene regions. Hence, until whole genome sequencing becomes less expensive, REA will continue to have an important role in the epidemiological study of adenoviruses.
In summary, a large collection of recently circulating clinical HAdV3s from diverse geographical areas within the United States was examined by molecular analyses. REA revealed temporal changes in the distribution of HAdV3 genome types and the emergence of novel subgenome types HAdV3a50 and HAdV3a51. Both strains were associated with an epidemic of clinical detections and at least one of these novel genome types was associated with more severe disease. This work and previous examples of virulent viral strain emergence and genetic HAdV diversity underscore the need for national and international HAdV surveillance and both serotypic and genotypic characterization.
Acknowledgments
The authors thank Dean D. Erdman and Xiaoyan Lu of the Centers for Disease Control and Prevention, Atlanta, Georgia, USA; Adriana E. Kajon of the Lovelace Respiratory Research Institute, Albuquerque, New Mexico; Margaret L. Chorazy and the numerous University of Iowa graduate students and laboratory interns who have contributed to the molecular study of the viral specimens. The authors also wish to thank the surveillance study collaborators who provided the clinical specimens: James D. Chappell, MD, PhD of the Departments of Pathology and Pediatrics, Vanderbilt University School of Medicine, Nashville, TN; Maria Vu and Leta Crawford Miksza, PhD of the California Department of Public Health; Jeffrey D. Dawson, ScD of the Department of Biostatistics, University of Iowa College of Public Health, Iowa City, IA; Gail J. Demmler, MD of the Department of Pediatrics, Baylor College of Medicine and Diagnostic Virology Laboratory, Texas Children’s Hospital, Houston, TX; Gary Doern, PhD from the University of Iowa College of Medicine; Christine C. Ginocchio, PhD of North Shore University Hospital and North Shore-LIJ Health System Laboratories, Manhasset, NY; Jennifer Goodrich, PhD of the University of North Carolina Hospitals, Chapel Hill, NC; Diane C. Halstead, PhD of Infectious Disease Laboratories, Baptist Medical Center, Jacksonville, FL; Sue C. Kehl PhD of the Department of Pathology, Medical College of Wisconsin, Milwaukee, WI; Deanna L. Kiska, PhD of the Department of Clinical Pathology, SUNY Upstate Medical University, Syracuse, NY; Diane S. Leland, PhD of Indiana University School of Medicine and Clarian Health Partners, Indianapolis, IN; Melissa B. Miller PhD of the Department of Pathology and Laboratory Medicine, University of North Carolina School of Medicine, Chapel Hill, NC; Christine C. Robinson, PhD of the Department of Pathology, The Children’s Hospital, Denver, CO; Kevin L. Russell, MD, MTM&H and David Metzgar, PhD of the Navy Respiratory Disease Laboratory, San Diego, CA; Michael A. Saubolle PhD of Laboratory Sciences of Arizona/Sonora Quest Laboratories, Tempe, AZ; Rangaraj Selvarangan, BVSc, PhD of the Department of Pathology and Laboratory Medicine, Children’s Mercy Hospital, Kansas City, MO; Gregory A. Storch, MD of St. Louis Children’s Hospital, St. Louis, Missouri; and Danielle M. Zerr, MD, MPH of the Department of Pediatrics, University of Washington and Children’s Hospital and Regional Medical Center, Seattle, WA. Thanks also to Kevin L. Knudtson, PhD of the University of Iowa’s DNA Facility for his much appreciated assistance in sequencing work and use of the facility’s NanoDrop spectrophotometer. This research has been conducted in compliance with all applicable Federal Regulations governing the protection of human subjects in research.
Financial support: National Institute of Allergy and Infectious Diseases, National Surveillance for Emerging Adenovirus Infections (NIH/NIAID R01 AI053034, G.C.G. principal investigator).
Footnotes
Potential conflicts of interest: G.C.G. served on the Data Monitoring Board for adenovirus vaccine trials sponsored by Duramed, Inc. No other authors have potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Itoh N, Tanaka K, Aoki K, Hinokuma R, Nakagawa H, Takeuchi S, et al. Four new genotypes of adenovirus type 3 isolated from patients with conjunctivitis in Japan. J Med Virol. 1999;59:73–7. doi: 10.1002/(sici)1096-9071(199909)59:1<73::aid-jmv12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 2.Jones MS, 2nd, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, et al. New adenovirus species found in a patient presenting with gastroenteritis. J Virol. 2007;81:5978–84. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YJ, Hong JY, Lee HJ, Shin SH, Kim YK, Inada T, et al. Genome type analysis of adenovirus types 3 and 7 isolated during successive outbreaks of lower respiratory tract infections in children. J Clin Microbiol. 2003;41:4594–9. doi: 10.1128/JCM.41.10.4594-4599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kajon AE, Murtagh P, Garcia Franco S, Freire MC, Weissenbacher MC, Zorzopulos J. A new genome type of adenovirus 3 associated with severe lower acute respiratory infection in children. J Med Virol. 1990;30:73–6. doi: 10.1002/jmv.1890300116. [DOI] [PubMed] [Google Scholar]
- 5.Gray GC, McCarthy T, Lebeck MG, Schnurr DP, Russell KL, Kajon AE, et al. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004–2006. Clin Infect Dis. 2007;45:1120–31. doi: 10.1086/522188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz H, Wigand R, Heinrich W. Worldwide epidemiology of human adenovirus infections. Am J Epidemiol. 1983;117:455–66. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- 7.Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587–602. doi: 10.1007/s00705-005-0722-7. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy T, Lebeck MG, Capuano AW, Schnurr DP, Gray GC. Molecular typing of clinical adenovirus specimens by an algorithm which permits detection of adenovirus coinfections and intermediate adenovirus strains. J Clin Virol. 2009;46:80–4. doi: 10.1016/j.jcv.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roe BA. Concentration of DNA by ethanol precipitation. [cited 2008 July 21, 2008]; Available from: http://mycoplasmas.vm.iastate.edu/lab_site/methods/DNA/precip.html.
- 10.Li QG, Wadell G. Comparison of 17 genome types of adenovirus type 3 identified among strains recovered from six continents. J Clin Microbiol. 1988;26:1009–15. doi: 10.1128/jcm.26.5.1009-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arens M, Dilworth V. Remarkably homogeneous population of adenovirus type 3 and 7 genome types. J Clin Microbiol. 1988;26:1604–8. doi: 10.1128/jcm.26.8.1604-1608.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wadell G. Molecular epidemiology of human adenoviruses. Curr Top Microbiol Immunol. 1984;110:191–220. doi: 10.1007/978-3-642-46494-2_7. [DOI] [PubMed] [Google Scholar]
- 13.Adrian T, Best B, Hierholzer JC, Wigand R. Molecular epidemiology and restriction site mapping of adenovirus type 3 genome types. J Clin Microbiol. 1989;27:1329–34. doi: 10.1128/jcm.27.6.1329-1334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang SY, Lee CN, Lin PH, Huang HH, Chang LY, Ko W, et al. A community-derived outbreak of adenovirus type 3 in children in Taiwan between 2004 and 2005. J Med Virol. 2008;80:102–12. doi: 10.1002/jmv.21045. [DOI] [PubMed] [Google Scholar]
- 15.Golovina GI, Zolotaryov FN, Yurlova TI. Sensitive analysis of genetic heterogeneity of adenovirus types 3 and 7 in the Soviet Union. J Clin Microbiol. 1991;29:2313–21. doi: 10.1128/jcm.29.10.2313-2321.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James L, Vernon MO, Jones RC, Stewart A, Lu X, Zollar LM, et al. Outbreak of human adenovirus type 3 infection in a pediatric long-term care facility--Illinois, 2005. Clin Infect Dis. 2007;45:416–20. doi: 10.1086/519938. [DOI] [PubMed] [Google Scholar]
- 17.Landry ML, Lebeck MG, Capuano AW, McCarthy T, Gray GC. Adenovirus type 3 outbreak in Connecticut associated with a novel variant. J Med Virol. 2009;81:1380–4. doi: 10.1002/jmv.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adrian T, Wigand R. Adenovirus 3–7, an intermediate strain of subgenus B. Intervirology. 1986;26:202–6. doi: 10.1159/000149702. [DOI] [PubMed] [Google Scholar]
- 19.Bailey AS, Richmond SJ. Genetic heterogeneity of recent isolates of adenovirus types 3, 4, and 7. J Clin Microbiol. 1986;24:30–5. doi: 10.1128/jcm.24.1.30-35.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brummitt CF, Cherrington JM, Katzenstein DA, Juni BA, Van Drunen N, Edelman C, et al. Nosocomial adenovirus infections: molecular epidemiology of an outbreak due to adenovirus 3a. J Infect Dis. 1988;158:423–32. doi: 10.1093/infdis/158.2.423. [DOI] [PubMed] [Google Scholar]
- 21.Cheng CC, Huang LM, Kao CL, Lee PI, Chen JM, Lu CY, et al. Molecular and clinical characteristics of adenoviral infections in Taiwanese children in 2004–2005. Eur J Pediatr. 2008;167:633–40. doi: 10.1007/s00431-007-0562-4. [DOI] [PubMed] [Google Scholar]
- 22.Choi EH, Kim HS, Park KH, Lee HJ. Genetic heterogeneity of the hexon gene of adenovirus type 3 over a 9-year period in Korea. J Med Virol. 2006;78:379–83. doi: 10.1002/jmv.20550. [DOI] [PubMed] [Google Scholar]
- 23.Deng fu Guo RS, Shinagawa M, Sato G, Aoki K, Sawada H. Genomic comparison of adenovirus type 3 isolates from patients with acute conjunctivitis in Japan, Australia, and the Phillipines. Microbiol Immunol. 1988;32:833–42. doi: 10.1111/j.1348-0421.1988.tb01444.x. [DOI] [PubMed] [Google Scholar]
- 24.Fujimoto T, Hamamoto I, Taniguchi K, Chikahira M, Okabe N. Molecular epidemiology of adenovirus type 3 detected from 1994 to 2006 in Hyogo Prefecture, Japan. Jpn J infect Dis. 2008;61:143–5. [PubMed] [Google Scholar]
- 25.Harley D, Harrower B, Lyon M, Dick A. A primary school outbreak of pharyngoconjunctival fever caused by adenovirus type 3. Commun Dis Intell. 2001;25:9–12. doi: 10.33321/cdi.2001.25.2. [DOI] [PubMed] [Google Scholar]
- 26.Itakura S, Aoki K, Sawada H, Shinagawa M. Analysis with restriction endonucleases recognizing 4- or 5-base-pair sequences of human adenovirus type 3 isolated from ocular diseases in Sapporo, Japan. J Clin Microbiol. 1990;28:2365–9. doi: 10.1128/jcm.28.10.2365-2369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajon AE, Portes SA, de Mello WA, Nascimento JP, Siqueira MM. Genome type analysis of Brazilian adenovirus strains of serotypes 1,2,3,5, and 7 collected between 1976 and 1995. J Med Virol. 1999;58:408–12. doi: 10.1002/(sici)1096-9071(199908)58:4<408::aid-jmv14>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 28.Li QG, Zheng QJ, Liu YH, Wadell G. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J Med Virol. 1996;49:170–7. doi: 10.1002/(SICI)1096-9071(199607)49:3<170::AID-JMV3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 29.Matsui K, Shimizu H, Yoshida A, Nagaoka E, Nishio O, Okuda K. Monitoring of adenovirus from conjunctival scrapings in Japan during 2005--2006. J Med Virol. 2008;80:997–1003. doi: 10.1002/jmv.21175. [DOI] [PubMed] [Google Scholar]
- 30.Mizuta K, Suzuki H, Ina Y, Yazaki N, Sakamoto M, Katsushima N, et al. Six-year longitudinal analysis of adenovirus type 3 genome types isolated in Yamagata, Japan. J Med Virol. 1994;42:198–202. doi: 10.1002/jmv.1890420218. [DOI] [PubMed] [Google Scholar]
- 31.Moraes MT, da Silva M, Leite JP, Nascimento JP. Genetic and antigenic analysis of adenovirus type 3 strains showing intermediate behavior in standard seroneutralization test. Mem Inst Oswaldo Cruz. 1998;93:231–5. doi: 10.1590/s0074-02761998000200019. [DOI] [PubMed] [Google Scholar]
- 32.Singh-Naz N, Brown M, Ganeshananthan M. Nosocomial adenovirus infection: molecular epidemiology of an outbreak. Pediatr Infect Dis J. 1993;12:922–5. [PubMed] [Google Scholar]
- 33.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J Clin Microbiol. 2004;42:3963–9. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W, McDonough MC, Erdman DD. Species-specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol. 2000;38:4114–20. doi: 10.1128/jcm.38.11.4114-4120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


