Abstract
OBJECTIVE
MicroRNAs (miRNAs) are a class of noncoding small RNAs that act as negative regulators of gene expression. The miRNAs exhibit tissue-specific expression patterns and changes in their expression may contribute to pathogenesis. The objectives of this study were to identify miRNAs expressed in articular chondrocytes, determine changes in osteoarthritic cartilage and address the function of miR-140.
METHODS
To identify miRNAs specifically expressed in chondrocytes, we performed gene expression profiling using miRNA microarrays and quantitative PCR with human articular chondrocytes compared to human mesenchymal stem cells (MSC). The expression pattern of miR-140 was monitored during chondrogenic differentiation of hMSC in pellet cultures and in human articular cartilage from normal and osteoarthritic knee joints. We tested effects of IL-1β on miR-140 expression. Double-strand (ds) miR-140 was transfected into chondrocytes to analyze changes in the expression of genes associated with osteoarthritis.
RESULTS
Microarray analysis showed that miR-140 has the largest difference in expression between chondrocytes and MSC. During chondrogenesis cultures of MSC miR-140 expression increased in parallel with Sox9 and Col2a1. Normal human articular cartilage expressed miR-140 and this was significantly reduced in OA tissue. In vitro treatment of chondrocytes with IL-1β suppressed miR-140 expression. Transfection of chondrocytes with ds-miR-140 downregulated IL-1β-induced ADAMTS-5 expression and rescued the IL-1β –dependent repression of Aggrecan gene expression.
CONCLUSION
This study shows that miR-140 has a chondrocyte differentiation-related expression pattern. The reduction in miR-140 expression in OA cartilage and in response to IL-1β may contribute to the abnormal gene expression pattern characteristic of OA.
Keywords: microRNA, chondrocytes, mesenchymal stem cells, cartilage, osteoarthritis
INTRODUCTION
Osteoarthritis (OA) is a chronic and highly prevalent degenerative joint disease. Approximately 40 million Americans are currently affected and this number is predicted to increase to 60 million within the next twenty years as a result of population aging and an increase in life expectancy (1, 2). Current treatment is limited to pain management and disease-modifying therapies are not available and in the late phase of the disease process where joint replacement surgery is often indicated. OA has been associated with age-related loss of the homeostatic balance between degradation and repair mechanisms. Cartilage cellularity in OA is reduced by chondrocyte death, and remaining chondrocytes are activated by cytokines and growth factors to a catabolic and abnormal differentiation that leads to degradation of extracellular matrix (3–6). Molecular mechanisms that govern articular chondrocyte differentiation during development and maintenance of articular cartilage are being characterized and this has the potential to lead to new therapeutic interventions.
MicroRNA (miRNAs) are a class of non-coding small RNAs that play roles in biological processes as negative regulators of gene expression by promoting mRNA degradation and/or repressing translation through sequence-specific interactions with the 3′ untranslated regions (UTRs) of specific mRNA targets (7–10). Hundreds of miRNAs have been found in various organisms, and many miRNAs are evolutionarily conserved. Moreover, one third of all mammalian mRNAs seem to be under miRNA regulation suggesting an essential role in regulating gene expression (11). Several miRNAs exhibit a tissue- or developmental stage-specific expression pattern and have been associated with diseases such as cancer, heart disease, diabetes and rheumatoid arthritis (12–16). Mice with limb or cartilage specific deletion of the miRNA processing enzyme Dicer showed a severe phenotype with reduced limb size but normal patterning (17, 18). As Dicer is indispensable for producing a functional, mature type of miRNA, this finding suggests that the presence of specific miRNAs plays a critical role in skeletal development. Although Tuddenham et al showed cartilage specific expression of miR-140 in mouse embryos (19), the role of tissue-specific miRNAs in articular cartilage has not been reported.
We hypothesized that miRNAs are novel regulators of cartilage homeostasis and changes in their expression and function play an important role in diseases affecting articular cartilage. The objectives of this study were to identify miRNAs expressed in articular chondrocytes, determine changes in osteoarthritic cartilage and address the function of the chondrocyte-specific miR-140.
MATERIALS AND METHODS
Human tissue samples, cell isolation and culture
Human articular cartilages from knee joints were obtained from 8 normal donors (38.22 ± 5.31 years of age, mean ± SD) and from 11 OA patients (79.36 ± 9.72 years of age, mean ± SD) undergoing total knee arthroplasty. Tissue collection was approved by the Scripps Human Subjects Committee. All samples were examined by Safranin O staining and graded according to a modified Mankin scale, with a score of less than 2 points being normal and a score of greater than 5 representing OA. RNA was isolated from fresh frozen cartilage by homogenizing the tissue in a freezer mill (Spex CertiPrep, Inc., Metuchen, NJ) and extracting the homogenate in Trizol (Invitrogen Corporation, Carlsbad, CA). Human chondrocytes were isolated and cultured as described previously (20). Experiments with chondrocytes were performed in passage 1–2. Human bone marrow-derived mesenchymal stem cells (MSCs) were isolated from iliac crest bone marrow obtained from normal adult donors (with the approval of Human Subjects Committee) and cultured as described previously (20, 21). Experiments with MSCs were performed in passage 3–6.
Microarray analysis
Small RNAs of less than 200 nucleotides in length were extracted from MSCs and chondrocytes with mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer’s protocol. Purified RNA was then labeled with Cy3 or Cy5 by mirVana miRNA labeling kit (Ambion). In brief, RNA was subjected to a tailing reaction with amine-modified nucleotide triphosphates by poly (A) polymerase, followed by amide formation using Cy dye ester. Labeled RNA was hybridized on slides, on which oligonucleotides against human miRNA had been arrayed (Hokkaido-System Science, Japan), and detected by a scanner (Agilent Technologies, Santa Clara, CA).
Chondrogenesis in human mesenchymal stem cells
Human bone marrow mesenchymal stem cells (MSC) were used to prepare pellets (5.0×105 cells/pellet) by centrifuging the cells at 500g in 15ml conical polypropylene tubes and cultured in chondrogenic medium (Lonza, Walkersville, MD) supplemented with BMP-2 (500ng/ml) and TGFs3 (10 ng/ml). Medium was changed every 2–3 days. To monitor miR-140 throughout chondrogenesis, MSCs were processed for RNA isolation on day 7 and day 14. Chondrogenesis was monitored via Sox9, Aggrecan and Col2a1 expression and Safranin O staining.
Treatment with IL-1β
Chondrocytes were maintained in 12-well plates, containing DMEM plus 10% calf serum and 1% penicillin/streptomycin. Following treatment with recombinant human IL-1β (5 ng/ml; PeproTech, Rocky Hill, NJ). Cells were washed with cold PBS, and total RNA was isolated with Trizol reagent. Quantitative PCR was performed with the TaqMan microRNA assay kit for mature miR-140 or Taqman Gene Expression Assay.
Transfection of double-stranded miR-140 into human articular chondrocytes
Double-strand (ds) RNA oligos representing mature sequences that mimic endogenous miR-140 were transfected into human chondrocytes at 80–90% confluence at 4nM concentration with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Synthesized RNA oligos 5′-CAGUGGUUUUACCCUAUGGUAG and 5′-ACCACAGGGUAGAACCACGGAC were annealed to obtain ds-miR-140. Silencer Negative Control siRNA #1 (Ambion, Austin, TX) at the same concentration as the specific miR-140 ds RNA was included in each experiment.
Quantitative real-time polymerase chain reaction (PCR)
Total RNA was isolated from cartilage tissues, monolayer or pellet cultures using Trizol (Invitrogen). Quantitative real time PCR (qPCR) for miRNAs was performed using the TaqMan MicroRNA reverse Transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Complementary DNA (cDNA) was produced using Ready To- GO You-Prime First-Strand Beads (GE Healthcare UK, UK) with total RNA 1μg and oligo (dT)18 primers. Quantitative real time PCR was performed using TaqMan Gene Expression Assay probe for Col2a1 (Hs00164004_m1), Aggecan Hs00202971_m1), ADAMTS-5 (Hs00199841_m1), MMP-13 (Hs00233992_m1), and GAPDH (Hs99999905_m1) (Applied Biosystems). The U18 and GAPDH genes were used as an internal control to normalize differences in each sample. Expression levels for each gene were assessed relative to U18 or GAPDH expression.
Statistical analysis
Statistically significant differences between two groups were determined with t tests. The results are reported as mean ± SEM. P values of less than 0.05 were considered significant.
RESULTS
miR-140 expression in articular chondrocytes and mesenchymal stem cells
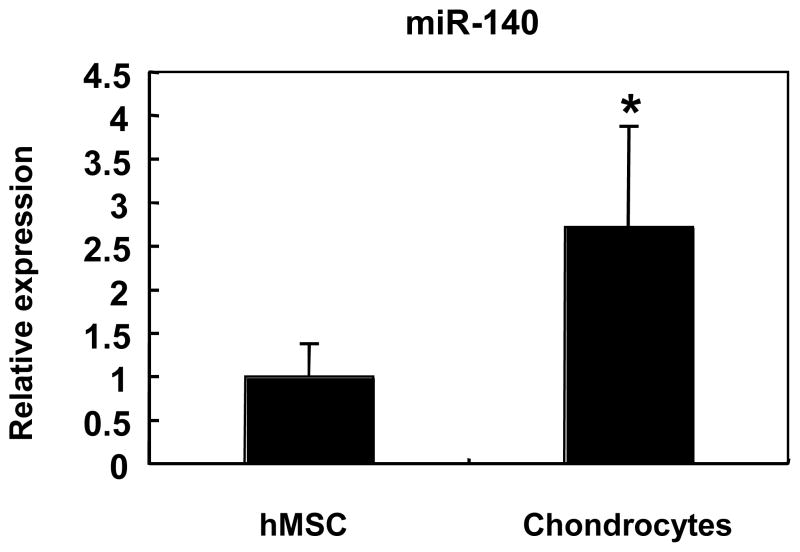
Chondrogenic differentiation of MSCs involves dynamic changes of various gene expression patterns including induced expression of chondrocytes specific genes, including Sox9 and Col2. In order to screen miRNAs specifically expressed in chondrocytes, we performed gene expression profiling using miRNA microarrays comparing primary chondrocytes from articular cartilage to MSCs. In primary articular chondrocytes, several miRNAs were more abundant as compared with undifferentiated MSCs. The largest difference was observed for miR-140 (Table 1). The high expression miR-140 in chondrocytes compared to MSCs was confirmed by qPCR (Figure 1).
Table 1.
miR expression in human articular chondrocytes and MSCs
| miRNA | Ratio (Chondrocyte/MSC) | * Chondrocyte | * hMSC |
|---|---|---|---|
| miR-140 | 2.12 | 540.89 | 254.377 |
| miR-197 | 1.88 | 5395.03 | 2869.38 |
| miR-148 | 1.78 | 644.35 | 361.58 |
| miR-328 | 1.70 | 2768.41 | 1632.95 |
| miR-27b | 1.63 | 6092.21 | 3746.00 |
| miR-16 | 1.59 | 4398.78 | 2764.09 |
| miR-222 | 1.55 | 6349.65 | 4087.27 |
| miR-15b | 1.55 | 668.29 | 430.56 |
| miR-505 | 1.54 | 1967.68 | 1273.77 |
| miR-23b | 1.52 | 8114.26 | 5334.24 |
RNA was isolated from articular chondrocytes and MSCs for microarray analysis of miRs. Differentially expressed (at least 1.5 fold difference) miRs are shown as ratio chondrocytes/MSCs.
raw signal intensity values.
Figure 1. The expression of miR-140 in articular chondrocytes and MSCs.
Array data for miR-140 were validated by qPCR on MSCs (n=3, different preparations from 2 different donors) and articular chondrocytes (n=8, different preparations from 8 different donors). miR-140 expression was significantly higher in chondrocytes compared to MSCs (*P=0.015). Values are mean ± SEM expressed as fold difference relative to expression level in MSCs.
Expression of miR-140 during chondrogenesis of MSCs
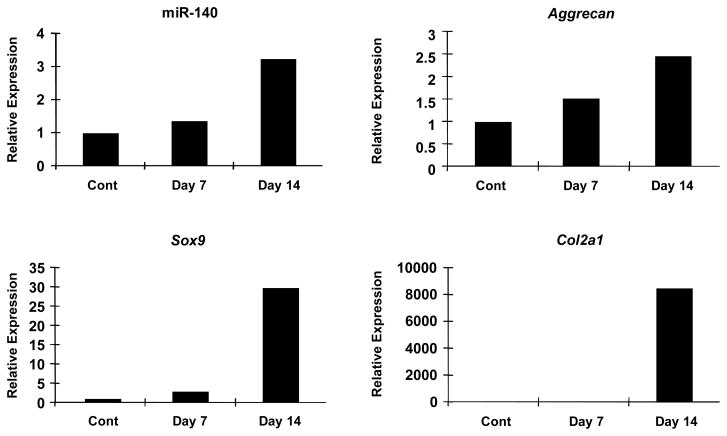
In vitro chondrogenesis assay using MSCs reflects, in part, in vivo skeletal formation. To examine dynamic expression pattern of miR140 during in vitro chondrogenesis, we performed Taqman-qPCR assay to analyze expression patterns of miR-140. Pellets of MSCs were strongly stained by safranin O after chondrogenesis induction for 14 day (data not shown). In this model, miR-140 expression gradually increased during chondrogenesis in parallel with Sox9, Aggrecan and Col2a1 expression (Figure 2). These data indicate that miR140 increases during chondrocytic differentiation of MSC and this is consistent with its high expression in chondrocytes.
Figure 2. Changes in miR-140 expression during chondrogenesis.
RNA was isolated from undifferentiated MSCs (cont) and from MSCs pellets cultures in chondrogenesis medium after 7 and 14 days. miR-140, Sox9 and Col2a1expression were analyzed by qPCR. The expression of miR-140 increased during chondrogenesis along with increased Sox9 and Col2a1. Results are shown as relative expression where expression in undifferentiated MSCs (cont) is defined as 1.
Expression of miR-140 in normal and OA cartilage
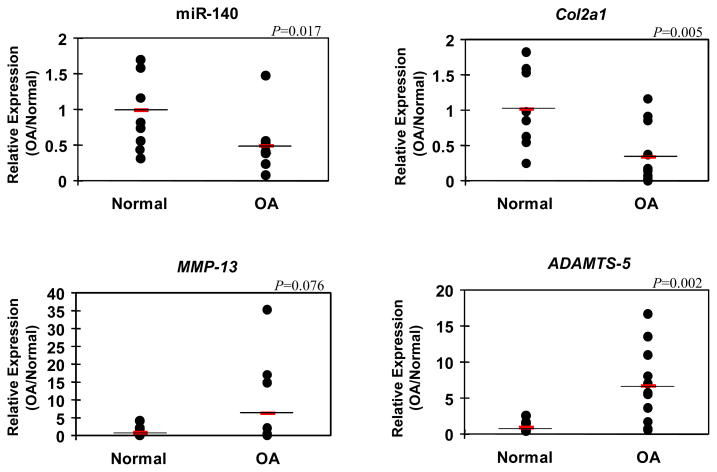
In OA pathogenesis, several chondrocyte specific genes, including Col2a1 and Sox9, are downregulated (22). On the other hand, cartilage degrading enzymes, including ADAMTS5 and MMP-13, are upregulated (23–25). To examine changes in miR-140 expression in OA articular cartilage, qPCR of miR-140 together with OA related marker genes was performed on 19 samples prepared from human knee articular cartilage (normal=8, OA=11). As expected, the expression of ADAMTS-5 was significantly increased in OA cartilage while the expression of Col2a1 was significantly lower than in normal cartilage (Figure 3). MiR-140 expression in articular cartilage from OA donors (65 to 93 years old; Mankin score: 5–10) was significantly lower than in normal cartilage (30 to 44 years old; Mankin score: 0–2) (Figure 3). These data demonstrate abnormally reduced miR-140 expression in OA cartilage, appears to correlate with increased ADAMTS-5 expression and reduced Col2a1 expression in the same samples.
Figure 3. miR-140 expression in normal and OA articular cartilage.
Full thickness cartilage was collected from normal (n=8) and OA (n=11) knee joints for RNA isolation. miR-140, Col2a1, ADAMTS-5, and Sox9 expression were determined by qPCR. miR140 (P=0.017) and Col2a1 (P=0.005) expression was significantly decreased and ADAMTS-5 (P=0.002) was significantly increased in OA cartilage compared with normal cartilage. Values are mean ± SEM expressed as fold difference relative to normal cartilage expression level.
Effect of IL-1β on miR-140 expression in articular chondrocytes
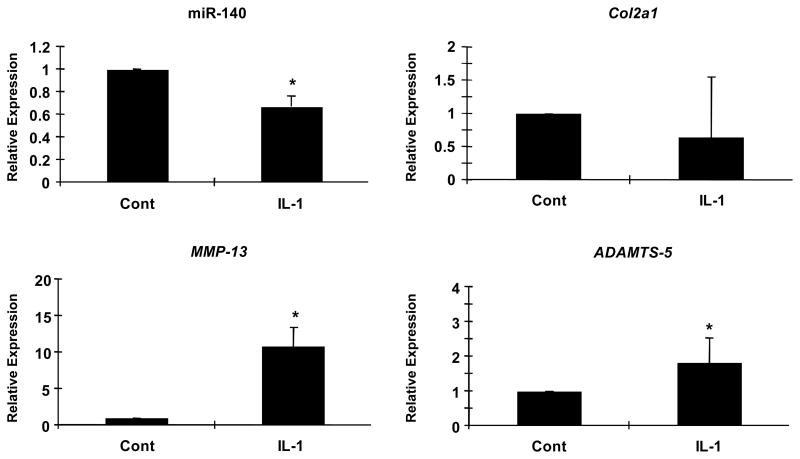
IL-1β is one of the critical mediators of osteoarthritis and IL-1β stimulation on chondrocytes causes similar gene expression patterns with OA cartilage (26, 27). To analyze effects of IL-1β on the expression of miR-140 in articular chondrocytes, we performed qPCR for miR-140 and Col2a1, Aggrecan, MMP-13 and ADAMTS-5. In response to IL-1β stimulation the expression of miR-140 was markedly decreased, while MMP-13 and ADAMTS-5 expression were significantly increased (Figure 4). Under the same experimental conditions expression of Col2a1 did not significantly change. Taken together, these results show reduced mi-R140 expression in the context of IL-1β induced OA-like changes in chondrocyte gene expression.
Figure 4. IL-1β suppresses miR-140 in vitro.
Articular chondrocytes (n=8 different preparations from 8 different donors) were treated with IL-1β (5 ng/ml) for 5 hours. miR-140, Col2a1, MMP-13 and ADAMTS-5 were analyzed by qPCR. IL-1β stimulation significantly decreased miR-140 expression and increased MMP-13 and ADAMTS-5 expression. Col2a1 expression did not significantly change. Values are mean ± SEM expressed as fold difference relative to control expression level. *Significant difference (p<0.05).
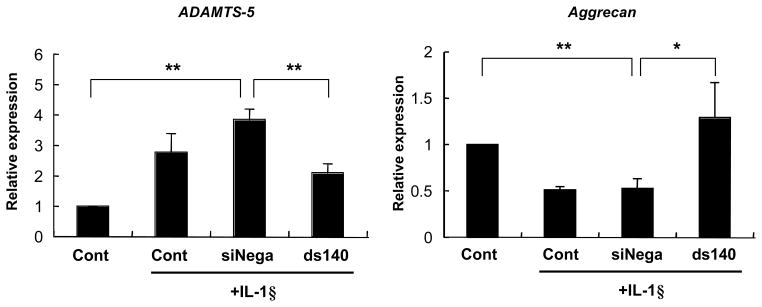
miR-140 modulates ADAMTS-5 and Aggrecan expression in articular chondrocytes
To investigate miR-140 function in chondrocytes, we examined whether expression of the above osteoarthritis related genes; ADAMTS-5, MMP-13, Col2a1 and Aggrecan, can be regulated by miR-140, when chondrocytes were stimulated with IL-1β with and without transfection of ds-miR-140. The ds-miR-140 significantly reduced ADAMTS-5 expression with IL-1β stimulation and conversely, Aggrecan expression with IL-1β stimulation was significantly increased by ds-miR-140 (Figure 5). In the absence of IL-1β, ds140 miRNA did not change the basal levels of aggrecan and we observed an increase in ADAMTS-5 mRNA with the ds140 miRNA as well as with the non-specific dsRNA. The expression of MMP-13 and Col2a1 were not significantly changed by ds-miR-140 (data not shown). These results demonstrated that miR-140 regulates genes encoding ADAMTS-5 and Aggrecan suggesting that miR-140 plays an important role in regulating the balance between extracellular matrix formation and degradation.
Figure 5. ADAMTS-5 and Aggrecan expression in articular chondrocytes by ds-miR-140.
Articular chondrocytes (n=3) were transfected with ds-miR-140. ADAMTS-5 and Aggrecan expression were analyzed by qPCR. (A) The ds-miR-140 significantly reduced ADAMTS-5 expression with and without IL-1β (5 ng/ml) stimulation for 5 hours. (B) The decreased Aggrecan expression with IL-1β stimulation was significantly increased by ds-miR-140. Values are mean ± SEM expressed as fold difference relative to siNegative control expression level. Significant difference (**=p<0.01, *= p<0.05).
DISCUSSION
This is the first study to identify miRs that are expressed in a differentiation-dependent pattern in mesenchymal stem cells and articular chondrocytes. We also show changes in expression of the selected miR-140 in OA cartilage and in response to IL-1β. Moreover, we demonstrate that ADAMTS-5, a critical proteinase in OA pathogenesis, is regulated by miR-140.
Previous studies using systematic whole mount in situ hybridization analysis for miRs in zebrafish revealed that many miRs are expressed in a tissue-specific pattern (28). From this database annotation, miR-140 was the only miR with a cartilage specific expression pattern. Zebrafish embryos injected with miR-140 duplex RNA had a profound facial phenotype, including cranial hemorrhaging and a hypoplastic roof of the mouth (29). Our miR-array screen detected several miRs that show large difference in expression in articular chondrocytes versus MSCs and this includes miR-140 with the largest expression difference between the two cell types. We also showed that during chondrogenesis, miR-140 expression increased in differentiated hMSC cells compared to undifferentiated MSC in parallel with Sox9 and Col2a1 expression. These findings suggest that miR-140 is a marker and possibly a regulator of chondrocytic differentiation.
The unique differentiation-related expression pattern of miR-140 is highlighted by our findings on miR-146 which is also expressed in chondrocytes. In contrast to miR-140, miR-146 has a broader tissue distribution, it is increased in response to IL-1, it is upregulated in OA and does not show changes related to chondrocyte differentiation (Arthritis Rheum, in press).
The ability of the chondrocytes to remodel and repair the cartilage ECM declines with aging and in OA and this is related to a decline in the anabolic activity of chondrocytes (30, 31). MiR-140 expression was reduced in OA cartilage and in the same samples expression of proteinases ADAMTS-5 increased and Col2a1 expression decreased. Thus, the abnormal expression pattern of miR-140 correlates with the imbalance of anabolic-catabolic responses in OA. Our observations of abnormal miR-140 expression in OA are consistent with one recent publication (32). IL-1β is one of the most prominent mediators of cartilage degradation and joint inflammation (33, 34). IL-1β induces a cascade of inflammatory and catabolic events in chondrocytes. It also changes chondrocyte anabolism by suppressing the synthesis of proteoglycans and collagens and by enhancing the production of matrix metalloproteinases (MMPs) (26, 27). miR-140 expression was down-regulated by IL-1β stimulation of chondrocytes in vitro. These data suggest that IL-1β may be at least one mechanism that is involved in the suppression of miR-140 in OA.
Our studies using dsRNA mimicking miR-140 suggest that miR-140 suppresses ADAMTS-5 mRNA levels. This observation is supported by preliminary observations of increased ADAMTS-5 expression in miR-140 knock out mice (in preparation).
The pathogenesis of osteoarthritis is associated with abnormal activation and differentiation of chondrocytes which overexpress inflammatory mediators and matrix degrading enzymes (3–6). Previous mechanisms examined in these abnormal cellular responses include chondrocyte stimulation by extracellular stimuli such as cytokines, growth factors, mechanical stress and matrix degradation products. Intracellularly these stimuli activate signaling cascades that lead to changes in gene expression (22, 35). This represents a new mechanism by which IL-1 changes gene expression in chondrocytes. MiR-140 represents novel additional control mechanism that is involved in the chondrocyte response to IL-1.
The present study is focused on miR-140 as it is the most cartilage specific miR. We performed searches in three databases (“TargetScan” http://www.targetscan.org/vert_50/, “ PicTar” http://pictar.mdc-berlin.de/, “miRanda” http://microrna.sanger.ac.uk/) and this yielded 223–975 potential miR-140 targets. Only 9 potential targets were identified in all three databases, and notably, this did not include ADAMTS-5. There remains uncertainty in regards to the rules for in silico miR target identification (36). At present the most conclusive target validation is the demonstration of changes in protein expression, cell function or phenotype in knock out or transgenic mice. Future studies are needed to determine the consequences of changes in the compete set of miR-140 targets for cartilage development and homeostasis. Currently ongoing studies with miR-140 knock out mice and miR-140 transgenic mice will provide information in this regard.
In conclusion, our study suggests that miR-140 is a chondrocyte differentiation-related miR. It may be novel regulator of cartilage homeostasis and changes in its expression and function play an important role in diseases affecting articular cartilage. Further studies on miR-140 have the potential to reveal important new regulatory pathways that control cartilage development and homeostasis and open a new insight on disease mechanisms and therapeutic interventions for OA.
Acknowledgments
This study was supported by NIH grants AR050631, AR056120, AG033409, AG07996, and Arthritis Foundation Fellowship (SO) and the Stein Endowment.
REFFERENCES
- 1.Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 2008;58(1):26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008;58(1):15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 3.Goldring MB. The role of the chondrocyte in osteoarthritis. Arthritis Rheum. 2000;43(9):1916–26. doi: 10.1002/1529-0131(200009)43:9<1916::AID-ANR2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 4.Lotz M. Cytokines in cartilage injury and repair. Clin Orthop Relat Res. 2001;391(Suppl):S108–15. doi: 10.1097/00003086-200110001-00011. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn K, D’Lima DD, Hashimoto S, Lotz M. Cell death in cartilage. Osteoarthritis Cartilage. 2004;12(1):1–16. doi: 10.1016/j.joca.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Goldring MB. Update on the biology of the chondrocyte and new approaches to treating cartilage diseases. Best Pract Res Clin Rheumatol. 2006;20(5):1003–25. doi: 10.1016/j.berh.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 10.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48):18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 15.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–92. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–9. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 17.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102(31):10898–903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105(6):1949–54. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–7. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 20.Maier R, Ganu V, Lotz M. Interleukin-11, an inducible cytokine in human articular chondrocytes and synoviocytes, stimulates the production of the tissue inhibitor of metalloproteinases. J Biol Chem. 1993;268(29):21527–32. [PubMed] [Google Scholar]
- 21.Grogan SP, Olee T, Hiraoka K, Lotz MK. Repression of chondrogenesis through binding of notch signaling proteins HES-1 and HEY-1 to N-box domains in the COL2A1 enhancer site. Arthritis Rheum. 2008;58(9):2754–63. doi: 10.1002/art.23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto M, Nakasa T, Hikata T, Asahara H. Molecular network of cartilage homeostasis and osteoarthritis. Med Res Rev. 2008;28(3):464–81. doi: 10.1002/med.20113. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PG, Magna HA, Reeves LM, Lopresti-Morrow LL, Yocum SA, Rosner PJ, et al. Cloning, expression, and type II collagenolytic activity of matrix metalloproteinase-13 from human osteoarthritic cartilage. J Clin Invest. 1996;97(3):761–8. doi: 10.1172/JCI118475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bau B, Gebhard PM, Haag J, Knorr T, Bartnik E, Aigner T. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 2002;46(10):2648–57. doi: 10.1002/art.10531. [DOI] [PubMed] [Google Scholar]
- 25.Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem. 2002;277(25):22201–8. doi: 10.1074/jbc.M200431200. [DOI] [PubMed] [Google Scholar]
- 26.Goldring MB, Birkhead J, Sandell LJ, Kimura T, Krane SM. Interleukin 1 suppresses expression of cartilage-specific types II and IX collagens and increases types I and III collagens in human chondrocytes. J Clin Invest. 1988;82(6):2026–37. doi: 10.1172/JCI113823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefebvre V, Peeters-Joris C, Vaes G. Modulation by interleukin 1 and tumor necrosis factor alpha of production of collagenase, tissue inhibitor of metalloproteinases and collagen types in differentiated and dedifferentiated articular chondrocytes. Biochim Biophys Acta. 1990;1052(3):366–78. doi: 10.1016/0167-4889(90)90145-4. [DOI] [PubMed] [Google Scholar]
- 28.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 29.Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40(3):290–8. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudhia J. Aggrecan, aging and assembly in articular cartilage. Cell Mol Life Sci. 2005;62(19–20):2241–56. doi: 10.1007/s00018-005-5217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aigner T, Soder S, Gebhard PM, McAlinden A, Haag J. Mechanisms of disease: role of chondrocytes in the pathogenesis of osteoarthritis--structure, chaos and senescence. Nat Clin Pract Rheumatol. 2007;3(7):391–9. doi: 10.1038/ncprheum0534. [DOI] [PubMed] [Google Scholar]
- 32.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 2008;3(11):e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldring MB. Osteoarthritis and cartilage: the role of cytokines. Curr Rheumatol Rep. 2000;2(6):459–65. doi: 10.1007/s11926-000-0021-y. [DOI] [PubMed] [Google Scholar]
- 34.Zwerina J, Redlich K, Polzer K, Joosten L, Kronke G, Distler J, et al. TNF-induced structural joint damage is mediated by IL-1. Proc Natl Acad Sci U S A. 2007;104(28):11742–7. doi: 10.1073/pnas.0610812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213(3):626–34. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44(1):47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]