Abstract
Activation of hepatic stellate cells (HSC) results in their proliferation and in the secretion of extracellular matrix (ECM) proteins, which leads to hepatic fibrosis. microRNAs (miRNAs) have been shown to regulate various cell functions, such as proliferation, differentiation, and apoptosis. Hence, we have analyzed the miRNAs that were differentially expressed in HSC isolated from sham-operated and bile duct-ligated rats. Expression of two miRNAs, miRNA-150 and miRNA-194, was reduced in HSC isolated from fibrotic rats compared with sham-operated animals. These two miRNAs were overexpressed in LX-2 cells, and their ability to inhibit cell proliferation, the expression of smooth muscle α-actin (SMA), a marker for activation, and collagen type I, a marker for ECM secretion, was determined. Overexpression of these two miRNAs resulted in a significant inhibition of proliferation (P < 0.05) and reduced SMA and collagen I levels compared with either untreated cells or nonspecific miRNA-expressing cells. Next, the protein targets of these two miRNAs were found using bioinformatics approaches. C-myb was found to be a target for miRNA-150, and rac 1 was found to be one of the targets for miRNA-194. Therefore, we studied the expression of these two proteins by overexpressing these two miRNAs in LX-2 cells and found that overexpression of miRNA-150 and miRNA-194 resulted in a significant inhibition of c-myb and rac 1 expression, respectively. We conclude that both miRNA-150 and miRNA-194 inhibit HSC activation and ECM production, at least in part, via inhibition of c-myb and rac 1 expression.
Keywords: c-myb, microRNA-150, microRNA-194, rac 1
the development of hepatic fibrosis is a result of a wound-healing response attributable to liver injury. Hepatic stellate cells (HSC) are the major cell type responsible for the progression of liver fibrosis. These cells reside in the space of Disse and store vitamin A in lipid droplets. Upon liver injury, they lose these droplets, proliferate, and differentiate into myofibroblast-like cells (22). The activated HSC undergo continuous proliferation and express activation markers such as smooth muscle α-actin (SMA) and plasminogen activator inhibitor-1. Several reports have shown that expression of matrix metalloproteinase (MMP)-1 is inhibited and that tissue inhibitor of metalloproteinase (TIMP)-1 expression is upregulated, resulting in an imbalance between MMP-1 and TIMP-1 and a net increase in extracellular matrix (ECM) accumulation during hepatic fibrosis (3). Several profibrogenic cytokines mediate effects via the activation of the signaling molecules such as c-myb and rac 1 (6, 7). Expression of all these genes is tightly regulated at different levels during hepatic fibrogenesis. We hypothesized that one of the regulatory mechanisms could be through microRNAs (miRNAs).
miRNAs are endogenous, small, noncoding 21–23-nucleotide RNAs that regulate gene expression by binding to the 3′ untranslated region (3′UTR) of the target gene mRNAs to repress translation or induce mRNA cleavage. Thus they play an important role in regulating various cell functions such as development, cell proliferation, differentiation, and apoptosis (4). Increasing evidence suggests that certain miRNAs may contribute to the maintenance of hepatocyte function (18). A recent study has shown that the miRNA-30 family is required for vertebrate hepatobiliary development (9). miRNA-122a has been shown to enhance hepatitis C viral replication in hepatocytes (10). Overexpression of miRNA-27a and 27b was shown to influence fat accumulation and cell proliferation during HSC activation (12). Although emerging evidence suggests a role of miRNAs in affecting liver function, there is a paucity of data on the role of miRNAs in hepatic fibrosis.
In this study, we have determined the differential expression of miRNAs in HSC isolated from both sham-operated and bile duct-ligated (BDL) rat livers. We selected two miRNAs, miRNA-150 and miRNA-194, whose expression was inhibited in HSC isolated from fibrotic livers. These miRNAs were overexpressed in an activated human HSC cell line, LX-2, and their roles in inhibiting proliferation, activation, and ECM production were determined.
MATERIALS AND METHODS
Animals, BDL, and HSC isolation.
Male Sprague-Dawley rats from Charles River Laboratories (Wilmington, MA) were used for BDL and HSC isolation. The animals were housed in facilities approved by the National Institutes of Health. All procedures were reviewed and approved by the Animal Welfare Committee of the University of California Davis. The BDL procedure was performed as described previously (13). Primary rat HSC were isolated from both sham-operated and BDL rats as previously described (13, 25) and used 3 days after isolation. HSC isolated from sham-operated rats were measured by vitamin A autofluorescence, and the HSC from BDL rats were judged by SMA expression. The purity of these isolated HSC was >90%. HSC were maintained in M199 medium supplemented with 20% fetal bovine serum, 100 U/ml penicillin, and 100 g/ml streptomycin and incubated at 37°C in a humidified atmosphere with 5% CO2.
miRNA isolation, purification, and microarray.
All kits and reagents were obtained from Applied Biosystems, Foster City, CA. Total RNA enriched with miRNAs was isolated from HSC using the mirVana miRNA isolation kit according to the manufacturer's instructions. miRNAs were purified using the FlashPAGE gel electrophoresis system and labeled with Alexa Fluor 555 followed by hybridization with mirVana miRNA bioarray slides according to the manufacturer's instructions. The slides were washed and then scanned with a Genepix 4000B scanner. Raw data were analyzed using Genepix Pro 6 software (Molecular Devices, Sunnyvale, CA). Normalization was performed by the expression of each miRNA replicate relative to a control miRNA (provided in the Bioarray essentials kit) added to each sample, thus allowing for comparisons among chips. Average values of the mean, intensity of each replicate in the two groups was analyzed.
Stem-loop real-time RT-PCR.
To confirm expression of miRNA-150 and miRNA-194, stem-loop real-time RT-PCR (SLqRT-PCR) was performed. Total RNA (10 ng) was used for first-strand cDNA synthesis using miRNA-150, miRNA-194-specific, stem-loop primer, or rnu-43 stem-loop primer, a control endogenous small nuclear RNA, (Applied Biosystems), followed by real-time PCR amplification with gene-specific forward primer and a reverse primer along with a probe, in an ABI Prizm 7500 PCR machine. The relative miRNA expression was calculated from three different experiments.
Cell culture and miRNA transfection.
The human immortalized HSC line, LX-2, was provided by Dr. S. L. Friedman, Mount Sinai Medical School, New York, NY. LX-2 cells exhibit typical features of HSC in primary culture such as expression of desmin, glial acidic fibrillary protein, and responsiveness to transforming growth factor-β. LX-2 cells were maintained in DMEM supplemented with 5% fetal bovine serum, 100 U/ml penicillin, and 100 g/ml streptomycin and incubated at 37°C in a humidified atmosphere with 5% CO2. On the day of transfection, these cells (1 × 105) were seeded on six-well plates and were transfected with the miRNAs [miRNA-150, miRNA-194, or nonspecific (NS)-miRNA] using siPORT NeoFX transfection agent (Applied Biosystems) according to the manufacturer's instructions. The cells were washed 24 h after transfection, replenished with new culture medium, and allowed to grow for 48 h.
Cell proliferation and TUNEL assay.
LX-2 cells were seeded either on 96-well plates or culture-treated glass slides and were transfected with the miRNAs. The cells seeded on 96-well plates were assayed for proliferation using the WST-1 reagent 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. For the apoptosis assay, the cells plated on glass slides were washed with ice-cold PBS three times for 5 min and fixed with 4% paraformaldehyde, followed with acetic acid/ethanol (1:3) for postfixation. The terminal deoxynucleotidyl transferase dUTP-mediated nick-end labeling (TUNEL) assay was carried out with the In Situ Cell Death Detection kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions. TUNEL-positive cells were identified as apoptotic, and the percentage of apoptotic cells was calculated by the ratio of TUNEL-positive cells to total cells in at least 10 different high-power fields of a fluorescent microscope in duplicate wells and in five different experiments.
Bioinformatics approaches.
The miRNA sequences were analyzed for their predicted target proteins using Targetscan 4.1, miRNADA, and Sanger's miRbase. The key proteins that were found to be common with great homology to the target sequences were selected for further analysis.
RNA isolation and qRT-PCR.
The cells were collected, total RNA was isolated using Qiagen total RNA isolation kit, and cDNA was synthesized according to manufacturer's instructions (Invitrogen, San Diego, CA). The c-myb, rac 1, and GAPDH primers were obtained from Santa Cruz Biotechnology, Santa Cruz, CA. First-strand cDNA was synthesized, followed by SYBRgreen qRT-PCR using ABI Prizm 7500. The relative expression was analyzed, and the data were collected from three different experiments, as reported by us previously (23).
ELISA and Western blot analysis.
Western blots were performed using anti-collagen-I, anti-SMA, and anti-β-actin antibodies (Santa Cruz Biotechnology). An ELISA for collagen-I was performed on the spent media collected from the transfected cells then concentrated fivefold using Centricon concentrator tubes (Thermo Scientific, Rockford, IL), and this medium was used to coat the ELISA plate overnight at 4°C. The next day, the plates were washed three times with PBS and then incubated with the anti-collagen-I antibody for 2 h followed by the horseradish peroxidase-conjugated secondary antibodies for 1 h. The plates were then washed and developed using 3,3′,5,5′-tetramethylbenzidine substrate, and the absorbance was measured spectrophotometrically.
Immunocytochemistry.
LX-2 cells were grown on chamber slides, and the transfection experiments were carried out as described before. The cells were fixed in 4% paraformaldehyde/PBS for 15 min at room temperature and washed with PBS three times. The cells were blocked with 2% bovine serum albumin in PBS for 1 h followed by incubation with anti-SMA antibody (1:200) for 16 h at 4°C (Santa Cruz Biotechnology). After washing with PBS, Alexa Fluor 488-labeled secondary antibody IgG (1:1,000) was applied and incubated for 60 min. After an additional washing, the cells were mounted and analyzed by fluorescence microscopy (20).
Statistical analysis.
All the experiments were performed in triplicate and at least three times. The data were expressed as means ± SEM and calculated using variance analysis and the Newman-Keuls test for multiple comparisons among groups. P < 0.05 was considered as statistically significant.
RESULTS
BDL causes differential expression of miRNAs.
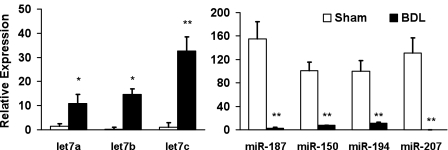
An established model of BDL was used to induce hepatic fibrosis in rats (13). The primary HSC were isolated from both sham-operated controls and BDL rats (n = 3 rats/group). The purity of isolated HSC from sham-operated rats was assessed by the autofluorescence of vitamin A droplets, and purity of the HSC from BDL rats was assessed by SMA expression. The purity of the isolated HSC was 95% or greater. The HSC were cultured for 3 days, and then differential expression of the miRNAs was determined by microarray analysis in HSC isolated from BDL rats compared with normal rats (Fig. 1). There was a significant increase in some of the let 7 family members such as let 7a, let 7b, and let 7c, whereas there was a significant decrease in miRNA-150, miRNA-194, miRNA-187, and miRNA-207 in HSC isolated from BDL rats. We hypothesized that the miRNAs that were inhibited in HSC isolated from BDL rats might play a role in inhibiting HSC activation and proliferation. On the basis of the bioinformatics approaches, we found that c-myb was a target for miRNA-150 and rac 1 for miRNA-194. Hence, we selected these two miRNAs for further experiments.
Fig. 1.
Bile duct ligation (BDL) causes the differential expression of microRNAs (miRNA, miR) in hepatic stellate cells (HSC). Liver fibrosis was induced by BDL in male Sprague-Dawley rats. The HSC were isolated and cultured for 3 days, and the miRNA microarray was performed. miRNAs were upregulated (left) or downregulated (right) in HSC isolated from BDL rats compared with sham-operated rats (n = 3; *P < 0.05; **P < 0.01).
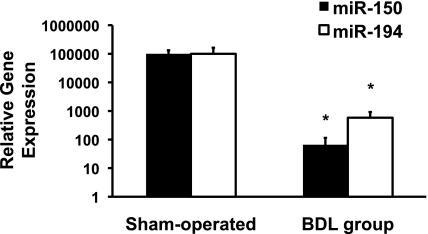
miRNA-150 and miRNA-194 expression was confirmed by the SLqRT-PCR technique using rnu-43 as an internal control. The results showed that there was a significant decrease in the expression of both miRNAs in HSC isolated from the BDL group compared with HSC isolated from the sham-operated control group (Fig. 2).
Fig. 2.
Stem-loop, real-time RT-PCR (SLqRT-PCR) of the HSC from BDL and sham-operated control rats. The purified miRNA isolated from HSC was used for SLqRT-PCR as described in materials and methods. The results confirmed that both miRNA-150 and miRNA-194 were significantly decreased in HSC isolated from the BDL group compared with the sham-operated control group (n = 3 in triplicates; *P < 0.001).
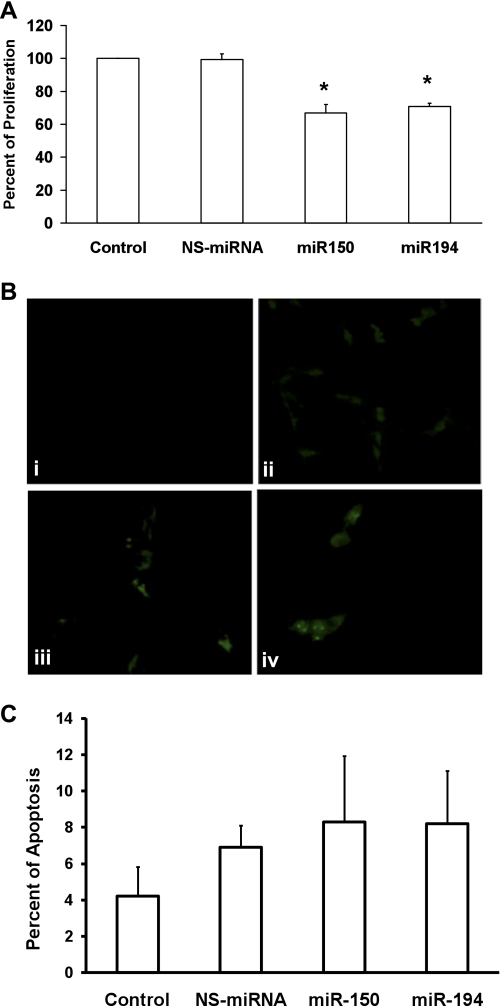
Overexpression of miRNA-150 and miRNA-194 causes inhibition of LX-2 proliferation.
The roles of these two miRNAs in modulating HSC proliferation and apoptosis were studied. An activated human hepatic cell line, LX-2, was selected for all the transfection experiments. Four groups of treatments were used, 1) untreated cells, 2) cells transfected with a NS-miRNA, an miRNA that has been shown not to inhibit any of the known mRNAs, 3) cells transfected with miRNA-150, and 4) cells transfected with miRNA-194. The cells were analyzed 72 h after transfection for proliferation using the WST-1 reagent. There was a 31% and 27% reduction of proliferation (n = 5, P < 0.05) in LX-2 cells transfected with miRNA-150 and miRNA-194, respectively (Fig. 3A). Next, their effect on induction of apoptosis was measured using the TUNEL assay (Fig. 3B). The number of apoptotic nuclei and the total number of cells were recorded in at least 10 different high-power fields. The percentage of apoptotic cells was calculated from five different experiments. There was no significant increase in apoptosis in the miRNA-transfected cells (Fig. 3C). These results indicate that, although these miRNAs could inhibit LX-2 cell proliferation, they did not influence cell survival.
Fig. 3.
Overexpression of miRNA-150 and miRNA-194 inhibits the proliferation of LX-2 cells. The activated human HSC line, LX-2, was cultured and treated in 4 groups, namely untreated cells (i), nonspecific (NS)-miRNA transfected cells (ii), miRNA-150-transfected cells (iii), and miRNA-194-transfected cells (iv). A: cells were incubated with the WST-1 reagent, and the proliferation assay was performed (n = 5; *P < 0.05). B: cells were stained for the induction of apoptosis using the terminal deoxynucleotidyl transferase dUTP-mediated nick-end labeling (TUNEL) assay kit. C: TUNEL-positive nuclei and the total number of cells were evaluated in at least 10 different high-power fields, and the percents of apoptosis were calculated from 5 different experiments.
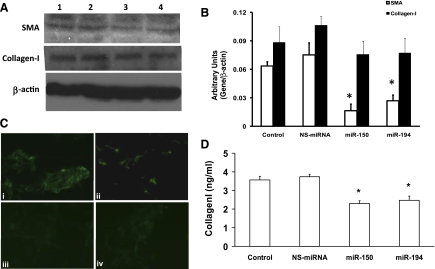
Overexpression of miRNA-150 and miRNA-194 inhibits SMA and collagen-I expression.
Next, the effects of overexpression of miRNA-150 and miRNA-194 on HSC activation and ECM protein production were evaluated. SMA is one of the markers of stellate cell activation, and it is expressed at high levels in LX-2 cells. Hence, the expression of SMA was analyzed, and we determined that overexpression of either of these two miRNAs resulted in a significant decrease in SMA levels compared with NS-miRNA-transfected or untreated cells, as determined by Western blots (Fig. 4A). The densitometry analysis of these Western blots showed that there was a significant inhibition of SMA expression in the miRNA-transfected cells (36.3 ± 2.7% and 22.1 ± 5.2% inhibition with miRNA-150 and miRNA-194, respectively; Fig. 4B). The immunocytochemical staining further verified the findings (42.3 ± 3.1% and 34.1 ± 7.5% inhibition with miRNA-150 and miRNA-194, respectively) (Fig. 4C). Collagen-Iα1 is the major ECM protein produced by the activated HSC. Hence, the effect of overexpression of these two miRNAs on the expression of collagen-I protein expression was also studied. Western blot analysis showed that the basal level of intracellular collagen-I levels in the cell lysates was not changed significantly but showed a trend toward being decreased (Fig. 4, A and B), whereas the collagen-I in the spent media was significantly less in the miRNA-transfected cells as measured by ELISA (P < 0.05) (Fig. 4D).
Fig. 4.
Overexpression of miRNA-150 and miRNA-194 inhibits SMA expression and collagen-I secretion. LX-2 cells were transfected with miRNAs as described in materials and methods. A: Western blots for smooth muscle α-actin (SMA), collagen-I, and β-actin were performed, and this is a representative of 3 experiments. Lane 1, untreated cells; lane 2, NS-miRNA-transfected cells; lane 3, miRNA-150-transfected cells; lane 4, miRNA-194-transfected cells. B: densitometry analysis of the Western blot data was performed, and the data were presented (n = 5; *P < 0.05). C: immunocytochemistry for SMA was performed, and this is representative of 5 experiments. i: Untreated cells. ii: NS-miRNA-transfected cells. iii: miRNA-150-transfected cells. iv: miRNA-194-transfected cells. D: spent media was collected from the cells, centrifuged to remove debris, and concentrated to 5-fold, and then ELISA for collagen-I was performed (n = 5; *P < 0.05).
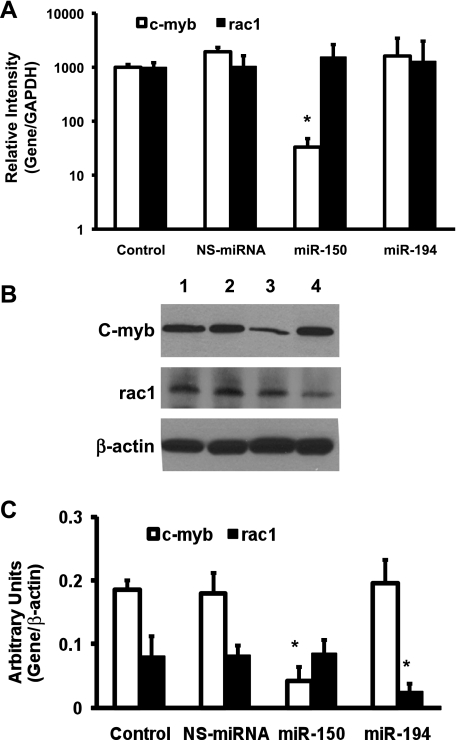
Overexpression of miRNA-150 and miRNA-194 inhibits c-myb and rac 1 expression in LX-2 cells.
On the basis of bioinformatics approaches using the Targetscan 4.1 and Sanger's database, c-myb was identified as a target gene of miRNA-150, and rac 1 was identified as one of the targets for miRNA-194 (Fig. 5). First, the effect of the miRNAs on c-myb and rac 1 mRNA expression was analyzed using qRT-PCR. The results showed that there was a significant inhibition of c-myb mRNA expression in miRNA-150-overexpressing cells but not in miRNA-194- or NS-miRNA-expressing cells (Fig. 6A). Overexpression of miRNA-194 did not result in significant change in rac 1 mRNA levels (Fig. 6A). Next, the protein expression levels of both c-myb and rac 1 were analyzed using Western blots. β-Actin was used as an internal control in all Western blots. Overexpression of miRNA-150 resulted in a significant inhibition of intracellular protein expression of c-myb, confirming the qRT-PCR results (Fig. 6B). Rac 1 protein expression was also inhibited significantly in miRNA-194-transfected cells but not in NS-miRNA- or miRNA-150-transfected cells (Fig. 6B). The Western blot results were quantified using densitometry in Fig. 6C. These results demonstrate that there was a significant inhibition of c-myb and rac 1 protein expression in LX-2 cells overexpressing miRNA-150 and miRNA-194, respectively. These data suggest that both miRNA-150 and miRNA-194 inhibit HSC activation and ECM expression, at least in part, via inhibiting c-myb and rac 1, respectively.
Fig. 5.
The predicted sequences for c-myb and rac 1 for miRNA-150 and miRNA-194. Three different bioinformatics approaches (Sanger's miRbase, Targetscan 4.1, and miRANDA) were used for the target prediction. C-myb had two matching sequences for miRNA-150, and rac 1 had one matching sequence as found in all 3 software programs. 3′UTR, 3′ untranslated region.
Fig. 6.
Effect of overexpression of miRNA-150 and miRNA-194 on the intracellular expression of c-myb and rac 1 in LX-2 cells. LX-2 cells were cultured and transfected with NS-miRNA, miRNA-150, or miRNA-194. A: cells were collected, total RNA was isolated, and the first-strand cDNA was synthesized, followed by SYBRgreen qRT-PCR amplification for c-myb, rac 1, and GAPDH. The relative expression of c-myb and rac 1 was compared with GAPDH expression (n = 3, *P < 0.05). B: total cellular protein was isolated, and equal amounts of protein were used to detect the intracellular expression of c-myb, rac 1, and β-actin by Western blots. This is representative of 3 experiments. Lane 1, untreated cells; lane 2, NS-miRNA-transfected cells; lane 3, miRNA-150-transfected cells; lane 4, miRNA-194-transfected cells. C: Western blot results from 3 experiments were quantified using densitometry analysis (n = 3, *P < 0.05).
DISCUSSION
Hepatic fibrosis is a result of a response to an injury process where HSC play an important role in its progression. Any chronic insult results in the activation and proliferation of HSC, and the activated HSC undergo proliferation and secrete excessive ECM proteins such as type-I collagen. We hypothesized that the change in the expression pattern of these proteins and the HSC behavior might be controlled, at least in part, via miRNAs. Several miRNAs have been shown to be involved in the development of nonalcoholic steatohepatitis (5) and hepatitis C virus infection (10), proliferation, and differentiation (1). Recently it has been shown that overexpression of miRNA-27 resulted in increased fat accumulation and proliferation of HSC (12). In this report, we used an established rat model of liver fibrosis induced by BDL and show that there was a differential expression of miRNAs in HSC from fibrotic livers compared with the sham-operated controls. Some of the let 7 family miRNAs were upregulated in fibrotic HSC. The let 7 family has been shown to control invasion and tumor formation in several cancer cell lines (2). At present, the role of these upregulated miRNAs is not known, and future studies will need to be directed to elucidate their role in HSC cell survival and differentiation.
We postulated that the miRNAs that are inhibited in the HSC isolated from BDL rats might play a role in inhibiting the activation of HSC. Using bioinformatics approaches, we determined the predicted target proteins for all the differentially expressed miRNAs in HSC from BDL rats. We found that, for the downregulated miRNA-150, one of the target proteins was c-myb, whereas, for miRNA-194, it was rac 1. Because these proteins play an essential role in liver fibrosis, we focused on these two miRNAs. LX-2 cells were selected for all the transfection experiments because of the following reasons: 1) they are highly activated and are similar to activated primary HSC (24); 2) these cells express high levels of SMA and collagen-I (24); and 3) 100% transfection efficiency can be achieved in these cells compared with a much lower percentage in primary HSC. The overexpression of miRNA-150 and miRNA-194 in LX-2 cells resulted in a significant inhibition of proliferation without affecting apoptosis.
During liver fibrogenesis, the expression of MMP-1 is suppressed (17) and TIMP-1 expression is stimulated (11). Because of this imbalance, there is an increased accumulation of collagen-I deposition in the space of Disse. Hence, we studied the overexpression of these two miRNAs on the expression of collagen-I, one of the major types of ECM secreted by HSC. Our data showed that there was a significant decrease in collagen-I in the media from the miRNA (-150 and -194)-transfected cells. Although there was a trend toward less intracellular collagen in the miRNA-transfected cells, the significant decrease was in the collagen in the media. The mechanism of this decrease is not clear. This decrease could be a result of increased degradation of collagen-I attributable to changes in MMP-1 and TIMP-1. Moreover, the activation of HSC results in increased levels of SMA. We analyzed SMA expression and found that the SMA protein levels were significantly decreased in both miRNA-150- and miRNA-194-overexpressing cells. The results demonstrate that these two miRNAs are involved in the inhibition of HSC proliferation, activation, and collagen deposition but did not affect apoptosis.
C-myb is a protooncogene that encodes a transcription factor involved in proliferation, differentiation, and survival of hematopoietic cells (19). There are reports showing that activated HSC also express c-myb and that its expression contributes to the development of fibrosis in animal models (14). In addition, oxidative stress has been shown to induce both collagen-I and SMA via c-myb in rat HSC (15). Rac 1, a member of the Rho family of small GTP-binding proteins, promotes proliferation and migration by activating the membrane-associated NADPH oxidase complex to produce superoxide anion, a signaling molecule involved in these processes. Various studies have shown that both reactive oxygen species and rac 1 are required for HSC proliferation and activation (8, 21). LX-2 cells express both c-myb and rac 1 constitutively. A recent study has shown that c-myb is targeted by miRNA-150 in breast cancer cells (16). Hence, we determined the role of miRNA-150 on c-myb expression and miRNA-194 on rac 1 expression in LX-2 cells. In agreement with the previous report (16), in our system miRNA-150 overexpression also resulted in decreased levels of c-myb mRNA and decreased c-myb protein expression. When the LX-2 cells were transfected with miRNA-194, there was no significant difference in rac 1 mRNA levels; however, the level of rac 1 protein was significantly inhibited, suggesting that the inhibition might be at a translational level.
In summary, our results suggest that two specific miRNAs (miRNA-150 and miRNA-194) might play a critical role in hepatic fibrogenesis. It appears that miRNA-150 and miRNA-194 regulate hepatic fibrosis, at least in part, via decreasing the expression of c-myb and rac 1.
GRANTS
The study was supported by an Innovative Development Award Program, UC Davis California (S. K. Venugopal), UC Davis Cancer Center and Clinical and Translational Sciences Center (S. K. Venugopal), NIH R01 DK075415 (M. A. Zern), and DK080715 (N. J. Torok).
DISCLOSURES
No conflicts of interest are declared by the author(s).
ACKNOWLEDGMENTS
We thank the China Scholarship Council for supporting Si-Si Wang, and the GlaxoSmithKline Research Fund of the Korean Association for the Study of the Liver for supporting Tae-Hun Kim.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature 431: 350–355, 2004 [DOI] [PubMed] [Google Scholar]
- 2.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 14: 400–409, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Cao Q, Mak KM, Lieber CS. Leptin represses matrix metalloproteinase-1 gene expression in LX2 human hepatic stellate cells. J Hepatol 46: 124–133, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev 16: 203–208, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 48: 1810–1820, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi SS, Sicklick JK, Ma Q, Yang L, Huang J, Qi Y, Chen W, Li YX, Goldschmidt-Clermont PJ, Diehl AM. Sustained activation of Rac1 in hepatic stellate cells promotes liver injury and fibrosis in mice. Hepatology 44: 1267–1277, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Eng FJ, Friedman SL. Transcriptional regulation in hepatic stellate cells. Semin Liver Dis 21: 385–395, 2001 [DOI] [PubMed] [Google Scholar]
- 8.Goldschmidt-Clermont PJ, Moldovan L. Stress, superoxide, and signal transduction. Gene Expr 7: 255–260, 1999 [PMC free article] [PubMed] [Google Scholar]
- 9.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology 136: 1081–1090, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300–3310, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbst H, Wege T, Milani S, Pellegrini G, Orzechowski HD, Bechstein WO, Neuhaus P, Gressner AM, Schuppan D. Tissue inhibitor of metalloproteinase-1 and -2 RNA expression in rat and human liver fibrosis. Am J Pathol 150: 1647–1659, 1997 [PMC free article] [PubMed] [Google Scholar]
- 12.Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett 583: 759–766, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Jiang JX, Mikami K, Shah VH, Torok NJ. Leptin induces phagocytosis of apoptotic bodies by hepatic stellate cells via a Rho guanosine triphosphatase-dependent mechanism. Hepatology 48: 1497–1505, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitada T, Seki S, Nakatani K, Kawada N, Kuroki T, Monna T. Hepatic expression of c-Myb in chronic human liver disease. Hepatology 26: 1506–1512, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Lee KS, Buck M, Houglum K, Chojkier M. Activation of hepatic stellate cells by TGF alpha and collagen type I is mediated by oxidative stress through c-myb expression. J Clin Invest 96: 2461–2468, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin YC, Kuo MW, Yu J, Kuo HH, Lin RJ, Lo WL, Yu AL. c-Myb is an evolutionary conserved miR-150 target and miR-150/c-Myb interaction is important for embryonic development. Mol Biol Evol 25: 2189–2198, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Milani S, Herbst H, Schuppan D, Grappone C, Pellegrini G, Pinzani M, Casini A, Calabro A, Ciancio G, Stefanini F, Ciancio AK, Surrenti C. Differential expression of matrix-metalloproteinase-1 and -2 genes in normal and fibrotic human liver. Am J Pathol 144: 528–537, 1994 [PMC free article] [PubMed] [Google Scholar]
- 18.Rogler CE. MicroRNAs make inroads into liver development. Gastroenterology 136: 770–772, 2009 [DOI] [PubMed] [Google Scholar]
- 19.Shen-Ong GL. The myb oncogene. Biochim Biophys Acta 1032: 39–52, 1990 [DOI] [PubMed] [Google Scholar]
- 20.Shirahashi H, Wu J, Yamamoto N, Catana A, Wege H, Wager B, Okita K, Zern MA. Differentiation of human and mouse embryonic stem cells along a hepatocyte lineage. Cell Transplant 13: 197–211, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, Goldschmidt-Clermont PJ, Finkel T. Regulation of reactive-oxygen-species generation in fibroblasts by Rac1. Biochem J 318: 379–382, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torok NJ. Recent advances in the pathogenesis and diagnosis of liver fibrosis. J Gastroenterol 43: 315–321, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Wege H, Le HT, Chui MS, Liu L, Wu J, Giri R, Malhi H, Sappal BS, Kumaran V, Gupta S, Zern MA. Telomerase reconstitution immortalizes human fetal hepatocytes without disrupting their differentiation potential. Gastroenterology 124: 432–444, 2003 [DOI] [PubMed] [Google Scholar]
- 24.Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut 54: 142–151, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, Wu J, Frizell E, Liu SL, Bashey R, Rubin R, Norton P, Zern MA. Rapamycin inhibits hepatic stellate cell proliferation in vitro and limits fibrogenesis in an in vivo model of liver fibrosis. Gastroenterology 117: 1198–1204, 1999 [DOI] [PubMed] [Google Scholar]