Abstract
In mammals, nicotinamide phosphoribosyltransferase (NAMPT) is responsible for the first and rate-limiting step in the conversion of nicotinamide to nicotinamide adenine dinucleotide (NAD+). NAD+ is an obligate cosubstrate for mammalian sirtuin-1 (SIRT1), a deacetylase that activates peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), which in turn can activate mitochondrial biogenesis. Given that mitochondrial biogenesis is activated by exercise, we hypothesized that exercise would increase NAMPT expression, as a potential mechanism leading to increased mitochondrial content in muscle. A cross-sectional analysis of human subjects showed that athletes had about a twofold higher skeletal muscle NAMPT protein expression compared with sedentary obese, nonobese, and type 2 diabetic subjects (P < 0.05). NAMPT protein correlated with mitochondrial content as estimated by complex III protein content (R2 = 0.28, P < 0.01), MRS-measured maximal ATP synthesis (R2 = 0.37, P = 0.002), and V̇o2max (R2 = 0.63, P < 0.0001). In an exercise intervention study, NAMPT protein increased by 127% in sedentary nonobese subjects after 3 wk of exercise training (P < 0.01). Treatment of primary human myotubes with forskolin, a cAMP signaling pathway activator, resulted in an ∼2.5-fold increase in NAMPT protein expression, whereas treatment with ionomycin had no effect. Activation of AMPK via AICAR resulted in an ∼3.4-fold increase in NAMPT mRNA (P < 0.05) as well as modest increases in NAMPT protein (P < 0.05) and mitochondrial content (P < 0.05). These results demonstrate that exercise increases skeletal muscle NAMPT expression and that NAMPT correlates with mitochondrial content. Further studies are necessary to elucidate the pathways regulating NAMPT as well as its downstream effects.
Keywords: nicotinamide phosphoribosyltransferase, pre-B cell colony-enhancing factor, visfatin, mitochondria, adenosine monophosphate-activated protein kinase, primary myotubes
nicotinamide phosphoribosyltransferase (NAMPT) was originally identified in humans as pre-B cell colony-enhancing factor (PBEF) (39) and later described as an adipokine named visfatin (13). Visfatin was originally shown to have insulin-mimetic effects (13); however, these results have proved to be irreproducible and in conflict with several other studies (1, 26, 38, 41), resulting in the retraction of the original report (12). “iNAMPT” and “eNAMPT” are now used to distinguish between the intracellular and extracellular forms of NAMPT/PBEF/visfatin (38). In mammals, iNAMPT is responsible for the first and rate-limiting step in the conversion of nicotinamide to nicotinamide dinucleotide (NAD+) in the NAD+ salvage pathway (36, 37). NAMPT protein is expressed ubiquitously in human tissues, including skeletal muscle (25), and provides NAD+ as a coenzyme for metabolic/energy-producing processes such as the TCA cycle and electron transport chain (ETC). In addition, NAD+ is an obligate cosubstrate for the enzymatic activity of mammalian sirtuin-1 (SIRT1) (22), a protein deacetylase that, among several other functions, activates peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) in skeletal muscle. PGC-1α is known to induce the transcription of mitochondrial genes as well as genes involved in fatty acid oxidation and reactive oxygen species scavenging (15) and is considered a potent stimulator of mitochondrial biogenesis. SIRT1 deacetylation of PGC-1α is essential for the upregulation of fatty acid oxidation in skeletal muscle under conditions of low glucose (15).
Perhaps the best-characterized cellular energy sensor to date is adenosine monophosphate-activated protein kinase (AMPK). AMPK is activated in response to metabolic stresses such as muscle contraction, glucose deprivation, hypoxia or ischemia (reviewed in Ref. 19). Upon activation, AMPK inhibits ATP-consuming pathways and stimulates ATP-producing pathways, such as glycolysis and fatty acid oxidation, in order to restore the energy charge of the cell (reviewed in Ref. 18). Fulco et al. (14) showed that mouse Nampt expression increased during glucose restriction in the mouse myoblast C2C12 cell line; importantly, this increase was dependent on AMPK. The authors also showed that SIRT1 activity was increased with glucose restriction or AMPK activation via 5-aminoimidazole-4-carboxamide-ribonucleoside (AICAR), implicating AMPK, NAMPT and SIRT1 in a skeletal muscle nutrient-sensing pathway (14). More recently, Cantó et al. (6) demonstrated that AMPK activation enhanced SIRT1 activity by increasing NAD+ levels, resulting in the deacetylation of PGC-1α in mouse skeletal muscle. On the basis of these findings, we hypothesized that the activation of AMPK via exercise would result in the induction of NAMPT in human skeletal muscle. Since AMPK is involved in the initiation of mitochondrial biogenesis via increasing expression of PGC-1α (42, 46) and SIRT1 deacetylation of PGC-1α is required for the transcription of mitochondrial fatty acid oxidation genes (15), we further hypothesized that NAMPT might play a role in skeletal muscle mitochondrial biogenesis. Indeed, in this pilot study we show that skeletal muscle NAMPT is upregulated in response to exercise training and correlates with mitochondrial content in humans, suggesting a potential role for NAMPT in the AMPK-mitochondrial biogenesis pathway. If true, NAMPT may be a high-priority target for increasing mitochondrial content in individuals who have type 2 diabetes (T2DM), where defects in mitochondrial content and function have been demonstrated (24, 30).
MATERIALS AND METHODS
Clinical studies.
Subjects were recruited into two distinct clinical investigations; the characteristics of these populations are presented in Table 1.
Table 1.
Cross-sectional study subject characteristics
| Athletes (A) | Nonobese FH− (B) | Nonobese FH+ (C) | Obese (D) | T2DM (E) | Significant Differences | |
|---|---|---|---|---|---|---|
| Age, yr | 22.8±4.0 | 23.8±2.2 | 25.2±4.3 | 30.8±3.7 | 48.2±12 | ‡E>A,B,C |
| †E>D | ||||||
| Body Weight, kg | 71.8±6.9 | 68.7±7.7 | 82.9±14.4 | 113.9±10.3 | 107.0±14.4 | ‡D>A,B |
| †D>C | ||||||
| ‡E>A,B | ||||||
| *E>C | ||||||
| BMI, kg/m2 | 22.9±2.9 | 22.6±1.9 | 27.0±3.1 | 37.1±5.4 | 35.5±3.4 | ‡D>A,B |
| †D>C | ||||||
| ‡E>A,B | ||||||
| †E>C | ||||||
| %Fat Mass | 12.0±2.5 | 20.1±4.8 | 25.1±4.6 | 32.3±5.1 | 32.2±3.7 | *A<B |
| ‡A<C,D,E | ||||||
| †B<D,E | ||||||
| Fasting glucose, mg/dl | 89.6±5.5 | 91.0±5.1 | 92.2±10.9 | 97.0±4.2 | 184.4±69.8 | †E>A,B,C,D |
| Glucose disposal rate, mg·kg FFM−1·min−1 | 0.21±0.04 | 0.18±0.07 | 0.11±0.02 | 0.05±0.02 | 0.05±0.02 | *A>C |
| ‡A>D,E | ||||||
| †B>D,E | ||||||
| ATPmax, mM/s | 1.04±0.20 | 0.60±0.08 | 0.59±0.10 | 0.63±0.09 | 0.49±0.20 | †A>B,C,D |
| ‡A>E | ||||||
| V̇o2max, ml·kg−1·min−1 | 50.9±3.1 | 32.1±3.4 | 29.9±6.3 | 24.9±3.2 | not measured | ‡A<B,C,D |
Values are means ± SD. FH−, subjects had no family history (FH) of type 2 diabetes mellitus (T2DM); FH+, subjects had ≥1 parent with T2DM. Glucose disposal rate is expressed per kg of fat free mass (FFM); V̇o2max is expressed per kg body wt per min. One-way ANOVA, Tukey's post hoc test:
P < 0.05,
P < 0.01,
P < 0.001.
Volunteers qualified and were enrolled in Study A (ACTIV; ClinicalTrials.gov ID NCT00401791) if they were age 18–30 yr, had a BMI >20 but <30kg/m2 (for athletes and nonobese subjects) or a BMI of >30kg/m2 (for healthy nondiabetic obese subjects). Body composition, insulin sensitivity, maximal ATP synthesis rate, and maximal aerobic capacity were determined at baseline in endurance-trained athletes as well as in sedentary subjects who were nonobese (with or without a family history of T2DM) or obese. Subjects consumed a standard diet (35% fat, 15% protein, 50% carbohydrate) for 2 days, were admitted to the metabolic unit overnight, and underwent a vastus lateralis biopsy using the Bergstrom technique after an overnight fast. A subset of five subjects from each group was analyzed for this study (subject characteristics of this subset are described in Table 1). After baseline testing, 13 nonobese sedentary subjects participated in an exercise training protocol consisting of alternating day sessions of a progressive 30- to 60-min interval protocol [75–85% maximum aerobic capacity (V̇o2max)] and a 50-min aerobic protocol (70% V̇o2max), both performed on a stationary bicycle. Subjects exercised on 13 days of a 3-wk period. Seven additional subjects acted as nonexercise controls. Insulin action, muscle biopsy, and laboratory parameters were obtained 2 days after the last exercise bout under identical conditions. Baseline characteristics of these subjects are described in Supplementary Table 1 (Supplementary materials are found in the online version this paper).
Volunteers qualified and were enrolled in Study B [TAKE TIME; ClinicalTrials.gov ID NCT00402012] if they had known T2DM, were “diet controlled” or were taking metformin, insulin, and/or sulfonylureas but not thiazolidinediones and were otherwise healthy. The overall purpose of this study was to examine the effects of pioglitazone on skeletal muscle metabolism and insulin sensitivity; the latter analyses are ongoing and have not yet been published. Body composition, insulin sensitivity, and maximal ATP synthesis rate were determined at baseline. Subjects were fed a standard diet (35% fat, 15% protein, 50% carbohydrate) for 2 days, admitted to the metabolic unit overnight, and underwent a vastus lateralis biopsy using the Bergstom technique after an overnight fast. A subset of five subjects was analyzed for this study (subject characteristics of this subset are described in Table 1).
Both protocols were approved by the PBRC Institutional Review Board, and all subjects gave written informed consent after being informed of the risks and benefits of participation.
Body composition.
Body fat mass and lean mass were measured on a Hologic dual energy X-ray absorptiometer (QDR 4500A; Hologic, Waltham, MA).
Euglycemic-hyperinsulinemic clamp.
The clamp was performed as previously described (10). Briefly, intravenous catheters were inserted in an antecubital vein for infusions and in a vein on the dorsum of the contralateral hand for sampling of arterialized blood. After baseline sampling, a primed continuous insulin infusion (80 mU·m−2·min−1) was continued for 2–4 h. The clamp was continued for at least 1 h after reaching a concentration of glucose ∼90–100 mg/dl. The mean rate of exogenous glucose infusion during steady state (last 30 min) was corrected for changes in glycemia and divided by fat-free mass (FFM) to assess insulin sensitivity.
Maximal ATP synthesis rate.
Maximal ATP synthesis rate (ATPmax) was determined as described previously (23) on a 3T GE Signa MNS magnet (GE, Milwaukee, WI) using a 4- or 6-cm 31P-tuned surface coil positioned over the distal vastus lateralis. Following the acquisition of a fully relaxed spectrum, 31P spectra were acquired every 6 s at rest (4 NEX) and continuously during a 24-, 30-, or 36-s ballistic exercise obtained by “kicking” against Velcro straps positioned tight across the leg and thigh. Exercise time and intensity were targeted to drop phosphocreartine (PCr) by 33–50% of basal PCr and to avoid a pH of <6.8, because lower pH inhibits oxidative phosphorylation and results in an artificially low ATPmax. ATPmax was calculated using the PCr recovery time constant (τ) and [PCr]rest: ATPmax = [PCr]rest/τ. Confirmation that ATPmax is a good measure of phosphorylation capacity comes from animal and human studies that have found this rate to vary in direct proportion to the oxidative enzyme activity of healthy muscle (29, 32). ATPmax has previously been validated against mitochondrial content (8, 9). The reproducibility of muscle ATPmax determinations has been published (2, 3), and repeated measures on the same subject at PBRC agree to within ±11%.
V̇o2max.
V̇o2max was determined using a standardized graded exercise testing protocol administered on a stationary bicycle ergometer (Lode Excalibur, Gronig, Netherlands). Participants started exercising at either 30 W (sedentary subjects) or 100 W (athletes). Workload and exercise load were increased by 20 W (sedentary subjects) or 30 W (athletes) every minute until the participant reached exhaustion. V̇o2, V̇co2, and respiratory exchange ratio (RER) were measured continuously throughout the exercise tests by using a Parvomedics True Max 2400 Metabolic Measurement Cart (Salt Lake City, UT). A V̇o2 peak was determined from the last minute of exercise, and it was considered V̇o2max if the participant met at least two of the following three criteria: 1) participant reached estimated maximum heart rate, 2) RER was above 1.15, and 3) the last two measurements of V̇o2 reached plateau.
DNA extraction and real-time PCR for mitochondrial DNA.
DNA was extracted from 11 to 33 mg of frozen muscle tissue by use of the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Relative amounts of mitochondrial DNA (mtDNA) and nuclear DNA were determined by quantitative real-time PCR as described previously (4, 20, 44). The sequences for the primer/probe sets used in the TaqMan (Applied Biosystems, Roche, Branchburg, NJ) analysis of mtDNA content for NADH dehydrogenase subunit 1 (ND1) and for lipoprotein lipase (LPL; acc. no. NM_000237) are listed in Supplementary Table 3. Mitochondrial content is expressed as number of copies of mtDNA per cell (ratio of ND1 to LPL).
Cell culture experiments.
Myoblast donors, participants in the ACTIV study, were lean, healthy, sedentary males aged 22–27 yr with BMIs ranging from 20.5 to 24.8 kg/m2. Human myoblasts were immunopurified and cultured as described previously (43). When myoblast cultures reached ∼80% confluence, they were differentiated for 5 days into mature myotubes. Differentiation media consisted of α-MEM medium supplemented with 2% fetal bovine serum, 2% PenStrep, and 0.5 mg/ml fetuin (Invitrogen-Gibco, Carlsbad, CA). For forskolin and ionomycin experiments, differentiation media also contained 30 μM palmitate. During the last 3 days of differentiation, myotubes were treated with 4 μM forskolin and/or 0.5 μM ionomycin for 60 min/day. For AMPK activation experiments, myotubes were treated with 0.5 mM AICAR (Cell Signaling Technology, Danvers, MA) for the last 8, 24, or 48 h of differentiation prior to harvest or for the entire duration of differentiation (5 days).
Western blotting.
Skeletal muscle homogenates were prepared by Polytron homogenization in RIPA buffer containing protease inhibitor and phosphatase inhibitor cocktails (Sigma, St. Louis, MO). Cultured myotube lysates were prepared in the same buffer by sonication. Protein content was quantified via bicinchoninic acid assay (Thermo Fisher Scientific, Wilmington, MA). Twenty-five micrograms of muscle homogenate or 20–70 μg of cell lysate was run on a 10% SDS-PAGE gel (Bio-Rad, Hercules, CA) and transferred to a PVDF membrane (Millipore, Billerica, MA). Membranes were incubated with antibodies to NAMPT (A300–372A; Bethyl, Montgomery, TX), complex III (MS304; MitoSciences, Eugene, OR), phospho-AMPKα (2531; Cell Signaling Technology, Danvers, MA), AMPKα (2532; Cell Signaling Technology), and/or GAPDH (4699-9555; Biogenesis, Poole, UK, or NB300-221; Novus Biologicals, Littleton, CO) overnight and then probed with goat anti-mouse IgG DyLight 680 or goat anti-rabbit IgG DyLight 800 (Thermo Fisher Scientific). Bands were visualized using an Odyssey 9120 Infrared Imaging System (LI-COR, Lincoln, NE) and quantified using Odyssey Application Software version 2.1 (LI-COR). Preliminary studies showed linearity of GAPDH-adjusted signal for a test sample from 10–70 μg of total muscle protein.
RNA extraction and qRT-PCR.
As described previously (40), muscle biopsy specimens were snap-frozen in liquid nitrogen at the bedside, and RNA was extracted via column purification using the Qiagen RNeasy Fibrous Mini Kit (Qiagen, Valencia, CA). RNA purity was determined on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), and RNA quantity was determined using an ND-1000 Nanodrop Spectrophotometer (Thermo Fisher Scientific). Primers and probes were designed using Primer Express version 2.1 (Applied Biosystems, Foster City, CA). Sequences of primer/probe sets are shown in Supplementary Table 2. The concentration of target mRNAs was determined by quantitative reverse transcriptase-PCR (qRT-PCR) using Taqman primers and fluorescent probes as the detection system. The qRT-PCR was performed on an ABI PRISM 7900 (Applied Biosystems, Foster City, CA) using the following parameters: one cycle of 48°C for 30 min, then 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All expression data were normalized to the ribosomal protein, large, P0 housekeeping gene (RPLP0).
Immunohistochemistry and confocal imaging.
Myotubes were stained as previously described (31), with minor modifications. Cells were fixed with 10% formalin and permeablized with 0.1% saponin prior to being incubated with the human total OXPHOS antibody cocktail (MS601; MitoSciences). Myotubes were then probed with donkey anti-mouse IgG conjugated to Alexa 680 (A21058; Invitrogen Molecular Probes). Nuclei were stained using DAPI (Invitrogen Molecular Probes). Images were taken using a confocal microscope (Zeiss 510 META; Carl Zeizz, Thornwood, NY). OXPHOS was quantified by measuring fluorescence intensity using a FlexStation II microplate reader (Molecular Devices, Sunnyvale, CA). Negative controls performed with the addition of a nonspecific mouse primary IgG did not present a fluorescent signal.
Statistical analysis.
Cross-sectional differences in clinical characteristics (Table 1), NAMPT/complex III protein expression, and mtDNA (Fig. 1) were analyzed via one-way ANOVA with Tukey's post hoc-adjusted P. A Shapiro-Wilk test was used to confirm the normality of the data. In cases where both parameters were distributed normally, a Pearson correlation was used (Fig. 2, B and C). In cases where one or both parameters were not normally distributed, a Spearman correlation was used (Figs. 2, A, D, and E, 3D, and 4, A and B). Pre- and postexercise training data were compared by paired Student's t-test (Fig. 3, A–C). Forskolin- and ionomycin-treated cells (IHC) and AICAR-treated cells were compared with control cells by paired Student's t-test (Figs. 5C and 6, B–D). Forskolin- and/or ionomycin-treated cells (complex III, NAMPT, pAMPK:AMPK) were compared with control cells by one-way ANOVA with Dunnett's post hoc-adjusted P (Figs. 5, D and E, and 6A). Differences in percent increase in NAMPT protein were analyzed by one-way ANOVA with Tukey's post hoc-adjusted P. P < 0.05 was considered significant.
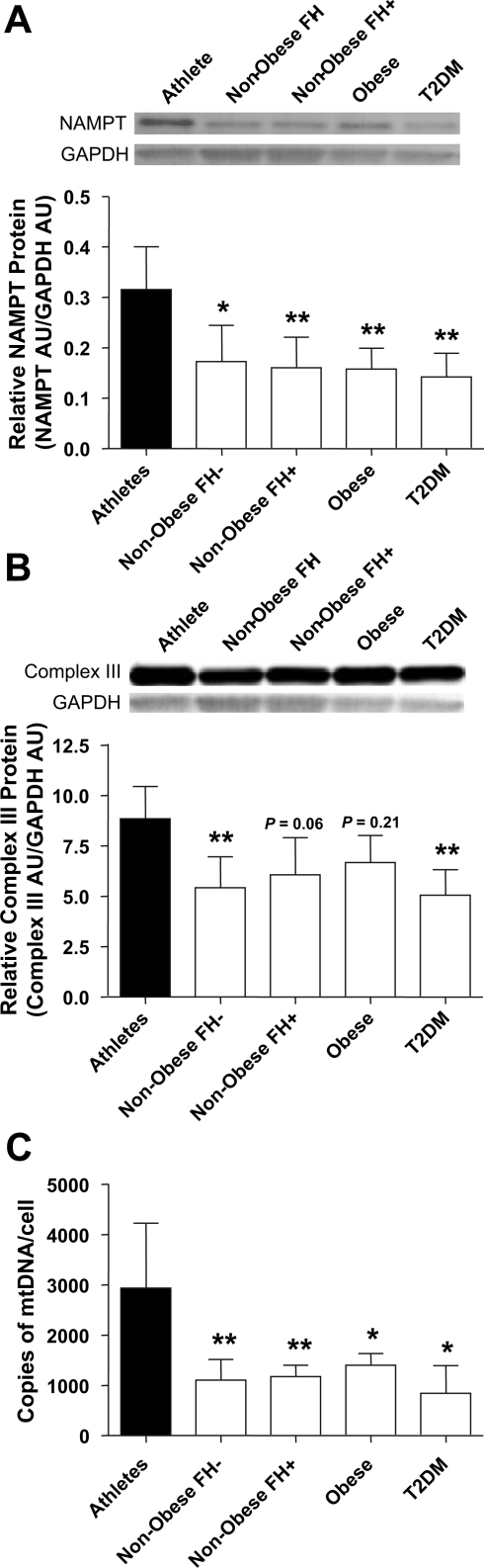
Fig. 1.
Skeletal muscle nicotinamide phosphoribosyltransferase (NAMPT) protein expression and mitochondrial content in metabolically distinct subjects. FH, nonobese sedentary subjects with or without family history of T2DM; AU, arbitrary units. A: NAMPT protein in skeletal muscle (means ± SD, n = 5, one-way ANOVA with Tukey's post hoc test, *P < 0.05, **P < 0.01 vs. Athletes). B: complex III protein in skeletal muscle (means ± SD, n = 5, one-way ANOVA with Tukey's post hoc test, **P < 0.01 vs. Athletes). C: mitochondrial DNA in skeletal muscle (means ± SD, n = 2–5, one-way ANOVA with Tukey's post hoc test, *P < 0.05, **P < 0.01 vs. Athletes).
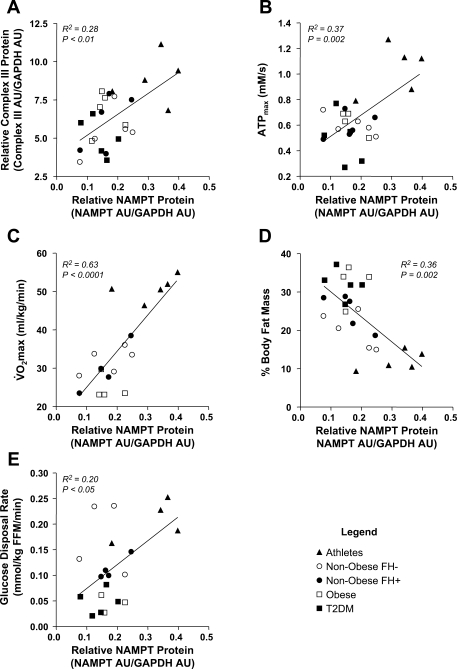
Fig. 2.
Relationship between skeletal muscle NAMPT protein and markers of mitochondrial content and function. A: Spearman correlation between NAMPT and complex III protein expression in skeletal muscle (n = 25, R2 = 0.28, P < 0.01). B: Pearson correlation between skeletal muscle NAMPT protein expression and ATPmax (n = 24, R2 = 0.37, P = 0.002). C: Pearson correlation between skeletal muscle NAMPT protein expression and V̇o2max (n = 18, R2 = 0.63, P < 0.0001). D: Spearman correlation between skeletal muscle NAMPT protein and %body fat mass (n = 24, R2 = 0.36, P = 0.002). E: Spearman correlation between skeletal muscle NAMPT protein and glucose disposal rate (n = 20, R2 = 0.20, P < 0.05).
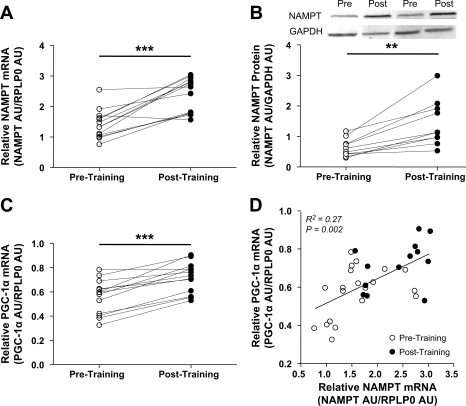
Fig. 3.
Skeletal muscle NAMPT and PPARγ coactivator-1α (PGC-1α) expression pre- and postexercise training. A: skeletal muscle NAMPT mRNA (means ± SD, n = 13, paired Student's t-test, ***P < 0.001). B: skeletal muscle NAMPT protein (means ± SD, n = 10, paired Student's t-test, **P < 0.01). C: skeletal muscle PGC-1α mRNA (means ± SD, n = 13, paired Student's t-test, ***P < 0.001). D: Spearman correlation between skeletal muscle NAMPT and PGC-1α mRNA (pretraining n = 20, posttraining n = 13, R2 = 0.27, P = 0.002).
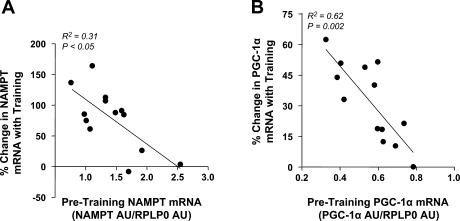
Fig. 4.
A: Spearman correlation between baseline level of skeletal muscle NAMPT mRNA and %change in NAMPT mRNA with exercise training (n = 13, R2 = 0.31, P < 0.05). B: Spearman correlation between baseline level of skeletal muscle PGC-1α mRNA and %change in PGC-1α mRNA with exercise training (n = 13, R2 = 0.62, P = 0.002).
Fig. 5.
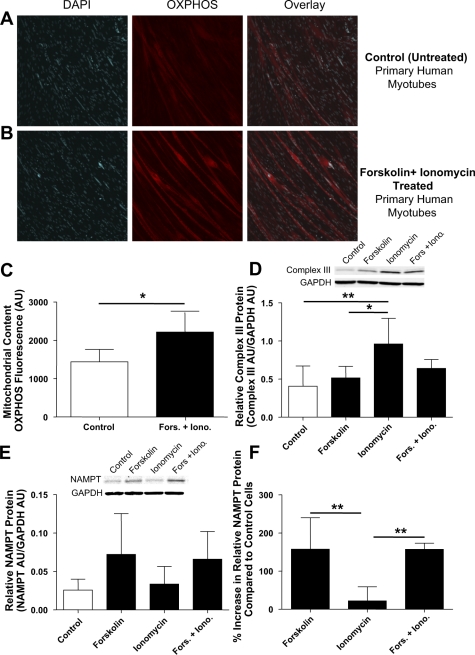
NAMPT protein expression and mitochondrial content in forskolin- and/or ionomycin-treated primary human myotubes. DAPI staining of nuclei and OXPHOS staining of mitochondria and overlay in control (untreated; A) and forskolin- and ionomycin-treated primary human myotubes (B). C: quantification of mitochondrial volume by OXPHOS fluorescence (means ± SD, n = 4, paired Student's t-test, P < 0.05) in control and forskolin- and ionomycin-treated cells. D: complex III protein in control and forskolin- and/or ionomycin-treated primary human myotubes (means ± SD, n = 5, one-way ANOVA with Dunnett post hoc test, *P < 0.05, **P < 0.01). E: NAMPT protein in control and forskolin- and/or ionomycin-treated primary human myotubes [means ± SD, n = 5, one-way ANOVA with Dunnett's post hoc test, not significant (NS)]. F: %increase in NAMPT protein vs. control cells (means ± SD, n = 5, one-way ANOVA with Tukey's post hoc test, **P < 0.01).
Fig. 6.
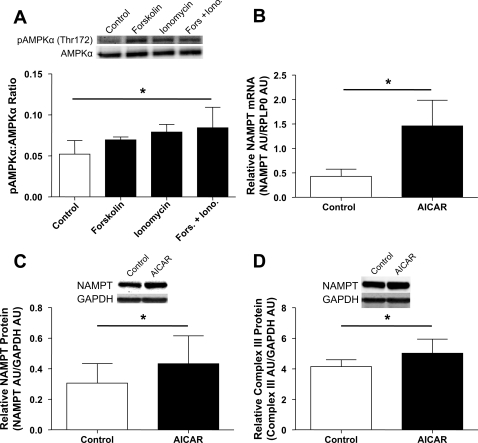
Effect of AMPK activation on NAMPT expression and mitochondrial content. A: phospho-AMPKα (Thr172)-to-AMPKα ratio in control and forskolin- and/or ionomycin-treated primary human myotubes (means ± SD, n = 4, one-way ANOVA with Dunnett's post hoc test, *P < 0.05). B: NAMPT mRNA in control and AICAR-treated primary human myotubes (means ± SD, n = 3, paired Student's t-test, *P < 0.05). C: NAMPT protein in control and AICAR-treated primary human myotubes (means ± SD, n = 4, paired Student's t-test, *P < 0.05). D: complex III protein in control and AICAR-treated primary human myotubes (means ± SD, n = 4, paired Student's t-test, *P < 0.05).
RESULTS
Athletes have higher skeletal muscle NAMPT protein expression.
NAMPT protein expression was examined in muscle homogenates obtained from vastus lateralis biopsies of five metabolically distinct groups of subjects: endurance-trained athletes, nonobese sedentary subjects with no family history of T2DM (FH−), nonobese sedentary subjects with at least one parent having T2DM (FH+), obese sedentary subjects and sedentary subjects with T2DM (Table 1). Endurance-trained athletes had approximately twofold higher skeletal muscle NAMPT protein expression than subjects who were nonobese (with or without a family history of T2DM, P < 0.05 and P < 0.01, respectively) or obese (P < 0.01) or who had T2DM (P < 0.01) (Fig. 1A). Athletes also had higher muscle mitochondrial content as measured by complex III protein (P < 0.01, nonobese FH− and T2DM subjects) and mtDNA (P < 0.01, nonobese FH− and FH+, P < 0.05, obese and T2DM; Fig. 1, B and C).
NAMPT is positively correlated with mitochondrial mass and function.
NAMPT protein expression was correlated with mitochondrial content (as measured by complex III protein, R2 = 0.28, P < 0.01; Fig. 2A); with ATPmax (R2 = 0.37, P = 0.002), an in vivo measurement of maximal mitochondrial ATP synthesis (Fig. 2B); and with V̇o2max measured on a stationary bicycle (R2 = 0.63, P < 0.0001; Fig. 2C). Interestingly, skeletal muscle NAMPT protein content was negatively correlated with percent body fat (R2 = 0.36, P = 0.002) and positively correlated with glucose disposal rate (R2 = 0.20, P < 0.05; Fig. 2, E and F).
Exercise training increases skeletal muscle NAMPT protein expression.
A subgroup of 13 nonobese subjects underwent an exercise training program (7 subjects acted as nonexercise controls). Baseline subject characteristics are described in Supplementary Table 1 (the effect of exercise training on these characteristics will be reported elsewhere). Skeletal muscle NAMPT mRNA increased by 68% (P < 0.001), and NAMPT protein increased by 127% (P < 0.01) with exercise training (Fig. 3, A and B), whereas PGC-1α mRNA, a potent stimulator of mitochondrial biogenesis, increased by 28% (P < 0.001; Fig. 3C). We found a positive correlation between NAMPT and PGC-1α mRNA (R2 = 0.27, P = 0.002; Fig. 3D). Interestingly, we observed a ceiling effect for both NAMPT and PGC-1α mRNA expression (R2 = 0.31, P < 0.05, and R2 = 0.62, P = 0.002, respectively): subjects with a high pretraining level of NAMPT or PGC-1α had a smaller increase with training (Figs. 4, E and F).
Treatment of human myotubes with forskolin increases NAMPT expression in vitro.
Primary human myotubes were treated daily with 60-min pulses of forskolin and ionomycin in vitro to simulate exercise training. Forskolin and ionomycin activate the cAMP- and calcium-signaling pathways, respectively, which are activated by exercise in skeletal muscle in vivo. Treatment with both forskolin and ionomycin increased mitochondrial volume by ∼50% (P < 0.05), as measured by quantitative confocal immunohistochemistry of the electron transport chain (Fig. 5, A–C). Increases in mitochondrial content as measured by complex III protein, however, appeared to be driven by ionomycin (P < 0.01; Fig. 5D). NAMPT protein content increased ∼2.5-fold compared with untreated cells when treated with forskolin and ionomycin or forskolin alone, whereas ionomycin did not affect NAMPT protein content (Fig. 5, E and F).
Treatment of human myotubes with AICAR increases NAMPT expression in vitro.
Treatment with ionomycin and chronic treatment with forskolin have previously been shown to activate AMPK in cells (11, 21). In our system, we observed small increases in the pAMPK/AMPK ratio after three daily forskolin or ionomycin pulse treatments, reaching significance when both forskolin and ionomycin were pulsed together (P < 0.05; Fig. 6A). To determine whether NAMPT could be upregulated by AMPK activation independently of the cAMP-signaling pathway, we treated cells with 0.5 mM AICAR during differentiation. Treatment of cells with AICAR for 8, 24, or 48 h had no effect on NAMPT mRNA or protein content or on mitochondrial content (data not shown). However, treatment with AICAR for the duration of differentiation (5 days) resulted in an ∼3.4-fold increase in NAMPT mRNA (P < 0.05; Fig. 6B) and a small but significant increase in NAMPT protein (P < 0.05; Fig. 6C). AICAR treatment also resulted in a small but significant increase in mitochondrial content as measured by complex III protein (P < 0.05; Fig. 6C).
DISCUSSION
NAMPT catalyzes the rate-limiting step in the conversion of nicotinamide to NAD+ in the mammalian NAD+ salvage pathway and is essential for the process of oxidative phosphorylation due to NAD+'s role as a shuttle for electrons between the TCA cycle and the ETC. NAD+ is also an obligate cosubstrate for the enzymatic activity of SIRT1 (22), an important protein deacetylase. In this pilot clinical study, we found that athletes have higher skeletal muscle NAMPT protein and that exercise training increases NAMPT protein expression in previously sedentary individuals. In the cross-sectional study, skeletal muscle NAMPT was correlated with mitochondrial content, maximal mitochondrial ATP synthesis and maximum aerobic capacity. Treatment of human myotubes with forskolin or AICAR was sufficient to increase NAMPT protein and, in the case of AICAR, mitochondrial content. These results support NAMPT's essential role in energy metabolism and suggest that NAMPT may be regulated in parallel with mitochondrial biogenesis or may perhaps play a role in the regulation of mitochondrial biogenesis in response to exercise. The pathways regulating mitochondrial biogenesis are of particular interest, as some studies have shown decreased skeletal muscle mitochondrial mass and abnormal mitochondrial morphology in individuals who have T2DM (24, 30, 33, 34). However, this area is controversial, as other studies have shown normal mitochondrial function in individuals with T2DM or have provided evidence supporting a lack of connection between mitochondria and insulin resistance (5, 17, 27, 35). Exercise is known to increase skeletal muscle mitochondrial content, although the pathways through which this occurs have not been fully elucidated. If NAMPT does indeed play a role in the regulation of mitochondrial biogenesis, NAMPT may represent a novel target for reversing decreased mitochondrial content in individuals with T2DM who exhibit this defect.
We have shown that athletes have approximately twofold higher skeletal muscle NAMPT protein compared with sedentary individuals and that NAMPT protein can be upregulated more than twofold in sedentary individuals following 3 wk of exercise training. Skeletal muscle NAMPT protein was correlated with mitochondrial content, maximal mitochondrial ATP synthesis, and maximum aerobic capacity. These preliminary data indicate that NAMPT is related not only to skeletal muscle mitochondrial mass and function but to overall aerobic fitness. The data are only correlative, however; therefore, further experiments will be required to determine whether NAMPT is a direct or an indirect player in (or simply upregulated in tandem with) the mitochondrial biogenesis pathway. It is interesting that the strongest correlation was found between NAMPT protein and maximal aerobic capacity (V̇o2max). It is generally believed that oxygen delivery by the cardiovascular system is the major determinant of V̇o2max; however, the strong correlation between skeletal muscle NAMPT and V̇o2max suggests that the production of sufficient NAD+ to shuttle electrons from the TCA cycle to the electron transport chain may also be an important factor. We observed a ceiling effect for both NAMPT and PGC-1α mRNA in response to exercise training: individuals having high baseline levels of NAMPT/PGC-1α showed a smaller percent increase posttraining. These data suggest that NAMPT and PGC-1α may be regulated in part by negative feedback loops.
Interestingly, we found that skeletal muscle NAMPT protein was negatively correlated with body fat and positively correlated with whole body insulin sensitivity. This is in opposition to what has been observed of circulating levels of eNAMPT, which have been reported to be increased in obesity (45) and T2DM (28) but reduced with exercise (7). The differences in eNAMPT and iNAMPT expression in obesity suggest independent regulation and possibly function.
We treated primary human myotubes in vitro with forskolin and/or ionomycin to establish whether this increase in NAMPT expression was a direct or indirect effect of exercise on muscle. Forskolin and ionomycin activate the cAMP- and calcium-signaling pathways, respectively. Treatment with forskolin (or forskolin + ionomycin) resulted in an ∼2.5-fold increase in NAMPT protein expression, whereas ionomycin alone had no effect on NAMPT protein. The stimulation of the cAMP-signaling pathway is therefore sufficient to increase NAMPT protein content in skeletal muscle. Previous studies have shown that long-term forskolin treatment of cells can activate AMPK (11); however, we did not achieve a significant increase in pAMPK/AMPK ratio with forskolin alone (although forskolin + ionomycin produced a significant increase). To test whether AMPK activation could upregulate NAMPT, we chronically treated cells with AICAR and found an increase in both NAMPT mRNA and protein, consistent with the findings of Fulco et al. (14) in C2C12 cells. Shorter-term AICAR exposure (8, 24, or 48 h) was not sufficient to affect NAMPT expression or mitochondrial content in our system, suggesting that the chronic stimulation of AMPK (or the “training effect”) is necessary to produce NAMPT upregulation. These data support the view that the increase in NAMPT protein seen with exercise is due, at least in part, to direct signaling effects within skeletal muscle, not to neural or endocrine effects of exercise training. Since NAD+ is an obligate cofactor for SIRT1, and SIRT1 deacetylates PGC-1α, it is tempting to hypothesize that an AMPK-NAMPT-SIRT1-PGC-1α pathway would stimulate mitochondrial biogenesis. Although Cantó et al. (6) have recently reported that SIRT1 mediates AMPK-induced PGC-1α deacetylation in mouse skeletal muscle, Gurd et al. (16) also recently showed that overexpression of SIRT1 in rodent muscle in fact decreased PGC-1α expression and mitochondrial content. Interestingly, although forskolin treatment resulted in increased NAMPT expression, we found that it had no effect on mitochondrial content as measured by complex III protein. AICAR treatment resulted in an increase in both NAMPT and mitochondrial content, whereas treatment with ionomycin increased mitochondrial content without increasing NAMPT. It is clear from the clinical data that exercise training increases both NAMPT expression and mitochondrial content, but whether these are coregulated by a single upstream system or act in series has yet to be elucidated. The forskolin/ionomycin results suggest that NAMPT and mitochondrial biogenesis are independently regulated, but the AICAR results suggest that both are regulated by AMPK. Further in vitro work is needed to dissect these signaling pathways.
In summary, skeletal muscle NAMPT is upregulated in response to exercise in humans and correlates with muscle PGC-1α mRNA, mitochondrial content, ATPmax, and V̇o2max. Combined with prior data implicating NAMPT in the regulation of SIRT1, this suggests that NAMPT may play a role in the regulation of mitochondrial biogenesis in response to exercise; however, a direct link has yet to be demonstrated. Future studies should focus on defining the downstream effects of NAMPT activation as well as its upstream regulators in skeletal muscle.
GRANTS
This work was supported by a Pennington Biomedical Research Center (PBRC) Pilot and Feasibility grant (S. R. Costford); National Institutes of Health (NIH) Grants R01 (1 R01 AG-030226-01A2, S. R. Smith); US Dept. of Agriculture (2005-34323-15741, S. R. Smith); and the PBRC Clinical Nutrition Research Unit (CNRU) (NIH 5 P30 DK-072476). This work utilized the facilities of the Cell Biology and Bioimaging Core facilities that are supported in part by COBRE (NIH P20 RR-021945) and CNRU (NIH 5 P30 DK-072476) grants from NIH.
DISCLOSURES
This work was also supported by an unrestricted research grant from Novartis, Novartis Clinical Innovation Fund (S. R. Smith), and an unrestricted research grant from Takeda Pharmaceuticals North America, Takeda Investigator-Initiated Trial (S. R. Smith).
ACKNOWLEDGMENTS
We acknowledge Xiaobing Fang and Dr. José Galgani for consultation on statistical analysis. We thank Dr. Conrad Earnest and Stephanie Anaya for their help with V̇o2max testing as well as Himanshu Chopra and Dr. Cedric Moro for helpful conversations. We also acknowledge the support and commitment of our research volunteers, who made this work possible.
REFERENCES
- 1.Arner P. Visfatin—a true or false trail to type 2 diabetes mellitus. J Clin Endocrinol Metab 91: 28–30, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Blei ML, Conley KE, Kushmerick MJ. Separate measures of ATP utilization and recovery in human skeletal muscle. J Physiol 465: 203–222, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blei ML, Conley KE, Odderson IB, Esselman PC, Kushmerick MJ. Individual variation in contractile cost and recovery in a human skeletal muscle. Proc Natl Acad Sci USA 90: 7396–7400, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogacka I, Xie H, Bray GA, Smith SR. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 54: 1392–1399, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Boushel R, Gnaiger E, Schjerling P, Skovbro M, Kraunsoe R, Dela F. Patients with type 2 diabetes have normal mitochondrial function in skeletal muscle. Diabetologia 50: 790–796, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–10560, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi KM, Kim JH, Cho GJ, Baik SH, Park HS, Kim SM. Effect of exercise training on plasma visfatin and eotaxin levels. Eur J Endocrinol 157: 437–442, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Conley KE, Esselman PC, Jubrias SA, Cress ME, Inglin B, Mogadam C, Schoene RB. Ageing, muscle properties and maximal O(2) uptake rate in humans. J Physiol 526: 211–217, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conley KE, Jubrias SA, Esselman PC. Oxidative capacity and ageing in human muscle. J Physiol 526: 203–210, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol Endocrinol Metab Gastrointest Physiol 237: E214–E223, 1979 [DOI] [PubMed] [Google Scholar]
- 11.Egawa M, Kamata H, Kushiyama A, Sakoda H, Fujishiro M, Horike N, Yoneda M, Nakatsu Y, Ying G, Jun Z, Tsuchiya Y, Takata K, Kurihara H, Asano T. Long-term forskolin stimulation induces AMPK activation and thereby enhances tight junction formation in human placental trophoblast BeWo cells. Placenta 29: 1003–1008, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, Watanabe E, Takagi T, Akiyoshi M, Ohtsubo T, Kihara S, Yamashita S, Makishima M, Funahashi T, Yamanaka S, Hiramatsu R, Matsuzawa Y, Shimomura I. Retraction. Science 318: 565, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, Watanabe E, Takagi T, Akiyoshi M, Ohtsubo T, Kihara S, Yamashita S, Makishima M, Funahashi T, Yamanaka S, Hiramatsu R, Matsuzawa Y, Shimomura I. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 307: 426–430, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 14: 661–673, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J 26: 1913–1923, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurd B, Yoshida Y, Lally J, Holloway G, Bonen A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis. J Physiol 587: 1817–18282009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock CR, Han DH, Chen M, Terada S, Yasuda T, Wright DC, Holloszy JO. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci USA 105: 7815–7820, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8: 774–785, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Hardie DG, Hawley SA, Scott JW. AMP-activated protein kinase–development of the energy sensor concept. J Physiol 574: 7–15, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He L, Chinnery PF, Durham SE, Blakely EL, Wardell TM, Borthwick GM, Taylor RW, Turnbull DM. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res 30: e68, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 280: 29060–29066, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Jubrias SA, Esselman PC, Price LB, Cress ME, Conley KE. Large energetic adaptations of elderly muscle to resistance and endurance training. J Appl Physiol 90: 1663–1670, 2001 [DOI] [PubMed] [Google Scholar]
- 24.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Kitani T, Okuno S, Fujisawa H. Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett 544: 74–78, 2003 [DOI] [PubMed] [Google Scholar]
- 26.Korner A, Garten A, Bluher M, Tauscher R, Kratzsch J, Kiess W. Molecular characteristics of serum visfatin and differential detection by immunoassays. J Clin Endocrinol Metab 92: 4783–4791, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Kruse SE, Watt WC, Marcinek DJ, Kapur RP, Schenkman KA, Palmiter RD. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab 7: 312–320, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Bermejo A, Chico-Julia B, Fernandez-Balsells M, Recasens M, Esteve E, Casamitjana R, Ricart W, Fernandez-Real JM. Serum visfatin increases with progressive beta-cell deterioration. Diabetes 55: 2871–2875, 2006 [DOI] [PubMed] [Google Scholar]
- 29.McCully KK. 31P-MRS of quadriceps reveals quantitative differences between sprinters and long-distance runners. Med Sci Sports Exerc 25: 1299–1300, 1993 [PubMed] [Google Scholar]
- 30.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273, 2003 [DOI] [PubMed] [Google Scholar]
- 31.Ohsaki Y, Maeda T, Fujimoto T. Fixation and permeabilization protocol is critical for the immunolabeling of lipid droplet proteins. Histochem Cell Biol 124: 445–452, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Paganini AT, Foley JM, Meyer RA. Linear dependence of muscle phosphocreatine kinetics on oxidative capacity. Am J Physiol Cell Physiol 272: C501–C510, 1997 [DOI] [PubMed] [Google Scholar]
- 33.Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300: 1140–1142, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 350: 664–671, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pospisilik JA, Knauf C, Joza N, Benit P, Orthofer M, Cani PD, Ebersberger I, Nakashima T, Sarao R, Neely G, Esterbauer H, Kozlov A, Kahn CR, Kroemer G, Rustin P, Burcelin R, Penninger JM. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131: 476–491, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Preiss J, Handler P. Enzymatic synthesis of nicotinamide mononucleotide. J Biol Chem 225: 759–770, 1957 [PubMed] [Google Scholar]
- 37.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 279: 50754–50763, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab 6: 363–375, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol 14: 1431–1437, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, Smith SR. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes 54: 1926–1933, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Stephens JM, Vidal-Puig AJ. An update on visfatin/pre-B cell colony-enhancing factor, an ubiquitously expressed, illusive cytokine that is regulated in obesity. Curr Opin Lipidol 17: 128–131, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun 296: 350–354, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Ukropcova B, McNeil M, Sereda O, de Jonge L, Xie H, Bray GA, Smith SR. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J Clin Invest 115: 1934–1941, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ukropcova B, Sereda O, de Jonge L, Bogacka I, Nguyen T, Xie H, Bray GA, Smith SR. Family history of diabetes links impaired substrate switching and reduced mitochondrial content in skeletal muscle. Diabetes 56: 720–727, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Zahorska-Markiewicz B, Olszanecka-Glinianowicz M, Janowska J, Kocelak P, Semik-Grabarczyk E, Holecki M, Dabrowski P, Skorupa A. Serum concentration of visfatin in obese women. Metabolism 56: 1131–1134, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA 99: 15983–15987, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]