Abstract
Transforming growth factor β (TGF-β) signals regulate multiple processes during development and in adult. We recently showed that tomoregulin-1 (TMEFF1), a transmembrane protein, selectively inhibits nodal but not activin in early Xenopus embryos. Here we report that TMEFF1 binds to the nodal coreceptor Cripto, but does not associate with either nodal or the type I ALK (activin receptor-like kinase) 4 receptor in coimmunoprecipitation assays. The inhibition of the nodal signaling by TMEFF1 in Xenopus ectodermal explants is rescued with wild-type but not mutant forms of Cripto. Furthermore, we show that the Cripto-FRL1-Cryptic (CFC) domain in Cripto, which is essential for its binding to ALK4, is also important for its interaction with TMEFF1. Our results demonstrate for the first time that nodal signaling can be regulated by a novel mechanism of blocking the Cripto coreceptor.
Keywords: Nodal, Cripto, TGF-β, tomoregulin-1
Members of the transforming growth factor β (TGF-β) family play pivotal roles in both invertebrate and vertebrate embryogenesis (for reviews, see Harland and Gerhart 1997; Schier and Shen 1999; Whitman 2001). During early vertebrate development, the activin/nodal/Vg1/GDF1 subfamily of ligands is involved in induction and patterning of the mesodermal and endodermal germ layers. Subsequently, these ligands participate in regulation of left-right axis formation. Removal of gene functions in the activin/nodal/Vg1/GDF1 pathways by mutations, gene knockout, or dominant negative components leads to aberrant development of vertebrate embryos that are defective in mesendodermal tissues and have left-right laterality defect (Harland and Gerhart 1997; Schier and Shen 1999; Whitman 2001).
Like other TGF-β ligands, activin/nodal/Vg1/GDF1 signals through two types of transmembrane serine/threonine kinase receptors. On ligand binding, the constitutively active type II receptor forms a complex with the type I receptor ALK (activin receptor-like kinase) 4 and 7, and activates the type I receptor through phosphorylation. The activated ALK then phosphorylates the cytoplasmic signal transducers Smad2 and Smad3, which form a hexameric complex with the common Smad, Smad4, and translocate into the nucleus to regulate gene expression in conjunction with other transcription factors (for reviews, see Massague 1998; Shi and Massague 2003). All of these ligands use the same type I receptor ALK4 and the type II receptors ActRIIA/IIB; however, nodal and Vg1/GDF1, but not activin, also require a membrane-associated EGF-CFC protein belonging to the Cripto family as a coreceptor in their signaling transduction (Gritsman et al. 1999; Reissmann et al. 2001; Yeo and Whitman 2001; Bianco et al. 2002; Yan et al. 2002; Cheng et al. 2003). Mutation in the Cripto family member One-eyed pinhead (Oep) in zebrafish leads to defective nodal/Vg1/GDF1 signaling, so that the resulting embryos mimic those with the mutations in the nodal ligands (Gritsman et al. 1999; Cheng et al. 2003). In mouse, nodal mutants display several defects in common with the mutants of Cripto or the related factor Cryptic (Conlon et al. 1994; Lowe et al. 2001; Brennan et al. 2002; Norris et al. 2002). Biochemical evidence suggests that Cripto members bind directly to nodal/Vg1/GDF1 ligands as well as the ALK4 receptor, thus facilitating the assembly of a functional receptor complex at the membrane (Reissmann et al. 2001; Yeo and Whitman 2001; Bianco et al. 2002; Yan et al. 2002; Cheng et al. 2003). Although Cripto members are not required for activin signaling, overexpression of Cripto does influence the activin activity by repressing the signal pathway (Gray et al. 2003). These results imply that Cripto may have a function in fine-tuning the signals through different TGF-β ligands in vivo.
TGF-β signals are regulated by multiple factors at different levels (for review, see Massague and Chen 2000; Shi and Massague 2003). While secreted factors modulate the binding of the TGF-β ligands to their cognate receptors, cytoplasmic and nuclear proteins regulate localization, degradation, and modification of the Smad signal transducers as well as the interaction of the Smads with other proteins, such as nuclear transcription factors. At the membrane level, TGF-β signaling can also be regulated either positively or negatively by receptor-like proteins, such as the type III TGF-β receptor betaglycan or the naturally occurring truncated receptor BAMBI (Lopez-Casillas et al. 1993; Onichtchouk et al. 1999). Recently, we showed that a transmembrane protein tomoregulin-1 (TMEFF1) can modulate signals through different TGF-β ligands in early Xenopus embryos (Chang et al. 2003). Although TMEFF1 has no effect on mesendodermal induction by activin in Xenopus ectodermal explants (animal caps), it inhibits both nodal and Vg1 activities in this assay (Chang et al. 2003). TMEFF1 contains two follistatin modules and an epidermal growth factor (EGF) motif in its extracellular domain, and a short conserved cytoplasmic tail following the transmembrane region (Eib and Martens 1996). Deletion analysis showed that the cytoplasmic domain of TMEFF1 is dispensable for its nodal inhibitory activity, a result that suggests that TMEFF1 blocks nodal signaling at the ligand or at the receptor level (Chang et al. 2003). To understand further the mechanism by which TMEFF1 inhibits nodal, we undertook biochemical studies. Here we report that TMEFF1 binds directly to the Cripto coreceptor, but does not interact with either nodal or the ALK4 type I receptor in coimmunoprecipitation assays. The inhibition of the nodal signaling by TMEFF1 is rescued with wild-type but not mutant forms of Cripto. Furthermore, we show that the Cripto-FRL1-Cryptic (CFC) domain in Cripto is important for the interaction of the two proteins. Our data thus demonstrate for the first time that, in addition to the extracellular, cytoplasmic, and nuclear regulation, nodal signaling can be modulated at the membrane by a nonreceptor protein, TMEFF1. Our discovery that Cripto interacts with TMEFF1 may also help to shed light on the studies of nodal-independent functions of Cripto in other cellular contexts.
Results and Discussion
TMEFF1 binds directly to Cripto, but does not associate with nodal or ALK4
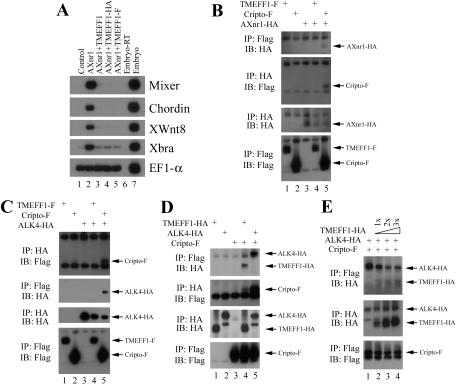
TMEFF1, a follistatin module-containing protein, selectively inhibits nodal and Vg1 but not activin (Chang et al. 2003). Because follistatin and the follistatin-related gene (FLRG) have been shown to inhibit activin through direct binding to the ligand (Kogawa et al. 1991; Schneyer et al. 2001; Tsuchida et al. 2001; Bartholin et al. 2002), it is possible that TMEFF1 uses a similar mechanism by which it selectively interacts with and blocks nodal but not activin. To test this possibility, we performed a biochemical study to assay for binding of nodal by TMEFF1. We first constructed a Flag-tagged TMEFF1 (TMEFF1-F) and examined its activity. A chimeric nodal ligand AXnr1-HA, consisting of the prodomain of activin and the HA-tagged mature region of Xnr1 (Xenopus nodal-related 1; Piccolo et al. 1999), was used in both the activity and the binding assays. As shown in Figure 1A, similar to the wild-type TMEFF1, TMEFF1-F blocks the induction of mesendodermal markers by AXnr1 in Xenopus animal caps (Fig. 1A, cf. lanes 3 and 5). The result suggests that TMEFF1-F retained its nodal inhibitory activity. When TMEFF1-F was coexpressed with AXnr1-HA in early Xenopus embryos and the embryonic extract was examined by a coimmunoprecipitation assay at early gastrula stages, we found that no TMEFF1-F was coprecipitated with AXnr1-HA by the anti-HA antibody; similarly, AXnr1-HA was not precipitated with TMEFF1-F by the anti-Flag antibody (Fig. 1B). The result implies that TMEFF1 may not bind to nodal. To confirm that our immunoprecipitation assay worked well, we analyzed the binding of nodal to Cripto under the same conditions, because it has been shown that Xenopus Xnr1 interacts with Cripto directly (Reissmann et al. 2001). As shown in Figure 1B, Cripto was coprecipitated with AXnr1 when a Flag-tagged Cripto (Cripto-F) was coexpressed with the HA-tagged AXnr1; the reverse coimmunoprecipitation also showed that AXnr1 was coprecipitated with Cripto (Fig. 1B). Our data thus demonstrate that, unlike Cripto, which binds to nodal but not activin (Reissmann et al. 2001; Gray et al. 2003), TMEFF1 does not selectively bind to nodal to inhibit its signaling. Our finding also helps to explain an earlier observation (Chang et al. 2003) that the soluble extracellular domain of TMEFF1, unlike the TMEFF1 mutant that lacks the cytoplasmic domain, does not block nodal signaling. This result, combined with the current finding, suggests that TMEFF1 may block nodal signaling at the membrane level.
Figure 1.
TMEFF1, a nodal inhibitor, binds directly to the nodal coreceptor Cripto, but does not associate with either nodal or ALK4. (A) The Flag- and the HA-tagged TMEFF1 inhibit nodal activity in Xenopus animal caps. (B) TMEFF1, unlike Cripto, does not bind to nodal. (C) TMEFF1 does not associate with the ALK4 receptor. (D) TMEFF1 binds directly to the Cripto coreceptor. (E) Binding of TMEFF1 to Cripto reduces the association of ALK4 with Cripto. In panels B-D, the RNAs encoding the tagged proteins (2 ng Cripto-F, 3 ng TMEFF1-HA/F, 3 ng ALK4-HA, and 4 ng AXnr1-HA) were injected into the animal poles of two-cell-stage embryos. In panel E, the doses of RNAs used are 1 ng Cripto-F; 2 ng ALK4-HA; and 2, 4, and 6 ng TMEFF1-HA. GFP RNA was used to keep the total amount of injected RNA constant. Protein extract made from early gastrula embryos was split into equal halves and subjected to IP with either anti-HA or anti-Flag antibodies. Western blot was performed with anti-HA or anti-Flag antibodies, as indicated. (IP) Immunoprecipitation; (IB) immunoblotting.
In addition to the type I receptor ALK4 and the type II receptors ActRIIA/IIB, which are shared among activin, nodal, and Vg1/GDF1 pathways, a membrane-associated EGF-CFC Cripto family protein is also required for nodal and Vg1/GDF1 signaling (Gritsman et al. 1999; Reissmann et al. 2001; Yeo and Whitman 2001; Bianco et al. 2002; Yan et al. 2002; Cheng et al. 2003). The selective inhibition of nodal and Vg1 but not activin by TMEFF1 may therefore result from the failure to form a functional receptor complex between Cripto and ALK4 in the presence of TMEFF1. This may occur if TMEFF1 directly associates with either the ALK4 receptor or the Cripto coreceptor. To examine whether either of these nodal receptors indeed interacts with TMEFF1, we performed coimmunoprecipitation experiments. We first assayed for potential binding of TMEFF1 to ALK4, using TMEFF1-F and an HA-tagged ALK4, which has been shown to be active in Xenopus embryos (Chang et al. 1997). As a positive control for binding to ALK4 (Yeo and Whitman 2001), we also examined the interaction of Cripto-F with ALK4-HA in parallel. Whereas we observed that Cripto coprecipitated with ALK4 and vice versa, we could not detect association of TMEFF1 with ALK4 under the same conditions (Fig. 1C). Although our results do not exclude the possibility that there may be a weak interaction between TMEFF1 and ALK4 that we cannot detect in our coimmunoprecipitation assays, our data suggest that TMEFF1 may not block nodal signaling by binding to the ALK4 receptor.
We next examined whether TMEFF1 could bind to the Cripto coreceptor. For this purpose, we constructed an HA-tagged TMEFF1. Functional analysis in Xenopus animal caps showed that, similar to TMEFF1-F, TMEFF1-HA retained its nodal inhibitory activity (Fig. 1A). When Cripto-F was coexpressed with TMEFF1-HA in early Xenopus embryos, we found that TMEFF1 was coprecipitated with Cripto by anti-Flag antibody; similarly, Cripto was coprecipitated with TMEFF1 by anti-HA antibody (Fig. 1D). Cripto or TMEFF1 alone cannot be precipitated by the antibodies that recognize the other epitope, suggesting that the coimmunoprecipitation is specific. In addition, we observed that TMEFF1 interacts directly with the Xenopus Cripto member FRL1 in our coimmunoprecipitation experiment (Supplementary Fig. 1). When compared with the positive control of Cripto binding to ALK4 (Yeo and Whitman 2001), we found that less protein was coprecipitated in the case of Cripto binding to TMEFF1 (Fig. 1D). Because no chemical cross-linker was used during coimmunoprecipitation, the result may imply that either the interaction of Cripto and TMEFF1 is weaker than that of Cripto and ALK4, or the protein complex is less stable. The discovery that TMEFF1 associates directly with Cripto but not nodal or ALK4, combined with the observation that the cytoplasmic domain is dispensable for the nodal inhibitory activity of TMEFF1 (Chang et al. 2003), strongly suggests that TMEFF1 inhibits nodal signaling through direct interaction with the Cripto coreceptor.
Because Cripto binds to both ALK4 and TMEFF1, it is possible that the two latter proteins compete for binding to available Cripto, and the exclusion of ALK4 from the Cripto complex leads to the inhibition of nodal signaling. To test this hypothesis, we performed a binding competition assay. Cripto-F was coexpressed with ALK4-HA in the absence or presence of increasing amounts of TMEFF1-HA in early Xenopus embryos, and protein extract from injected embryos at gastrula stages was immunoprecipitated with an anti-Flag antibody. As shown in Figure 1E, whereas the level of ALK4-HA expression was constant in all samples, the amount of ALK4-HA coprecipitated with Cripto-F declined gradually in the presence of increasing doses of TMEFF1 (Fig. 1E, cf. lanes 2-4 and 1). The data suggest that TMEFF1 may inhibit nodal signaling by an interaction with Cripto that prevents the access of ALK4 to Cripto.
Inhibition of nodal signal by TMEFF1 is cell-autonomous and is rescued by wild-type but not mutant forms of Cripto
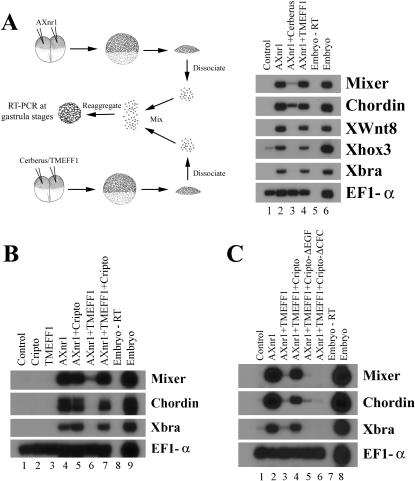
Our biochemical data suggest that TMEFF1 may block nodal pathway through its interaction with Cripto. If this is true, then, unlike secreted nodal antagonists, TMEFF1 may only inhibit nodal signaling in a cell-autonomous manner. To test this, we performed a cell-mixing experiment. Animal caps expressing AXnr1 were dissociated at blastula stages and mixed with dissociated ectodermal cells that expressed Cerberus or TMEFF1. The mixed cells were reaggregated immediately and incubated to early gastrula stages for reverse transcriptase PCR (RT-PCR) assay (Fig. 2A). In this experiment, the secreted nodal inhibitor Cerberus was able to block nodal activity in a non-cell-autonomous fashion, so that the mesendodermal marker induction by AXnr1 was either completely blocked or greatly reduced (Fig. 2A). In contrast, though TMEFF1 inhibits nodal when coexpressed with AXnr1 in animal caps, it could not confer its nodal inhibitory activity to neighboring cells, so that the markers induced by AXnr1 were still present in the cell-mixing experiment (Fig. 2A, lane 4). The results suggest that TMEFF1 blocks nodal pathway cell autonomously.
Figure 2.
Inhibition of nodal activity by TMEFF1 is cell-autonomous and is rescued by Cripto. (A) Unlike Cerberus, TMEFF1 inhibits nodal in a cell-autonomous fashion. (B,C) Nodal activity inhibited by TMEFF1 is rescued by wild-type (B) but not the mutant forms (C) of Cripto. RNAs encoding AXnr1 (0.2 ng), TMEFF1 (1 ng), Cerberus (1 ng), and wild-type or the mutant forms of Cripto (0.5-1 ng) were injected into the animal poles of two-cell-stage embryos. Animal caps were explanted at blastula stages and incubated to early gastrula before total RNA was extracted for RT-PCR assay. For the cell-mixing experiment in A, animal caps were dissociated, mixed, and reaggregated immediately as indicated, and the reaggregated cell mixtures were processed at gastrula stages, similar to other samples.
If TMEFF1 inhibits nodal activities through its interaction with Cripto, then overexpression of Cripto may overcome the inhibition and rescue the nodal signaling. We thus examined this possibility in animal cap assays. As described previously, TMEFF1 inhibited mesendodermal marker induction by AXnr1 (Fig. 2B, cf. lanes 6 and 4). When Cripto was coinjected with AXnr1 and TMEFF1, the transcription of marker genes was restored (Fig. 2B, cf. lanes 7 and 6). In addition, a constitutively active ALK4, which could stimulate nodal signaling independent of Cripto (Gritsman et al. 1999), also rescued the marker induction by AXnr1 (data not shown). These results therefore support our model that TMEFF1 inhibits nodal signaling through blocking the activation of the nodal receptor complex.
To further analyze whether the functional rescue by Cripto is specific, we attempted to rescue the nodal activity with several Cripto mutants. Cripto family proteins contain two homologous regions among all members, a divergent EGF domain and a conserved CFC motif (Schier and Shen 1999; Shen and Schier 2000; Whitman 2001). It has been shown that both domains are required for nodal signaling. Deletion of the EGF domain or point mutation of conserved residues in either domain leads to defective nodal signaling in early Xenopus embryos (Yeo and Whitman 2001). In our assays, when we coexpressed AXnr1 and TMEFF1 with Cripto mutants that lack the EGF or the CFC domain, or contain point mutations in these domains, we found that the nodal signaling could not be rescued (Fig. 2C; data not shown). These results demonstrate that rescue of TMEFF1's effect on nodal signaling requires a wild-type Cripto.
The CFC domain in Cripto is important for its interaction with TMEFF1
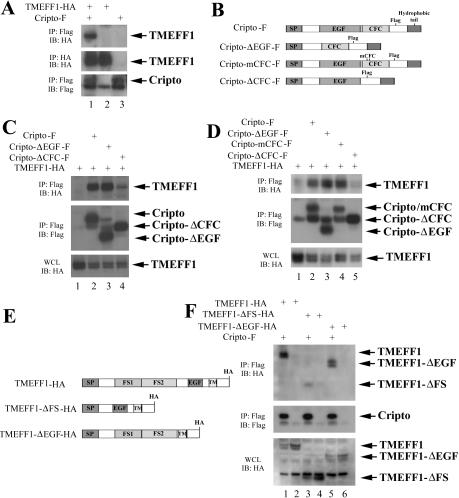
Cripto binds to ALK4 through its conserved CFC motif, and the EGF domain may be involved in its binding to nodal (Yeo and Whitman 2001; Yan et al. 2002). The interaction of TMEFF1 with Cripto may potentially mask either the EGF or the CFC motif so that Cripto cannot associate with either nodal or ALK4 to form a functional ligand/receptor complex. To address which domain is involved in the binding of Cripto to TMEFF1, we analyzed the interaction of TMEFF1 with the Cripto deletion mutants. For this experiment, we used the cell culture system. As shown in Figure 3A, similar to the situation in early Xenopus embryos, TMEFF1-HA was coprecipitated with Cripto-F when the plasmids encoding these genes were cotransfected into Chinese hamster ovary (CHO) cells, suggesting that the two proteins also interact in mammalian cell culture (Fig. 3A). When we used Flag-tagged CriptoΔEGF, which lacks the EGF domain (Fig. 3B; Yeo and Whitman 2001), we observed that the level of coprecipitated TMEFF1 was comparable to that when wild-type Cripto was used (Fig. 3C). In contrast, when we coexpressed TMEFF1 with CriptoΔCFC, which lacks the CFC motif (Fig. 3B), we observed a consistent reduction of the coprecipitated TMEFF1 (Fig. 3C). The data reveal that the CFC domain, which is critical in physical interaction with ALK4, is also important in binding of Cripto to TMEFF1. Two conserved residues in the CFC domain are essential for Cripto binding to ALK4; when these residues are mutated, the resulting mCFC mutant no longer binds to ALK4 (Fig. 3B; Yeo and Whitman 2001). To determine whether these amino acids are also involved in TMEFF1 binding, we coexpressed Cripto mCFC with TMEFF1 and assayed for their interaction by coimmunoprecipitation. We observed no reduction of coprecipitated TMEFF1 using this mutant (Fig. 3D). The result suggests that though both ALK4 and TMEFF1 bind to the CFC motif, they may contact different residues in this domain.
Figure 3.
Cripto and TMEFF1 associate in CHO cells, and this association is attenuated by deletion of the CFC domain of Cripto. (A) TMEFF1 binds to Cripto in CHO cells. (B) Schematic diagrams of wild-type and mutant Cripto. (C) Deletion of the CFC domain in Cripto attenuates the interaction between Cripto and TMEFF1. (D) The Cripto mCFC mutant with two point mutations of the conserved residues in the CFC domain, unlike the CFC deletion mutant, does not affect the binding of Cripto to TMEFF1. (E) Schematic diagrams of wild type and the deletion mutants of TMEFF1. (F) The TMEFF1 deletion mutants that lack either the follistatin modules or the EGF motif still bind to Cripto. CHO cells were transfected with the mutants or the wild-type Flag-tagged Cripto and/or HA-tagged TMEFF1, as indicated. Cell lysates were immunoprecipitated with anti-HA (IP: HA) or anti-Flag (IP:Flag) antibodies. One percent of each sample was run as whole-cell lysate (WCL). Bound protein was detected by anti-HA (IB:HA) or anti-Flag (IB:Flag) antibodies on Western blot. (SP) Signal peptide; (EGF) epidermal growth factor-like domain; (CFC) Cripto-FRL1-Cryptic domain; (FS) follistatin domain; (TM) transmembrane region.
To determine which region in TMEFF1 may be involved in its association with Cripto, we also examined the different domains of TMEFF1 using deletion mutants. There are two highly conserved regions in all TMEFF family members, which contain two follistatin modules and an EGF motif, respectively. When HA-tagged TMEFF1 mutants that lack either the follistatin modules (TMEFF1-ΔFS) or the EGF motif (TMEFF1-ΔEGF; Fig. 3E) were cotransfected with Cripto-F, we found that similar levels of wild-type or mutant forms of TMEFF1 were coprecipitated with Cripto, though we occasionally observed a slight reduction in the level of TMEFF1-ΔFS (Fig. 3F). The data suggest that the sequence outside of the follistatin and the EGF domains may be involved in direct binding of TMEFF1 to Cripto. This biochemical result is also consistent with our previous observation that both TMEFF1-ΔFS and TMEFF1-ΔEGF can still inhibit nodal activity (Chang et al. 2003).
Nodal signal plays essential roles during vertebrate embryogenesis. Strict regulation of nodal activity is therefore important to ensure correct development of vertebrate body plans. Nodal functions can be regulated by multiple factors. Cerberus, for example, blocks nodal signaling through direct binding to the ligand (Piccolo et al. 1999). Lefty, on the other hand, may bind to the nodal receptors to prevent the access of nodal to its receptors (Sakuma et al. 2002). Bone morphogenetic proteins (BMPs) can also inhibit nodal activity by forming heterodimers with nodal (Yeo and Whitman 2001; Eimon and Harland 2002). In this study, we show for the first time that a membrane protein can interact directly with the Cripto coreceptor to block nodal signaling, thus providing a novel mechanism by which nodal activity can be regulated. Interestingly, we find that the CFC domain in Cripto, which is critical for its interaction with ALK4, is also important for its interaction with TMEFF1, but the crucial residues required for association with ALK4 and TMEFF1 may differ. Together with the competition results, our data suggest that TMEFF1 and ALK4 compete for binding to Cripto, and association of TMEFF1 with Cripto may physically exclude binding of Cripto to ALK4, thus leading to the inhibition of the nodal signaling. In addition to its function during early development, Cripto has also been found to be amplified in several carcinomas (for review, see Normanno et al. 2001; Adamson et al. 2002). Cripto may signal in both ALK4-dependent and ALK4-independent fashion in these carcinoma cells to activate receptor tyrosine kinase and the downstream mitogen-activated protein (MAP) kinase pathway (Bianco et al. 1999, 2002, 2003). Because TMEFF1 directly associates with Cripto in both Xenopus and in mammalian cell culture, it is possible that TMEFF1 may directly participate in mediating Cripto signaling or modulate the nodal-independent signal transduction by Cripto in these cell contexts. Although these issues are currently under investigation, our studies reveal a key connection between TMEFF1 and Cripto function, and may provide important clues to our understanding of molecular mechanisms underlying a variety of activities mediated by Cripto and/or TMEFF family members, including influencing neuronal cell function and the formation and progression of cancers (Bianco et al. 1999, 2002, 2003; Horie et al. 2000; Glynne-Jones et al. 2001; Gery et al. 2002, 2003; Siegel et al. 2002).
Materials and methods
Plasmid construction for tagged and mutant proteins
PCR-based strategy was used to generate all constructs. For TMEFF1-HA, an N-terminal primer X7365-N(RI) (5′-GGGGAATTCACCATGGATG GATTGCACCCT-3′) and two overlapping C-terminal primers, HAC1(XbaI) (5′-GCTCTAGACTACACAGCATAGTCAGGCACGTCGT ACGG-3′) and HA-C2(7365) (5′-AGGCACGTCGTACGGATACACCAT CCGGGAAGAAGT-3′), were used for PCR over the pCS2++TMEFF1 template (Chang et al. 2003). The PCR product was digested and inserted into the EcoRI/XbaI sites of pCS2++ vector. TMEFF1ΔFS-HA and TMEFF1ΔEGF-HA were made by replacing the extracellular domain sequence in pCS2++TMEFF1-HA with that from pCS2++ TMEFF1-ΔFS and pCS2++ TMEFF1-ΔEGF, respectively (Chang et al. 2003). TMEFF1-Flag was made by PCR with the N-terminal primer X7365-N(RI) and two C-terminal primers, Flag-C1(XbaI) (5′-GCTCTAGACTACACCTTGT CATCGTCATCCTTGTAGTC-3′), and Flag-C2(7365) (5′-GTCATCCT TGTAGTCGCCCACCATCCGGGAAGAAGT-3′), on the pCS2++TMEFF1 template. The PCR product was digested and inserted into EcoRI/XbaI sites of pCS2++ vector. For CriptoΔCFC-Flag, two PCR reactions were performed with the N-terminal fragment amplified with the primers Cripto-N(SpeI) (5′-GGACTAGTCACCATGGGGTACTTCTCA-3′) and N3′(BamHI) (5′-GCGGATCCGTGCTCTTTGCGAACATC-3′), and the C-terminal fragment amplified with the primers C5′(BamHI) (5′-CGG GATCCGGTCACGTGATGGACCAG-3′) and C3′(XhoI) (5′-AGGCTC GAGAGGCCTTGAATT-3′), on pCS2+Cripto-3Flag template. The PCR products were digested with SpeI/BamHI and BamHI/XhoI, respectively, and inserted into the SpeI/XhoI sites of pCS2+Cripto-3Flag vector. Sequencing of the construct confirmed that the CFC domain, containing amino acids 99-134, was eliminated. Other Cripto constructs were kindly provided by Drs. T. Keller and M. Whitman (Harvard Medical School, Boston, MA; Yeo and Whitman 2001).
RT-PCR assay
Plasmids were linearized (AscI for pCS2++-based plasmids, NotI for pCS2+-based plasmids) and RNAs were synthesized with mMessage mMachine kit (Ambion) as described (Chang et al. 1997). The RNAs encoding AXnr1 (0.2 ng), TMEFF1/HA/Flag (1 ng), Cripto/ΔEGF/ΔCFC/mEGF1/mEGF2/mCFC (0.5-1 ng), and Cerberus (1 ng) were injected into both animal poles of two-cell-stage embryos. Animal caps were then cut at blastula stages 8.5-9, either dissociated and reaggregated immediately as described (Chang et al. 1997) or left intact, and incubated to early gastrula stages (stages 10-11) before total RNA was extracted for RT-PCR assay for gene expression. The primers used in the RT-PCR experiments were as described (Chang et al. 1997).
Coimmunoprecipitation analysis
Coimmunoprecipitation with Xenopus protein embryonic extract was performed as described (Yeo and Whitman 2001), with the exception that no chemical cross-linker was used. Basically, RNAs encoding Cripto-F (2 ng), TMEFF1-HA/F (3 ng), ALK4-HA (3 ng), or AXnr1-HA (4 ng) were injected alone or in combination into two-cell-stage Xenopus embryos. The doses of RNA used for the competition experiment were as indicated in the Figure 1 legend. Protein extract was made at early gastrula stages (stage 10+) and split into two halves. One half was immunoprecipitated with an anti-Flag antibody (Sigma), and the other half was precipitated with an anti-HA antibody (Covance). The precipitated samples were separated on 10% PAGE and transferred to Immobilon P membrane (Millipore). Western blot was subsequently performed to detect coprecipitated proteins.
For cell culture assay, CHO cells maintained in Dulbecco's modified eagle medium (DMEM) with 10% fetal bovine serum were transfected at 60%-70% confluence with the plasmids encoding TMEFF1/TMEFF1ΔEGF-HA (4-7 µg), TMEFF1ΔFS-HA (18 µg), and/or Cripto/CriptoΔEGF/CriptoΔCFC-Flag (2-3µg), using FuGENE 6 reagent (Roche). Two days after transfection, the cells were lysed in the coimmunoprecipitation buffer (Yeo and Whitman 2001) in the presence of protease inhibitor cocktail (Roche), 2 µg/mL aprotinin, 2 µg/mL leupeptin, and 1 mM phenylmethylsulfonyl fluoride. One percent of cell lysate was set aside for use as whole cell lysate. The remainder was precipitated with the anti-Flag antibody (Sigma) and analyzed by Western blot as described earlier.
Acknowledgments
We thank Drs. Tracy Keller, Malcolm Whitman, Eva Reissman, and Makoto Asashima for kindly providing plasmids used in this study, and Dr. Fannie Lin for technical advice on Western blot. P.W.H. is partly supported by the MSTP program, and C.C. is supported by HHMI Institutional Career Development Award and NIH grant HD43345.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1127703.
Supplemental material is available at http://www.genesdev.org.
References
- Adamson E.D., Minchiotti, G., and Salomon, D.S. 2002. Cripto: A tumor growth factor and more. J. Cell. Physiol. 190: 267-278. [DOI] [PubMed] [Google Scholar]
- Bartholin L., Maguer-Satta, V., Hayette, S., Martel, S., Gadoux, M., Corbo, L., Magaud, J.P., and Rimokh, R. 2002. Transcription activation of FLRG and follistatin by activin A, through Smad proteins, participates in a negative feedback loop to modulate activin A function. Oncogene 21: 2227-2235. [DOI] [PubMed] [Google Scholar]
- Bianco C., Kannan, S., De Santis, M., Seno, M., Tang, C.K., Martinez-Lacaci, I., Kim, N., Wallace-Jones, B., Lippman, M.E., Ebert, A.D., et al. 1999. Cripto-1 indirectly stimulates the tyrosine phosphorylation of erb B-4 through a novel receptor. J. Biol. Chem. 274: 8624-8629. [DOI] [PubMed] [Google Scholar]
- Bianco C., Adkins, H.B., Wechselberger, C., Seno, M., Normanno, N., De Luca, A., Sun, Y., Khan, N., Kenney, N., Ebert, A., et al. 2002. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial cells. Mol. Cell. Biol. 22: 2586-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C., Strizzi, L., Rehman, A., Normanno, N., Wechselberger, C., Sun, Y., Khan, N., Hirota, M., Adkins, H., Williams, K., et al. 2003. A Nodal- and ALK4-independent signaling pathway activated by Cripto-1 through Glypican-1 and c-Src. Cancer Res. 63: 1192-1197. [PubMed] [Google Scholar]
- Brennan J., Norris, D., and Robertson, E.J. 2002. Nodal activity in the node governs left-right asymmetry. Genes & Dev. 16: 2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Wilson, P.A., Mathews, L.S., and Hemmati-Brivanlou, A. 1997. A Xenopus type I activin receptor mediates mesodermal but not neural specification during embryogenesis. Development 124: 827-837. [DOI] [PubMed] [Google Scholar]
- Chang C., Eggen, B.J.L., Weinstein, D.C., and Brivanlou, A.H. 2003. Regulation of nodal and BMP signaling by tomoregulin-1 (X7365) through novel mechanisms. Dev. Biol. 255: 1-11. [DOI] [PubMed] [Google Scholar]
- Cheng S.K., Olale, F., Bennett, J.T., Brivanlou, A.H., and Schier, A.F. 2003. EGF-CFC proteins are essential coreceptors for the TGF-β signals Vg1 and GDF1. Genes & Dev. 17: 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon F.L., Lyons, K.M., Takaesu, N., Barth, K.S., Kispert, A., Herrmann, B., and Robertson, E.J. 1994. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120: 1919-1928. [DOI] [PubMed] [Google Scholar]
- Eib D.W. and Martens, G.J.M. 1996. A novel transmembrane protein with epidermal growth factor and follistatin domains expressed in the hypothalamo-hypophysial axis of Xenopus laevis. J. Neurochem. 67: 1047-1055. [DOI] [PubMed] [Google Scholar]
- Eimon P.M. and Harland, R.M. 2002. Effects of heterodimerization and proteolytic processing on Derrier and Nodal activity: Implications for mesoderm induction in Xenopus. Development 129: 3089-3113. [DOI] [PubMed] [Google Scholar]
- Gery S., Sawyers, C.L., Agus, D.B., Said, J.W., and Koeffler, H.P. 2002. TMEFF2 is an androgen-regulated gene exhibiting antiproliferative effects in prostate cancer cells.Oncogene 21: 4739-4746. [DOI] [PubMed] [Google Scholar]
- Gery S., Yin, D., Xie, D., Black, K.L., and Koeffler, H.P. 2003. TMEFF1 and brain tumors. Oncogene 22: 2723-2727. [DOI] [PubMed] [Google Scholar]
- Glynne-Jones E., Harper, M.E., Seery, L.T., James, R., Anglin, I., Morgan, H.E., Taylor, K.M., Gee, J.M., and Nicholson, R.I. 2001. TENB2, a proteoglycan identified in prostate cancer that is associated with disease progression and androgen independence. Int. J. Cancer 94: 178-184. [DOI] [PubMed] [Google Scholar]
- Gray P.C., Harrison, C.A., and Vale, W. 2003. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proc. Natl. Acad. Sci. 100: 5193-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman K., Zhang, J., Cheng, S., Heckscher, E., Talbot, W.S., and Schier, A.F. 1999. The EGF-CFC protein One-eyed pinhead is essential for nodal signaling. Cell 97: 121-132. [DOI] [PubMed] [Google Scholar]
- Harland R.M. and Gerhart, J. 1997. Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13: 611-667. [DOI] [PubMed] [Google Scholar]
- Horie M., Mitsumoto, Y., Kyushiki, H., Kanemoto, N., Watanabe, A., Taniguchi, Y., Nishino, N., Okamoto, T., Kondo, M., Mori, T., et al. 2000. Identification and characterization of TMEFF2, a novel survival factor for hippocampal and mesencephalic neurons. Genomics 67: 146-152. [DOI] [PubMed] [Google Scholar]
- Kogawa K., Nakamura, T., Sugino, K., Takio, K., Titani, K., and Sugino, H. 1991. Activin-binding protein is present in pituitary. Endocrinology 128: 1434-1440. [DOI] [PubMed] [Google Scholar]
- Lopoez-Casillas F., Wrana, J.L., and Massague, J. 1993. Betaglycan presents ligand to the TGFβ signaling receptor. Cell 73: 1435-1444. [DOI] [PubMed] [Google Scholar]
- Lowe L.A., Yamada, S., and Kuehn, M.R. 2001. Genetic dissection of nodal function in patterning the mouse embryo. Development 128: 1831-1843. [DOI] [PubMed] [Google Scholar]
- Massague J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67: 753-791. [DOI] [PubMed] [Google Scholar]
- Massague J. and Chen, Y.-G. 2000. Controlling TGF-β signaling. Genes & Dev. 14: 627-644. [PubMed] [Google Scholar]
- Normanno N., Bianco, C., De Luca, A., and Salomon, D.S. 2001. The role of EGF-related peptides in tumor growth. Front. Biosci. 6: d685-d707. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Brennan, J., Bikoff, E.K., and Robertson, E.J. 2002. The foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129: 3455-3468. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D., Chen, Y.G., Dosch, R., Gawantka, V., Delius, H., Massague, J., and Niehrs, C. 1999. Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature 401: 480-485. [DOI] [PubMed] [Google Scholar]
- Piccolo S., Agius, E., Leyns, L., Bhattacharyya, S., Grunz, H., Bouwmeester, T., and De Robertis, E.M. 1999. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397: 707-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann E., Jornvall, H., Blokzijl, A., Andersson, O., Chang, C., Minchiotti, G., Persico, M.G., Ibanez, C.F., and Brivanlou, A.H. 2001. The orphan receptor ALK7 and the activin receptor ALK4 mediate signaling by nodal proteins during vertebrate development. Genes & Dev. 15: 2010-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma R., Ohnishi, Y.-I., Meno, C., Fujii, H., Juan, H., Takeuchi, J., Ogura, T., Li, E., Miyazono, K. and Hamada, H. 2002. Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells. 7: 401-412. [DOI] [PubMed] [Google Scholar]
- Schier A.F. and Shen, M.M. 1999. Nodal signaling in vertebrate development. Nature 403: 385-389. [DOI] [PubMed] [Google Scholar]
- Schneyer A., Tortoriello, D., Sidis, Y., Keutmann, H., Matsuzaki, T., and Holms, W. 2001. Follistatin-related protein (FSRP):A new member of the follistatin gene family. Mol. Cell. Endocrinol. 180: 33-38. [DOI] [PubMed] [Google Scholar]
- Shen M.M. and Schier, A.F. 2000. The EGF-CFC gene family in vertebrate development. Trends Genet. 16: 303-309. [DOI] [PubMed] [Google Scholar]
- Shi Y. and Massague, J. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113: 685-700. [DOI] [PubMed] [Google Scholar]
- Siegel D.A., Huang, M.K., and Becker, S.F. 2002. Ectopic dendrite initiation: CNS pathogenesis as a model of CNS development. Int. J. Dev. Neurosci. 20: 373-389. [DOI] [PubMed] [Google Scholar]
- Tsuchida K., Matsuzaki, T., Yamakawa, N., Liu, Z., and Sugino, H. 2001. Intracellular and extracellular control of activin function by novel regulatory molecules. Mol. Cell. Endocrinol. 180: 25-31. [DOI] [PubMed] [Google Scholar]
- Whitman M. 2001. Nodal signaling in early vertebrate embryos: Themes and variations. Dev. Cell 1: 605-617. [DOI] [PubMed] [Google Scholar]
- Yan Y.-T., Liu, J.-J., Luo, Y.E.C., Haltiwanger, R.S., Abate-Shen, C., and Shen, M.M. 2002. Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol. Cell. Biol. 22: 4439-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C. and Whitman, M. 2001. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell 7: 949-957. [DOI] [PubMed] [Google Scholar]