Abstract
Asymmetric dimethylarginine (ADMA) has been implicated in the progression of cardiovascular disease as an endogenous inhibitor of nitric oxide synthase. The regulation of dimethylarginine dimethylaminohydrolase (DDAH), the enzyme responsible for metabolizing ADMA, is poorly understood. The transcription factor sterol response element binding protein (SREBP) is activated by statins via a reduction of membrane cholesterol content. Because the promoters of both DDAH1 and DDAH2 isoforms contain sterol response elements, we tested the hypothesis that simvastatin regulates DDAH1 and DDAH2 transcription via SREBP. In cultured endothelial cells, simvastatin increased DDAH1 mRNA expression compared with vehicle. In an ADMA loading experiment, simvastatin treatment resulted in a decrease in ADMA content, an indication of increased DDAH activity. The knockdown of SREBP1c protein led to an increase in DDAH1 mRNA expression and activity, whereas the knockdown of SREBP2 led to a decrease in DDAH1 mRNA expression. The role of SREBP2 in the activation of the DDAH1 was supported by chromatin immunoprecipitation studies demonstrating increased binding of SREBP2 to the DDAH1 promoter upon simvastatin stimulation. These data indicate that SREBP1c might act as a repressor and SREBP2 as an activator of DDAH transcription and activity. This study describes a novel mechanism of reciprocal regulation by the SREBP family members of the DDAH-ADMA system, which represents a potential link between cellular cholesterol content and endothelial dysfunction observed in cardiovascular disease.
Keywords: asymmetric dimethylarginine, dimethylarginine dimethylaminohydrolase, endothelial nitric oxide synthase
a decrease in the bioavailability of the vasodilator nitric oxide (NO) is a hallmark of cardiovascular diseases including hypertension, atherosclerosis, and metabolic syndrome (6, 9, 44). One mechanism thought to be partially responsible for the reduction in NO and resultant endothelial dysfunction in these diseases is an increase in the levels of the endogenous inhibitor of NO synthase (NOS), asymmetric dimethylarginine (ADMA) (8, 21, 30, 33, 58). Increased plasma levels of ADMA are observed in patients with various forms of cardiovascular disease and, because of their correlation with adverse cardiovascular outcomes, have been suggested to be diagnostic biomarkers (29, 31, 42, 64).
Free ADMA is generated via protein methylation and subsequent protein hydrolysis (23) and is metabolized to citrulline and dimethylamine by dimethylarginine dimethylaminohydrolase (DDAH) (27, 28, 54). Because the metabolism of ADMA by DDAH isoforms 1 and 2 is the primary route of clearance of ADMA from the circulation (1, 37, 38, 51), changes in the levels of DDAH expression and/or activity represent a possible mechanism leading to the accumulation of ADMA and the progression of endothelial dysfunction in cardiovascular disease. Indeed, a genetic deficiency in DDAH leads to increased plasma ADMA, endothelial dysfunction, and elevated blood pressure in mice (26), whereas an overexpression of DDAH lowers plasma ADMA, improves endothelial function, and lowers blood pressure (10, 11, 17, 19, 47, 49). While a growing body of literature supports a role for DDAH in regulating the levels of ADMA and, consequently, endothelial function, the mechanisms of transcriptional regulation of DDAH expression are not completely understood.
An analysis of the regulatory sequences of the DDAH genes identifies the response elements for nuclear hormone receptors including retinoid X receptor-α, peroxisome proliferator-activated receptor-γ, and farnesoid X receptor (FXR). A stimulation of endothelial cells with retinoic acid, pioglitazone, and the FXR agonist GW-4064 has been reported to increase DDAH promoter activity (2) and expression (18, 60), with differential regulation of DDAH1 and DDAH2 promoters by both FXR and retinoid X receptor-α agonists. We observed that the stimulation of human umbilical vein endothelial cells (HUVECs) with simvastatin also leads to an increased expression of DDAH isoforms. One of the established mechanisms of simvastatin action is the activation of sterol response element binding proteins (SREBPs), and we found that the promoter regions of both DDAH1 and DDAH2 contain recognition sites for SREBP binding, with DDAH1 containing a higher number of SREBP response elements (SREs) than DDAH2. Since SREBP transcription factors have been shown to be disregulated both in endothelial dysfunction and metabolic disorders, we became interested in investigating their involvement in the regulation of DDAH expression. The purpose of the present study was to test the hypothesis that SREBP1c and SREBP2, the major isoforms of SREBP expressed in endothelial cells, mediate the effects of simvastatin on the expression of DDAH1 and DDAH2.
MATERIALS AND METHODS
Cell culture.
HUVECs were purchased from Lonza (Walkersville, MD) and cultured in EGM-2 media (Lonza) on Collagen I BioCoat cellware (BD Biosciences, Rockville, MD). EGM-2 media contains 0.3 mmol/l of l-arginine. Cells of passages 3–6 were used for all experiments. For statin studies, HUVECs were seeded at 1.5 × 105 cells per well of a gelatin-coated 24-well plate, allowed to recover in EGM-2 overnight, and then treated with activated simvastatin (1 μmol/l) or vehicle for 24 h. Short hairpin RNA (shRNA) lentivirus for SREBP1c and SREBP2 or nonsilencing control (NSC)-enhanced green fluorescent protein (EGFP) lentivirus, with the shRNA constructs by The RNAi Consortium (Open Biosystems, Huntsville, AL), were used for gene-silencing experiments. To determine the optimal treatment conditions, cells were transduced with increasing multiplicity of infection (MOI) of NSC-EGFP lentivirus in serum-free media overnight and then grown in EGM-2 for 48 h. At 5 MOI, 85–95% of cells were EGFP positive, and this MOI was used for all experiments.
Cellular DDAH activity.
To determine the level of DDAH activity, ADMA concentration was quantified using high-performance liquid chromatography (HPLC) following the exogenous addition of ADMA to cells. After treatment with either statin or lentivirus, the cells were rinsed with PBS and incubated with ADMA (100 μmol/l) for 2 h, and the residual ADMA was then removed by PBS washes. The cells were scraped in PBS, sonicated on ice, and centrifuged to pellet cellular debris. Crude lysates were treated with NG-monomethyl-l-arginine (l-NMMA; Sigma) as an internal standard and subjected to solid-phase extraction and HPLC according to the methods of Teerlink et al. (52). Data are reported as the peak area ratio of ADMA to l-NMMA, normalized to the protein concentration of each sample. The protein concentration of the crude lysates was determined using the bicinchoninic acid method (Pierce, Rockford, IL). The measurement of cellular total nitrate/nitrite (NOx) was performed using a colorimetric Griess reagent kit (Cayman Chemical, Ann Arbor, MI).
Western blot analysis.
After lentiviral or simvastatin stimulus treatment, the cells were lysed in radioimmunoprecipitation assay buffer containing 25 mmol/l Tris·HCl (pH 7.6), 150 mmol/l NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS, supplemented with Halt protease inhibitor cocktail (Pierce). Samples (5–15 μg protein) were subjected to SDS-PAGE, followed by a transfer to polyvinylidene difluoride membrane (Bio-Rad, Cambridge, MA). The membranes were probed with polyclonal antibodies against SREBP1c or SREBP2 (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:1,500, and goat polyclonal antibody against DDAH1 (Calbiochem, EMD Chemicals, Gibbstown, NJ) or DDAH2 (Abcam, Cambridge, MA) at a dilution of 1:500, followed by appropriate horseradish peroxidase-conjugated secondary antibodies (Pierce) at a dilution of 1:2,000, all in Tris-buffered saline (TBS) with 3% milk and 0.2% Tween-20. After an incubation with Pico Sensitivity signal reagent (Pierce), the bands were detected using Kodak film and quantified using Bio-Rad Quantity One software. The stripped membranes were reprobed with β-actin, GP3DH, or β-tubulin antibodies (1:3,000; Santa Cruz Biotechnology) for signal normalization.
Quantitative real-time PCR.
After transduction and/or stimulation, the cells were lysed and the RNA was isolated according to Qiagen RNeasy protocol (Qiagen, Valencia, CA). cDNA was synthesized from 1 μg of total RNA using random hexamers and MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA). Quantitative real-time PCR was carried out using a HT7000 cycler (Applied Biosystems). Primer/probe sets for DDAH1, DDAH2, protein arginine N-methyltransferase 1 (PRMT1), PRMT2, endothelial NOS (eNOS), SREBP1c, SREBP2, and hydroxymethylglutaryl-CoA reductase were purchased from Applied Biosystems, and samples were run in multiplex reactions with primer-limited GAPDH (See Table 1). For each sample, the level of expression of these transcripts was determined relative to the expression of GAPDH.
Table 1.
Primer sets and chromatin immunoprecipitation primer sequences
| Gene | Primer Set Catalog no. (Applied Biosystems) |
|---|---|
| qRT-PCR | |
| DDAH1 | Hs00201707_m1 |
| DDAH2 | Hs00203889_m1 |
| NOS3 (eNOS) | Hs00167166_m1 |
| SREBP1 | Hs00231674_m1 |
| SREBP2 | Hs00190237_m1 |
| Gene | Primer Sequence (5′ to 3′) | |
|---|---|---|
| ChIP | ||
| DDAH1 | Fwd | ACATGGTGAAACCCTGTCTCTGCT |
| Rvs | TGGCACGATCTCGGCTTACTTCAA | |
| DDAH2 | Fwd | TGGAGTGTGTCCATTGGGTAGCAA |
| Rvs | GCAAGTGGCAAAGGCCCTCAAATA | |
| ACC-1 | Fwd | TGCACGCCTGTCAGCCATC |
| Rvs | CCTCACCTCACCTCAGGGT | |
| LDLR | Fwd | TGGGAATCAGAGCTTCACGGGTTA |
| Rvs | GGGCCCACGTCATTTACAGCATTT |
qRT-PCR, quantitative RT-PCR; ChIP, chromatin immunoprecipitation; DDAH, dimethylarginine dimethylaminohydrolase; eNOS, endothelial nitric oxide synthase; SREBP, sterol response element binding protein; ACC-1, acetyl-CoA carboxylase 1; LDLR, LDL receptor; Fwd, foward; Rvs, reverse.
Chromatin immunoprecipitation.
HUVECs were grown in 100-mm plates, in triplicate for each condition, and treated as described. After being cross-linked with 1% formaldehyde for 10 min at 37°C, the samples were dissolved in chromatin immunoprecipitation lysis buffer (Upstate, Charlottesville, VA), and chromatin immunoprecipitation was performed using a commercially available kit as per the manufacturer's instructions (Upstate) with the following modifications. DNA was sheared using sonification (13 pulses of 15 s on/30 s off each at 50% power on ice) to the length of ∼300–600 base pairs, verified by agarose gel electrophoresis. Samples were immunoprecipitated with SREBP1c (C-20), SREBP2 (H-164), polymerase II (PolII; H-224), or rabbit IgG antibodies (Santa Cruz Biotechnology). Quantitative RT-PCR (qRT-PCR) was performed with phenol-chloroform extracted/ethanol-precipitated DNA using SYBR green chemistry. The results were normalized to PCR with DNA from nonspecific IgG pull-down reactions. Specificity was determined using primers that amplified regions of DNA distal to the regions of interest. Sequences of primers to the hydroxymethylglutaryl-CoA reductase, LDL receptor (LDLR), and DDAH1 and DDAH2 promoters used are shown in Table 1. The levels of genes were measured in parallel for specific antibody and PolII-precipitated samples to determine whether binding occurred concomitantly with the recruitment of the transcriptosome.
Data analysis.
Data are presented as means ± SE of triplicate replicates performed on a minimum of three independent occasions. Statistical analyses were performed by ANOVA for multiple comparisons and unpaired two-tailed Student's t-test for two-group comparisons. The value for P < 0.05 was considered significant.
RESULTS
Simvastatin treatment increases DDAH expression and activity.
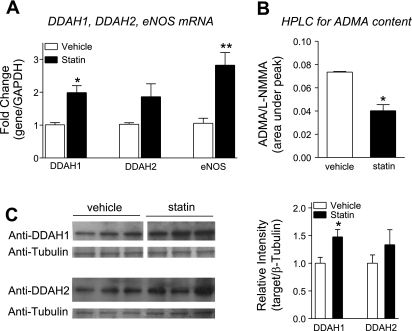
Activated simvastatin in the concentration range of 100 nm to 3 μM increased DDAH expression in a dose-dependent manner. The treatment of HUVECs for 24 h with physiologically relevant (1 μmol/l) concentration of activated simvastatin resulted in 2.0- and 1.9-fold increase in mRNA expression of DDAH1 and DDAH2, respectively, and a 2.8-fold increased expression of eNOS (Fig. 1A). The increase in DDAH expression was associated with an increase in DDAH activity, evidenced by a 49% reduction in ADMA content (Fig. 1B). Increased protein levels of DDAH1 (1.5-fold) and DDAH2 (1.3-fold) were observed upon statin treatment (Fig. 1C).
Fig. 1.
Simvastatin increases dimethylarginine dimethylaminohydrolase 1 (DDAH1) and DDAH2 expression, protein levels, and DDAH activity. Human umbilical vein endothelial cells (HUVECs) were treated with simvastatin (1 μM, 24 h). A: quantitative RT-PCR (qRT-PCR) for expression of DDAH1, DDAH2, and endothelial nitric oxide synthase (eNOS). B: HPLC for residual asymmetric dimethylarginine (ADMA) content in simvastatin-treated HUVECs incubated with ADMA (200 μM, 2 h post-statin treatment). l-NMMA, NG-monomethyl-l-arginine. C: DDAH1 and DDAH2 protein levels in response to simvastatin treatment, Western blot analysis (WB), and quantification. *P < 0.05 and **P < 0.01. vs. vehicle.
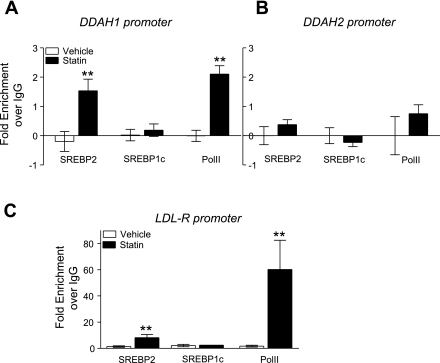
Simvastatin stimulates recruitment of SREBP2 but not SREBP1c to the DDAH1 promoter.
The prediction of transcription factor response elements (BioBase; Transfac) in the DDAH1 promoter identified four SREBP binding sites (SREs) on the plus strand: three upstream of the transcription start site (TSS) and one directly downstream. An analysis of the DDAH2 promoter identified one SRE on the minus strand upstream of the TSS. The functional significance of these putative sites and the impact of the difference in the number of SREs on expression was probed by chromatin immunoprecipitation. Figure 2A shows that treatment with 1 μM simvastatin, relative to vehicle control, increased SREBP2 binding to the DDAH1 promoter 1.5-fold over the control baseline zero (data normalized to PCR with primers to distal regions of DNA and samples are adjusted for individual IgG pull-downs), whereas SREBP1c binding was unchanged. At the same time, statin treatment increased the binding of PolII to the DDAH1 promoter 2.0-fold, suggesting that there is transcriptosome recruitment and that the promoter is active at the time of SREBP2 binding. PCR analysis of the DDAH2 promoter SRE binding sites did not show significant SREBP2 or PolII recruitment after statin treatment (Fig. 2B). To verify the results obtained with the SREBP antibodies, we looked at the SRE sites in the promoter of SREBP target genes, LDLR for SREBP2, and acetyl-CoA carboxylase 1 (ACC-1) for SREBP1c (3). Statin treatment increases binding of SREBP2 to the SRE in the LDLR promoter, indicating antibody specificity (Fig. 2C). The specificity of the SREBP1c antibody was confirmed by PCR of the SRE sites proximal to the TSS of the ACC-1 promoter. Simvastatin increased the association of SREBP1c with the SRE binding site proximal to the TSS of the ACC-1 gene (data not shown).
Fig. 2.
Simvastatin treatment increases sterol response element binding protein 2 (SREBP2), but not SREBP1c recruitment, to the DDAH1 promoter SREBP binding sites. HUVECs were treated with simvastatin (1 μM) or vehicle. After 24 h, cells were cross-linked with formaldehyde and chromatin immunoprecipitation (ChIP) analysis was performed. ChIP with SREBP2, SREBP1c, and polymerase II (PolII) antibodies were normalized to IgG pull-down. A: qRT-PCR with primers amplifying the region of 2 SREBP response elements in the DDAH1 promoter. B: qRT-PCR with primers amplifying the region of the SREBP response element proximal to the transcription start site in the DDAH2 promoter. C: qRT-PCR with primers amplifying the region of proximal SREBP response element in the LDL receptor (LDLR) promoter. **P < 0.01. vs. vehicle.
Knockdown of SREBP1c increases and knockdown of SREBP2 decreases DDAH1 expression.
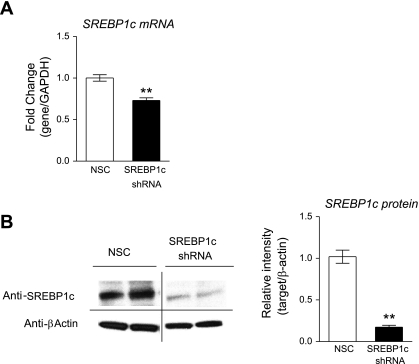
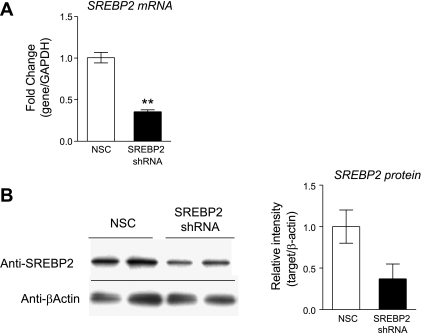
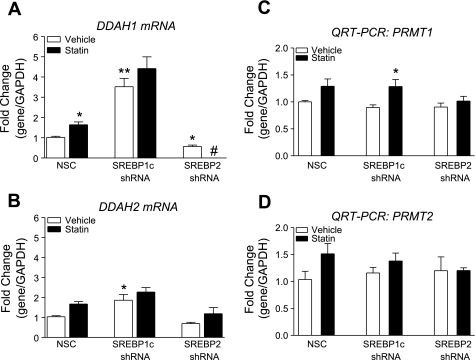
Both SREBP1c and SREBP2 are detectable by qRT-PCR in HUVECs, with SREBP2 message being 2.0- to 4.0-fold more abundant then SREBP1c message (data not shown) and GeneLogic database (Gaithersburg, MD). HUVECs were transduced with lentiviral shRNA targeting either SREBP1c or SREBP2. We screened five shRNA constructs for SREBP1c and eight shRNA constructs for SREBP2 and identified two constructs that led to the greatest decrease in SREBP1c and SREBP2 protein levels, respectively. Since shRNA has effects on both transcription and translation, the decrease in mRNA and protein levels do not always correlate. A treatment with shRNA targeting SREBP1c caused a 30% decrease in SREBP1c mRNA (Fig. 3A) and a 80% decrease in protein relative to NSC-transduced cells (Fig. 3B). A treatment with shRNA targeting SREBP2 caused a 65% decrease in SREBP2 mRNA (Fig. 4A) and a 63% decrease in protein relative to NSC-transduced cells (Fig. 4B). SREBP1c knockdown was associated with a 3.5-fold increase in DDAH1 mRNA, and simvastatin treatment in SREBP1c knockdown cells did not result in a significant increase in DDAH1 expression. SREBP2 knockdown was associated with a 44% reduction in DDAH1 mRNA, and simvastatin treatment in SREBP2 knockdown completely removed DDAH1 message, resulting in “undetermined” threshold cycle readings (Fig. 5A). qRT-PCR with DDAH2 primers revealed a somewhat similar pattern: SREBP1c knockdown resulted in a 1.9-fold increase in DDAH2 mRNA, and SREBP2 knockdown resulted in a 31% decrease in DDAH2 mRNA. Simvastatin treatment after SREBP1c led to a 2.2-fold increased DDAH2 levels, not significantly different from the SREBP1c knockdown alone. However, after SREBP2 silencing, simvastatin treatment resulted in a 1.2-fold increase in DDAH2 levels (not significantly different from the knockdown alone) (Fig. 5B). PRMT1 or PRMT2 levels were not significantly changed (Fig. 5, C and D).
Fig. 3.
Lentiviral shRNA knockdown of SREBP1c in HUVECs. Lentiviral transductions with SREBP1c shRNA or nonsilencing control (NSC) lentivirus. Forty-eight hours after transduction, qRT-PCR or WB were performed as described. A: qRT-PCR for SREBP1c. B: SREBP1c protein levels, WB, and quantification. **P < 0.01 vs. NSC.
Fig. 4.
Lentiviral short hairpin RNA (shRNA) knockdown of SREBP2 in HUVECs. Lentiviral transductions with SREBP2 shRNA or NSC lentivirus. Forty-eight hours after transduction, qRT-PCR or WB were performed. A: qRT-PCR for SREBP2. B: SREBP2 protein levels, WB, and quantification. **P < 0.01 vs. NSC.
Fig. 5.
Lentiviral shRNA knockdown of SREBP1c leads to increased expression of DDAH1, and lentiviral knockdown of SREBP2 leads to decreased expression of DDAH1. Effect of simvastatin on DDAH1 and protein arginine N-methyltransferase-1 (PRMT1) and -2 expression in the knockdowns are shown. Lentiviral transductions with SREBP1c shRNA, SREBP2 shRNA, or nonsilencing control lentivirus and treatment with 300 nM simvastatin for 24 h are shown. A: qRT-PCR for DDAH1. B: qRT-PCR for DDAH2. C: qRT-PCR for PRMT1. D: qRT-PCR for PRMT2. *P < 0.05 and **P < 0.01 vs. vehicle in NSC. #5/6 samples had no detectable DDAH1 mRNA.
Lentiviral shRNA-mediated knockdown of SREBP1c results in increased DDAH activity and increased nitrite content.
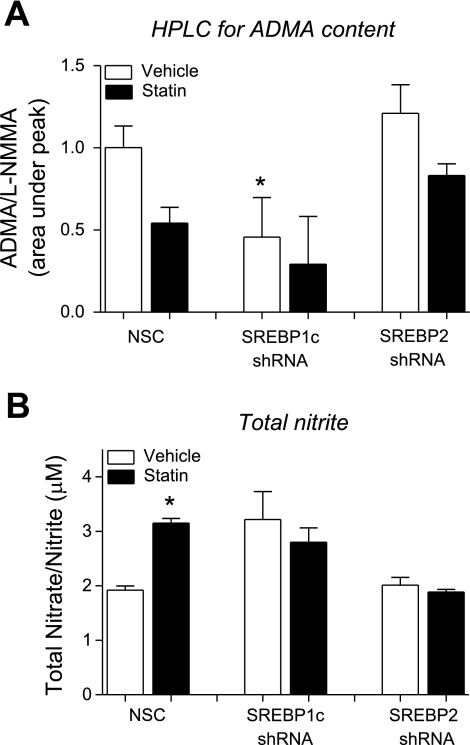
To determine whether changes in DDAH1 message translated to functional changes in DDAH cellular activity, ADMA levels were measured using HPLC. This measure provides an estimate of DDAH activity. SREBP1c knockdown alone was associated with a 54% reduction in cellular ADMA compared with NSC-transduced cells, suggesting an increased DDAH activity. SREBP1c knockdown cells treated with simvastatin had an additional decrease in ADMA content (70%) relative to NSC. SREBP2 knockdown alone did not lead to significant changes in DDAH activity. In the SREBP2 knockdown cells treated with simvastatin, there was a trend toward a decreased ADMA content, but it did not reach statistical significance (Fig. 6A). The levels of nitrites are an indication of the activity of the NOS isoforms. SREBP1c knockdown was associated with a 1.67-fold increase in total NOx levels compared with NSC, similar to the degree of increase observed after simvastatin treatment in NSC samples. SREBP1c knockdown followed by simvastatin treatment did not lead to a further increase in the NOx levels. SREBP2 knockdown was not associated with significant changes in NOx levels. Interestingly, SREBP2 knockdown abrogated the simvastatin-induced increase in NOx (Fig. 6B).
Fig. 6.
Lentiviral shRNA-mediated knockdown of SREBP1c in HUVECs results in decreased cellular ADMA content and increased nitrite content. HUVECs transduced with lentiviral shRNA targeting SREBP1c or SREBP2 and treated with simvastatin (300 nM, 24 h) were assayed for changes in the ADMA metabolism by HPLC and for nitric oxide synthase (NOS) activity by total nitrite quantification with Griess reagent. A: HPLC of ADMA, estimated DDAH activity. B: total nitrite content, an estimate of NOS activity. *P < 0.05. vs. NSC.
DISCUSSION
Recent studies have underscored the importance of the DDAH-ADMA system in regulating the bioavailability of NO and vascular endothelial function. Increased activity of DDAH is correlated with lower ADMA levels in mouse models of DDAH overexpression (10, 19). Elevated levels of plasma ADMA are detected in a number of pathologies, including hypercholesterolemia, type II diabetes, hypertension, chronic heart failure, and renal disease and, as such, have been heralded as an independent cardiovascular risk factor (4, 24, 29, 32).
Intracellular and plasma ADMA levels are a sum of its production in the course of normal protein turnover by PRMTs and its disposal either by renal clearance or conversion to citrulline and methylamines by the DDAH (56). However, it is unclear whether the higher plasma levels in human disease states correlate with higher intracellular levels and are, as such, a “spillover.” Studies examining statin effects in vivo have reported endothelial protection without overtly affecting plasma ADMA levels (12, 20, 39), although tissue levels of ADMA were not examined (41, 45, 59, 62). Statins are known to have antiatherosclerotic and antiapoptotic effects (7, 13, 15, 43, 48, 53, 57) and improve endothelial dysfunction by multiple mechanisms, including the increases in NO-mediated vasorelaxation, decreases in levels of circulating proinflammatory cytokines, and decreases in oxidative stress, among others (14, 34, 50). The data from the current study demonstrate that, in addition, statins may increase intracellular DDAH activity and ADMA metabolism, suggesting yet another mechanism contributing to their overall beneficial effects on endothelial dysfunction.
In the current study, the metabolism of exogenously applied ADMA was used as an index of DDAH activity. With SREBP1c knockdown, higher levels of DDAH message and protein were observed, consistent with the removal of a repressor influence on the DDAH1 promoter. The moderate increase in DDAH levels with SREBP1c knockdown was paralleled by an increase in its activity, estimated both by a reduction in ADMA content, following cellular loading of ADMA, and an increase in cellular NOx, an index of NOS activity (36). With SREBP2 knockdown, DDAH1 levels decreased, and it was expected that ADMA metabolism would also decrease. However, after the transduction with SREBP2 shRNA and ADMA loading, the cellular ADMA content was not increased as expected. Because the cells were loaded with 100 μmol/l ADMA, this excess may have masked any potential decrease in DDAH activity. Furthermore, NOx levels did not change with SREBP2 knockdown, which could be due to limitations in the sensitivity of the Griess reagent-based detection. We demonstrated that simvastatin treatment of HUVECs results in a lowered ADMA content and increased NOx levels and followed up on these experiments by investigating the effects of SREBP silencing in this context. SREBP1c silencing did not significantly alter the effects of simvastatin on ADMA content and NOx, perhaps indicating we have reached an upper limit of DDAH activity. SREBP2 silencing appeared to block simvastatin-induced increased in NOx levels. This supports the basic conclusion that SREBP2 mediates the positive effects of simvastatin on the ADMA-DDAH-NOS axis.
Levels of ADMA have been shown to be induced by oxidized LDL via changes in the expression of type-1 PRMTs (5, 63). Therefore, we also considered the possibility that the observed simvastatin effect on ADMA may be partially due to decreases in the expression of PRMT1 and PRMT2. Neither the simvastatin treatment nor the modulation of SREBP levels had significant effects on the expression of PRMTs. Still, a possibility remains that the activity of PRMTs is affected by simvastatin treatment, and the effects of simvastatin in the context of oxidized LDL-induced increases in ADMA warrant further investigation.
Our data suggest that SREBP1c and SREBP2 have reciprocal activity at the DDAH1 promoter. At baseline, SREBP1c is a transcriptional repressor, whereas SREBP2 is a transcriptional activator. Upon changes in the sterol levels, the regulation of the DDAH1 promoter becomes more complex as statins activate both SREBP1c and SREBP2. Because the net effect of statin treatment is increased DDAH1 expression, we predict that in endothelial cells, statins may activate SREBP2 to a greater extent than SREBP1c. Since a decrease in SREBP1c leads to a disinhibition of the DDAH1 promoter, we expected that simvastatin, by stimulating only SREBP2, would further increase DDAH1 expression. However, after SREBP1c knockdown, simvastatin treatment did not result in additional increases in the levels of DDAH1. The lack of an increase suggests that with SREBP1c knockdown alone, optimal activity of the promoter has been achieved. As it is commonly seen with eNOS, where most of the physiological and pathophysiological stimuli only lead to a 2.0- to 3.0-fold induction of the message (46), an upper limit of DDAH expression may have been reached. After SREBP2 knockdown, we expected simvastatin treatment to lead to a further decrease in the DDAH1 expression since only SREBP1c would be stimulated. In fact, the disappearance of the DDAH1 message suggests that this is indeed the case.
The three observations that 1) statins induced DDAH2 to a lesser extent then DDAH1, 2) SREBP1c knockdown resulted in a greater induction of DDAH1 compared with DDAH2, and 3) SREBP2 bound to the promoter of DDAH1 but not DDAH2 suggest that SREBPs probably play a stronger role in regulating DDAH1 expression than DDAH2. A condition-dependent differential regulation of the two DDAH isoforms has been shown by others (16, 40, 55, 61). The preferential regulation of DDAH1 versus DDAH2 by SREBPs may be due to the greater number of SREBP binding sites in the DDAH1 promoter. Other transcription factors (heat shock factor 1; ELK-1) may play a role in the regulation of DDAH2 promoter by statin, and in the analysis of DDAH2 promoter, a number of candidates have been identified (22). A stabilization of the DDAH2 mRNA after statin treatment, similar to what happens to eNOS (25, 35), may also account for the changes we observed. The close proximity of the SREBP binding sites in the DDAH1 promoter makes it difficult to determine the precise response element to which the transcription factor binds, and promoter deletion studies would be required to resolve the binding sites. Further studies are also required to identify other components of the SREBP complex, such as coactivators or corepressors associated with the DDAH promoter. Such studies may provide clues to effective and specific manipulation of DDAH expression.
The apparent reciprocal regulation of DDAH1 expression by SREBP1c and SREBP2 suggests a potential sensitivity of the DDAH-ADMA system to the metabolic state of the organism. Because SREBP1c and SREBP2 are involved in, and respond to, changes in lipogenesis and cholesterol synthesis, the results from the current study suggest that DDAH may be yet another link between altered lipid and cholesterol metabolism, as might occur in metabolic disease and vascular endothelial dysfunction. If a disruption of the DDAH-ADMA system in the course of metabolic diseases is in part modulated by SREBPs, then treatments that activate or inhibit these transcription factors may exert additional effects on vascular contractility. An increased understanding of the mechanism by which metabolic disregulation leads to the development of cardiovascular pathologies may lead to the development of better strategies for therapeutic intervention.
GRANTS
This study was supported by GlaxoSmithKline.
DISCLOSURES
None.
ACKNOWLEDGMENTS
Current address of D. G. Johns: Merck Research Laboratories, Atherosclerosis, RY80T-A185, 126 E. Lincoln Ave., PO Box 2000, Rahway, NJ 07065.
REFERENCES
- 1.Achan V, Broadhead M, Malaki M, Whitley G, Leiper J, MacAllister R, Vallance P. Asymmetric dimethylarginine causes hypertension and cardiac dysfunction in humans and is actively metabolized by dimethylarginine dimethylaminohydrolase. Arterioscler Thromb Vasc Biol 23: 1455–1459, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Achan V, Tran CT, Arrigoni F, Whitley GS, Leiper JM, Vallance P. all-trans-Retinoic acid increases nitric oxide synthesis by endothelial cells: a role for the induction of dimethylarginine dimethylaminohydrolase. Circ Res 90: 764–769, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Bennett MK, Seo YK, Datta S, Shin DJ, Osborne TF. Selective binding of SREBP isoforms and co-regulatory proteins to promoters for lipid metabolic genes in liver. J Biol Chem 283: 15628–15637, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boger RH. Asymmetric dimethylarginine (ADMA) and cardiovascular disease: insights from prospective clinical trials. Vasc Med 10, Suppl 1: S19–S25, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Boger RH, Sydow K, Borlak J, Thum T, Lenzen H, Schubert B, Tsikas D, Bode-Boger SM. LDL cholesterol upregulates synthesis of asymmetrical dimethylarginine in human endothelial cells: involvement of S-adenosylmethionine-dependent methyltransferases. Circ Res 87: 99–105, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Bonetti PO, Lerman LO, Lerman A. Endothelial dysfunction: a marker of atherosclerotic risk. Arterioscler Thromb Vasc Biol 23: 168–175, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Bruegel M, Teupser D, Haffner I, Mueller M, Thiery J. Statins reduce macrophage inflammatory protein-1alpha expression in human activated monocytes. Clin Exp Pharmacol Physiol 33: 1144–1149, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Cooke JP. Does ADMA cause endothelial dysfunction? Arterioscler Thromb Vasc Biol 20: 2032–2037, 2000 [DOI] [PubMed] [Google Scholar]
- 9.Cooke JP. NO and angiogenesis. Atheroscler Suppl 4: 53–60, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Dayoub H, Achan V, Adimoolam S, Jacobi J, Stuehlinger MC, Wang BY, Tsao PS, Kimoto M, Vallance P, Patterson AJ, Cooke JP. Dimethylarginine dimethylaminohydrolase regulates nitric oxide synthesis: genetic and physiological evidence. Circulation 108: 3042–3047, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Dayoub H, Rodionov R, Lynch C, Cooke JP, Arning E, Bottiglieri T, Lentz SR, Faraci FM. Overexpression of dimethylarginine dimethylaminohydrolase inhibits asymmetric dimethylarginine-induced endothelial dysfunction in the cerebral circulation. Stroke 39: 180–184, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Eid HM, Eritsland J, Larsen J, Arnesen H, Seljeflot I. Increased levels of asymmetric dimethylarginine in populations at risk for atherosclerotic disease. Effects of pravastatin. Atherosclerosis 166: 279–284, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Enomoto S, Sata M, Fukuda D, Nakamura K, Nagai R. Rosuvastatin prevents endothelial cell death and reduces atherosclerotic lesion formation in ApoE-deficient mice. Biomed Pharmacother 63: 19–26, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Guven GS, Atalar E, Yavuz B, Beyazit Y, Kekilli M, Kilicarslan A, Sahiner L, Oz G, Ozer N, Aksoyek S, Haznedaroglu IC, Sozen T. Simvastatin treatment improves endothelial function and increases fibrinolysis in patients with hypercholestrolemia. J Natl Med Assoc 98: 627–630, 2006 [PMC free article] [PubMed] [Google Scholar]
- 15.Hartung D, Sarai M, Petrov A, Kolodgie F, Narula N, Verjans J, Virmani R, Reutelingsperger C, Hofstra L, Narula J. Resolution of apoptosis in atherosclerotic plaque by dietary modification and statin therapy. J Nucl Med 46: 2051–2056, 2005 [PubMed] [Google Scholar]
- 16.Hasegawa K, Wakino S, Tanaka T, Kimoto M, Tatematsu S, Kanda T, Yoshioka K, Homma K, Sugano N, Kurabayashi M, Saruta T, Hayashi K. Dimethylarginine dimethylaminohydrolase 2 increases vascular endothelial growth factor expression through Sp1 transcription factor in endothelial cells. Arterioscler Thromb Vasc Biol 26: 1488–1494, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa K, Wakino S, Tatematsu S, Yoshioka K, Homma K, Sugano N, Kimoto M, Hayashi K, Itoh H. Role of asymmetric dimethylarginine in vascular injury in transgenic mice overexpressing dimethylarginie dimethylaminohydrolase 2. Circ Res 101: e2–e10, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Hu T, Chouinard M, Cox AL, Sipes P, Marcelo M, Ficorilli J, Li S, Gao H, Ryan TP, Michael MD, Michael LF. Farnesoid X receptor agonist reduces serum asymmetric dimethylarginine levels through hepatic dimethylarginine dimethylaminohydrolase-1 gene regulation. J Biol Chem 281: 39831–39838, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Jacobi J, Sydow K, von Degenfeld G, Zhang Y, Dayoub H, Wang B, Patterson AJ, Kimoto M, Blau HM, Cooke JP. Overexpression of dimethylarginine dimethylaminohydrolase reduces tissue asymmetric dimethylarginine levels and enhances angiogenesis. Circulation 111: 1431–1438, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Jang JJ, Ho HK, Kwan HH, Fajardo LF, Cooke JP. Angiogenesis is impaired by hypercholesterolemia: role of asymmetric dimethylarginine. Circulation 102: 1414–1419, 2000 [DOI] [PubMed] [Google Scholar]
- 21.Jin JS, D'Alecy LG. Central and peripheral effects of asymmetric dimethylarginine, an endogenous nitric oxide synthetase inhibitor. J Cardiovasc Pharmacol 28: 439–446, 1996 [DOI] [PubMed] [Google Scholar]
- 22.Jones LC, Tran CT, Leiper JM, Hingorani AD, Vallance P. Common genetic variation in a basal promoter element alters DDAH2 expression in endothelial cells. Biochem Biophys Res Commun 310: 836–843, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Kakimoto Y, Akazawa S. Isolation and identification of N-G,N-G- and N-G,N′-G-dimethyl-arginine, N-epsilon-mono-, di-, and trimethyllysine, and glucosylgalactosyl- and galactosyl-delta-hydroxylysine from human urine. J Biol Chem 245: 5751–5758, 1970 [PubMed] [Google Scholar]
- 24.Kielstein JT, Zoccali C. Asymmetric dimethylarginine: a novel marker of risk and a potential target for therapy in chronic kidney disease. Curr Opin Nephrol Hypertens 17: 609–615, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Kosmidou I, Moore JP, Weber M, Searles CD. Statin treatment and 3′ polyadenylation of eNOS mRNA. Arterioscler Thromb Vasc Biol 27: 2642–2649, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Leiper J, Nandi M, Torondel B, Murray-Rust J, Malaki M, O'Hara B, Rossiter S, Anthony S, Madhani M, Selwood D, Smith C, Wojciak-Stothard B, Rudiger A, Stidwill R, McDonald NQ, Vallance P. Disruption of methylarginine metabolism impairs vascular homeostasis. Nat Med 13: 198–203, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Leiper J, Vallance P. Biological significance of endogenous methylarginines that inhibit nitric oxide synthases. Cardiovasc Res 43: 542–548, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Leiper JM, Santa Maria J, Chubb A, MacAllister RJ, Charles IG, Whitley GS, Vallance P. Identification of two human dimethylarginine dimethylaminohydrolases with distinct tissue distributions and homology with microbial arginine deiminases. Biochem J 343: 209–214, 1999 [PMC free article] [PubMed] [Google Scholar]
- 29.Leong T, Zylberstein D, Graham I, Lissner L, Ward D, Fogarty J, Bengtsson C, Bjorkelund C, Thelle D. Asymmetric dimethylarginine independently predicts fatal and nonfatal myocardial infarction and stroke in women: 24-year follow-up of the population study of women in Gothenburg. Arterioscler Thromb Vasc Biol 28: 961–967, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Lin KY, Ito A, Asagami T, Tsao PS, Adimoolam S, Kimoto M, Tsuji H, Reaven GM, Cooke JP. Impaired nitric oxide synthase pathway in diabetes mellitus: role of asymmetric dimethylarginine and dimethylarginine dimethylaminohydrolase. Circulation 106: 987–992, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Maas R, Dentz L, Schwedhelm E, Thoms W, Kuss O, Hiltmeyer N, Haddad M, Kloss T, Standl T, Boger RH. Elevated plasma concentrations of the endogenous nitric oxide synthase inhibitor asymmetric dimethylarginine predict adverse events in patients undergoing noncardiac surgery. Crit Care Med 35: 1876–1881, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Maas R, Schulze F, Baumert J, Lowel H, Hamraz K, Schwedhelm E, Koenig W, Boger RH. Asymmetric dimethylarginine, smoking, and risk of coronary heart disease in apparently healthy men: prospective analysis from the population-based Monitoring of Trends and Determinants in Cardiovascular Disease/Kooperative Gesundheitsforschung in der Region Augsburg study and experimental data. Clin Chem 53: 693–701, 2007 [DOI] [PubMed] [Google Scholar]
- 33.MacAllister RJ, Parry H, Kimoto M, Ogawa T, Russell RJ, Hodson H, Whitley GS, Vallance P. Regulation of nitric oxide synthesis by dimethylarginine dimethylaminohydrolase. Br J Pharmacol 119: 1533–1540, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marketou ME, Zacharis EA, Nikitovic D, Ganotakis ES, Parthenakis FI, Maliaraki N, Vardas PE. Early effects of simvastatin versus atorvastatin on oxidative stress and proinflammatory cytokines in hyperlipidemic subjects. Angiology 57: 211–218, 2006 [DOI] [PubMed] [Google Scholar]
- 35.McQuillan LP, Leung GK, Marsden PA, Kostyk SK, Kourembanas S. Hypoxia inhibits expression of eNOS via transcriptional and posttranscriptional mechanisms. Am J Physiol Heart Circ Physiol 267: H1921–H1927, 1994 [DOI] [PubMed] [Google Scholar]
- 36.Moncada S, Palmer RM, Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev 43: 109–142, 1991 [PubMed] [Google Scholar]
- 37.Nijveldt RJ, Teerlink T, Siroen MP, van Lambalgen AA, Rauwerda JA, van Leeuwen PA. The liver is an important organ in the metabolism of asymmetrical dimethylarginine (ADMA). Clin Nutr 22: 17–22, 2003 [DOI] [PubMed] [Google Scholar]
- 38.Nijveldt RJ, Teerlink T, van Guldener C, Prins HA, van Lambalgen AA, Stehouwer CD, Rauwerda JA, van Leeuwen PA. Handling of asymmetrical dimethylarginine and symmetrical dimethylarginine by the rat kidney under basal conditions and during endotoxaemia. Nephrol Dial Transplant 18: 2542–2550, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Paiva H, Laakso J, Lehtimaki T, Isomustajarvi M, Ruokonen I, Laaksonen R. Effect of high-dose statin treatment on plasma concentrations of endogenous nitric oxide synthase inhibitors. J Cardiovasc Pharmacol 41: 219–222, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. Am J Physiol Heart Circ Physiol 293: H3227–H3245, 2007 [DOI] [PubMed] [Google Scholar]
- 41.Panichi V, Mantuano E, Paoletti S, Santi S, Manca Rizza G, Cutrupi S, Pizzini P, Spoto B, Tripepi G, Zoccali C. Effect of simvastatin on plasma asymmetric dimethylarginine concentration in patients with chronic kidney disease. J Nephrol 21: 38–44, 2008 [PubMed] [Google Scholar]
- 42.Perticone F, Sciacqua A, Maio R, Perticone M, Maas R, Boger RH, Tripepi G, Sesti G, Zoccali C. Asymmetric dimethylarginine, l-arginine, and endothelial dysfunction in essential hypertension. J Am Coll Cardiol 46: 518–523, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Rubba P. Effects of atorvastatin on the different phases of atherogenesis. Drugs 67, Suppl 1: 17–27, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Rudic RD, Shesely EG, Maeda N, Smithies O, Segal SS, Sessa WC. Direct evidence for the importance of endothelium-derived nitric oxide in vascular remodeling. J Clin Invest 101: 731–736, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santilli F, Bucciarelli L, Noto D, Cefalu AB, Davi V, Ferrante E, Pettinella C, Averna MR, Ciabattoni G, Davi G. Decreased plasma soluble RAGE in patients with hypercholesterolemia: effects of statins. Free Radic Biol Med 43: 1255–1262, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Searles CD. Transcriptional and posttranscriptional regulation of endothelial nitric oxide synthase expression. Am J Physiol Cell Physiol 291: C803–C816, 2006 [DOI] [PubMed] [Google Scholar]
- 47.Sydow K, Mondon CE, Schrader J, Konishi H, Cooke JP. Dimethylarginine dimethylaminohydrolase overexpression enhances insulin sensitivity. Arterioscler Thromb Vasc Biol 28: 692–697, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahara N, Kai H, Ishibashi M, Nakaura H, Kaida H, Baba K, Hayabuchi N, Imaizumi T. Simvastatin attenuates plaque inflammation: evaluation by fluorodeoxyglucose positron emission tomography. J Am Coll Cardiol 48: 1825–1831, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Tanaka M, Sydow K, Gunawan F, Jacobi J, Tsao PS, Robbins RC, Cooke JP. Dimethylarginine dimethylaminohydrolase overexpression suppresses graft coronary artery disease. Circulation 112: 1549–1556, 2005 [DOI] [PubMed] [Google Scholar]
- 50.Tawfik HE, El-Remessy AB, Matragoon S, Ma G, Caldwell RB, Caldwell RW. Simvastatin improves diabetes-induced coronary endothelial dysfunction. J Pharmacol Exp Ther 319: 386–395, 2006 [DOI] [PubMed] [Google Scholar]
- 51.Teerlink T. ADMA metabolism and clearance. Vasc Med 10, Suppl 1: S73–S81, 2005 [DOI] [PubMed] [Google Scholar]
- 52.Teerlink T, Nijveldt RJ, de Jong S, van Leeuwen PA. Determination of arginine, asymmetric dimethylarginine, and symmetric dimethylarginine in human plasma and other biological samples by high-performance liquid chromatography. Anal Biochem 303: 131–137, 2002 [DOI] [PubMed] [Google Scholar]
- 53.Thakur NK, Hayashi T, Sumi D, Kano H, Tsunekawa T, Iguchi A. HMG-CoA reductase inhibitor stabilizes rabbit atheroma by increasing basal NO and decreasing superoxide. Am J Physiol Heart Circ Physiol 281: H75–H83, 2001 [DOI] [PubMed] [Google Scholar]
- 54.Tojo A, Welch WJ, Bremer V, Kimoto M, Kimura K, Omata M, Ogawa T, Vallance P, Wilcox CS. Colocalization of demethylating enzymes and NOS and functional effects of methylarginines in rat kidney. Kidney Int 52: 1593–1601, 1997 [DOI] [PubMed] [Google Scholar]
- 55.Tran CT, Fox MF, Vallance P, Leiper JM. Chromosomal localization, gene structure, and expression pattern of DDAH1: comparison with DDAH2 and implications for evolutionary origins. Genomics 68: 101–105, 2000 [DOI] [PubMed] [Google Scholar]
- 56.Tran CT, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atheroscler Suppl 4: 33–40, 2003 [DOI] [PubMed] [Google Scholar]
- 57.Tsuchiya A, Nagotani S, Hayashi T, Deguchi K, Sehara Y, Yamashita T, Zhang H, Lukic V, Kamiya T, Abe K. Macrophage infiltration, lectin-like oxidized-LDL receptor-1, and monocyte chemoattractant protein-1 are reduced by chronic HMG-CoA reductase inhibition. Curr Neurovasc Res 4: 268–273, 2007 [DOI] [PubMed] [Google Scholar]
- 58.Vallance P, Leone A, Calver A, Collier J, Moncada S. Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 339: 572–575, 1992 [DOI] [PubMed] [Google Scholar]
- 59.Vladimirova-Kitova L, Deneva T, Angelova E, Nikolov F, Marinov B, Mateva N. Relationship of asymmetric dimethylarginine with flow-mediated dilatation in subjects with newly detected severe hypercholesterolemia. Clin Physiol Funct Imaging 28: 417–425, 2008 [DOI] [PubMed] [Google Scholar]
- 60.Wakino S, Hayashi K, Tatematsu S, Hasegawa K, Takamatsu I, Kanda T, Homma K, Yoshioka K, Sugano N, Saruta T. Pioglitazone lowers systemic asymmetric dimethylarginine by inducing dimethylarginine dimethylaminohydrolase in rats. Hypertens Res 28: 255–262, 2005 [DOI] [PubMed] [Google Scholar]
- 61.Wang D, Gill PS, Chabrashvili T, Onozato ML, Raggio J, Mendonca M, Dennehy K, Li M, Modlinger P, Leiper J, Vallance P, Adler O, Leone A, Tojo A, Welch WJ, Wilcox CS. Isoform-specific regulation by N(G),N(G)-dimethylarginine dimethylaminohydrolase of rat serum asymmetric dimethylarginine and vascular endothelium-derived relaxing factor/NO. Circ Res 101: 627–635, 2007 [DOI] [PubMed] [Google Scholar]
- 62.Young JM, Strey CH, George PM, Florkowski CM, Sies CW, Frampton CM, Scott RS. Effect of atorvastatin on plasma levels of asymmetric dimethylarginine in patients with non-ischaemic heart failure. Eur J Heart Fail 10: 463–466, 2008 [DOI] [PubMed] [Google Scholar]
- 63.Zhang WZ, Venardos K, Finch S, Kaye DM. Detrimental effect of oxidized LDL on endothelial arginine metabolism and transportation. Int J Biochem Cell Biol 40: 920–928, 2008 [DOI] [PubMed] [Google Scholar]
- 64.Zoccali C, Bode-Boger S, Mallamaci F, Benedetto F, Tripepi G, Malatino L, Cataliotti A, Bellanuova I, Fermo I, Frolich J, Boger R. Plasma concentration of asymmetrical dimethylarginine and mortality in patients with end-stage renal disease: a prospective study. Lancet 358: 2113–2117, 2001 [DOI] [PubMed] [Google Scholar]