Abstract
Arabidopsis COP1 is a constitutive repressor of photomorphogenesis that interacts with photomorphogenesis-promoting factors such as HY5 to promote their proteasome-mediated degradation. SPA1 is a repressor of phytochrome A-mediated responses to far-red light. Here we report that COP1 acts as part of a large protein complex and interacts with SPA1 in a light-dependent manner. We further demonstrate the E3 ubiquitin ligase activity of COP1 on HY5 in vitro and the alteration of that activity by SPA1. Thus, the COP1-SPA1 interaction defines a critical step in coordinating COP1-mediated ubiquitination and subsequent degradation of HY5 with PHYA signaling.
Keywords: Arabidopsis, photomorphogenesis, phytochrome, protein degradation, ubiquitination, protein complex
Plants show a high degree of developmental plasticity in the control of seedling development in response to light. Arabidopsis monitors the ambient light conditions by using multiple wavelength-specific photoreceptors, including the red (600-700 nm) and far-red (FR; 700-750 nm) light-absorbing phytochromes (phyA to phyE) and the blue/ultraviolet-A (320-500 nm) light-absorbing cryptochromes (Cry1 and Cry; Wang and Deng 2002). Studies have shown that the abundance of HY5, a bZIP transcription factor that promotes photomorphogenic (light-grown) development (Oyama et al. 1997), is correlated with the extent of photomorphogenesis and is primarily regulated at the level of protein degradation via the proteasome pathway (Osterlund et al. 2000). The control of HY5 protein stability requires the COP/DET/FUS group of constitutive repressors, which define four biochemical entities [COP1, COP10, DET1, and the COP9 signalosome (CSN); Wang and Deng 2002].
Of these constitutive photomorphogenic repressors, only COP1 is known to directly interact with HY5 and target it for proteasome-mediated degradation (Osterlund et al. 2000). COP1 encodes a 76-kD protein containing a RING-finger domain, followed by a coiled-coil and a WD40 domain (Deng et al. 1992). RING-finger domains are commonly conserved in a subclass of ubiquitin-protein ligases (E3s; Pickart 2001). Protein ubiquitination requires the sequential action of three enzymes, a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and an E3. The E3 confers the substrate specificity by direct recognition of the substrate proteins (Pickart 2001). All these results suggest a model in which COP1 acts as an E3 in the ubiquitin-proteasome pathway within the nucleus and promotes the destabilization of HY5 and other substrates. Recently, COP1 has been shown to mediate the ubiquination of a MYB transcription factor, LAF1 (Seo et al. 2003).
Distinct light-signaling pathways initiated by multiple photoreceptors eventually converge on COP1 and lead to its inactivation (Wang and Deng 2002). However, little information is available regarding the molecular link between the phytochrome-mediated FR/red light signaling and COP1. Of the five phytochromes found in Arabidopsis, phyA is mainly responsible for continuous FR (FRc)-mediated responses (Wang and Deng 2002). SPA1 has been identified as a nuclear-localized repressor of phyA-mediated signaling (Hoecker et al. 1999). SPA1 contains a protein kinase-like domain, a coiled-coil region, and a C-terminal WD40 domain that is highly related in structure to that of COP1. Moreover, interaction of COP1 with SPA1 through their coiled-coil regions (Hoecker and Quail 2001) raises the possibility that SPA1 and COP1 may work together to target substrate proteins for degradation in relating to phyA-signaling. Indeed, it has been shown recently that the coiled-coil domain of SPA1 is able to enhance the E3 ligase activity of COP1 on the transcription factor LAF1 (Seo et al. 2003). As an attempt to further understand the molecular link of PHYA-signaling with COP1, here we have characterized functional interactions between COP1 and SPA1 at both the genetic and biochemical level.
Results and Discussion
COP1 acts as part of large protein complex(es)
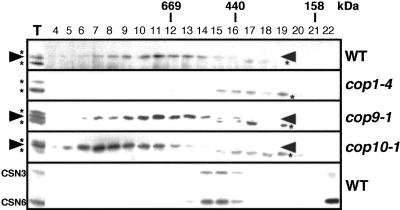
Many RING-type E3 ubiquitin ligases form protein complexes in vivo (Pickart 2001). To examine if this is the case for COP1, we carried out gel filtration analyses (Fig. 1). Because of the low activity of our COP1 antibodies, the band corresponding to COP1 was located between two nonspecific cross-reacting bands (Fig. 1). As expected, the COP1 band was absent in extracts from cop1-4 seedlings, whereas the two flanking nonspecific bands remained largely unchanged (Fig. 1). The majority of COP1 is present in an ∼700-kD protein complex(es) in dark-grown wild-type plants and is clearly different from the profiles of the CSN (Fig. 1) and the COP10 complex (Suzuki et al. 2002). Previous experiments have demonstrated that COP1 is capable of homodimerization (Torii et al. 1998). However, in the current experimental conditions, no detectable amounts of the COP1 protein were seen in the fractions corresponding to the size of its homodimer (150-160 kD). Thus, it is likely that COP1 constitutes the complex(es) in heterogeneous associations with other factors. Further analyses revealed that COP1 is also present in the same size complex(es) in white light (data not shown), as well as in the absence of CSN (Fig. 1, cop9-1). Therefore, light regulation of COP1 must include control mechanisms other than simple association-dissociation of COP1 from the rest components of the complex. Interestingly, the peak of the COP1 complex(es) is shifted to larger fractions by loss of COP10 (Fig. 1, cop10-1). This may represent an unknown inactive state of the COP1 complex(es), which requires COP10 for conversion to a smaller active form.
Figure 1.
COP1 acts as part of large protein complex. Gel filtration fractions of protein extracts from 4-day-old dark-grown wild-type [Wassilewskija (Ws) ecotype], cop1-4, cop9-1, and cop10-1 seedlings were subjected to immunoblot analyses. The arrowhead and asterisk indicate the positions of COP1 and cross-reacting bands, respectively. The fraction numbers and molecular weight are indicated above. T represents total soluble extracts used for gel filtration. An immunoblot probed with anti-CSN3 and anti-CSN6 antibodies is also shown.
Synergistic enhancement of the cop1 and spa1 mutations
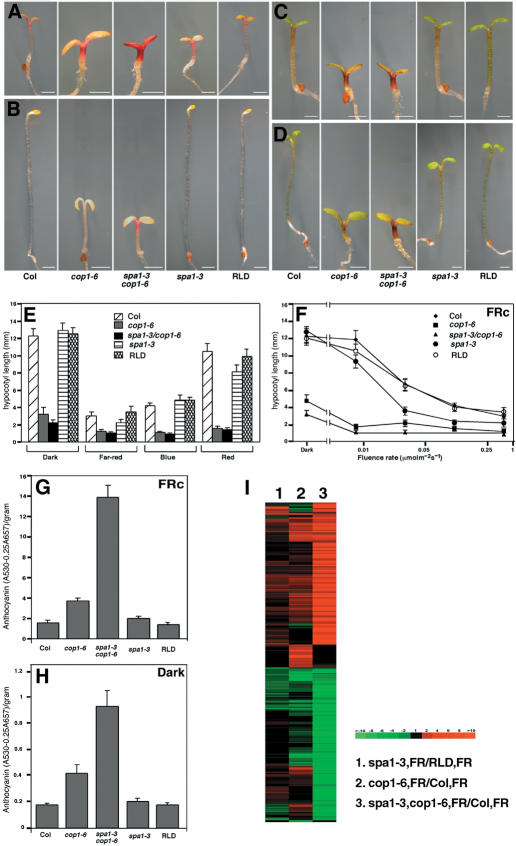
To obtain genetic evidence for the functional interaction between COP1 and SPA1, we constructed a double mutant between a weak cop1 mutant allele, cop1-6 (McNellis et al. 1994), and a severe spa1 mutant allele, spa1-3 (Hoecker et al. 1999). Under FRc, the spa1-3/cop1-6 double mutant possesses slightly shorter hypocotyls and shows higher accumulation of anthocyanin compared with those of the single parental mutants (Fig. 2A,E,G), suggesting that these mutations have a synergistic effect in PHYA signaling. This effect was observed over a range of FRc fluence rates, although it was less pronounced under high fluence rate irradiations (Fig. 2F). Notably, the double mutant shows a synergistic phenotype even in darkness, in which no obvious phenotype is detected in the spa1-3 mutant (Fig. 2B,E,H). This suggests that COP1 and SPA1 may functionally interact even in the absence of FR. A cDNA microarray analysis focusing on FR-regulated genes (Wang et al. 2002) revealed a similar synergism in the gene expression profiles affected by these two mutations (Fig. 2I).
Figure 2.
Synergistic enhancement of cop1-6 and spa1-3 mutations. (A-D) Morphology of spa1-3, cop1-6, a spa1-3/cop1-6 double mutant, and their wild-type ecotype (cop1-6 Col, spa1-3 RLD) 4-day-old seedlings grown under 0.5 µM m-2 s-1 continuous far-red (A), darkness (B), 30 µM m-2 s-1 continuous blue light (C), and 4 µM m-2 s-1 continuous red light (D). Bars, 1 mm. (E,F) Graphs compare hypocotyl length under various light conditions (E) and under continuous far-red with different fluence rates (F). (E-H) Error bars represent the standard deviation of >20 plants each. Bottom panels show anthocyanin content under continuous far-red light (G) and darkness (H). (I) Comparison of the global gene expression profiles of 5-day-old continuous far-red-light-grown plants focusing on the core group (total 696) of FR-responsive genes (Wang et al. 2002). Lane 1 shows ratio of spa1-3 compared with wild-type (RLD) seedlings (spa1.3 FR/RLD FR). Lane 2 shows cop1-6 compared with wild-type (Col) seedlings (cop1.6 FR/Col FR). Lane 3 shows the spa1-3/cop1-6 double mutant compared with wild-type (Col) seedlings (spa1.3,cop1.6 FR/Col FR).
SPA1 controls the protein level of HY5 but not of COP1
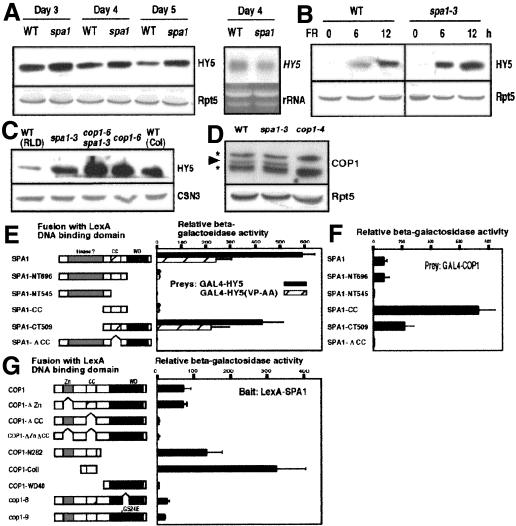
Because COP1 promotes the destabilization of HY5 (Osterlund et al. 2000),we tested whether SPA1 contributes to the down-regulation of HY5 abundance. We compared HY5 levels between wild-type and the spa1-3 mutant seedlings under FRc. During the time period examined, HY5 accumulates to high levels in spa1-3 without detectable increase in the mRNA level (Fig. 3A). Upon transfer of the seedlings from darkness to FRc, HY5 begins to accumulate more rapidly in spa1-3 than in the wild type (Fig. 3B). We also found a slight synergistic effect of the spa1-3 and cop1-6 mutations on HY5 abundance (Fig. 3C). However, no detectable change in the COP1 expression level was found in spa1-3 (Fig. 3D). These results indicate that the hyperphotomorphogenic phenotype of spa1-3 is caused, at least in part, through high-level accumulation of HY5, rather than through decreased expression of COP1.
Figure 3.
SPA1 promotes down-regulation of HY5 protein levels without affecting COP1 expression. Anti-HY5 immunoblot analyses of seedlings grown under continuous far-red (A,C) and 3-day-old seedlings transferred from darkness to far-red light (B) for indicated times. Immunoblots probed with anti-Rpt5 or anti-CSN3 antibodies are shown as loading controls. A Northern blot was probed with the 32P-labeled HY5 cDNA. (D) COP1 levels in the 4-day-old continuous far-red-grown seedlings are indicated by the arrowhead. The asterisks indicate the positions of cross-reacting bands by the COP1 antibodies. The wild-type (WT) ecotype used (A,B,D) is RLD. Yeast two-hybrid analyses of the SPA1-HY5 interactions (E) and the SPA1-COP1 interactions (F,G). The constructs used for analysis are shown on the left. (Zn) RING-finger domain; (CC) coiled-coil domain; (WD40) WD40 repeat region; (SPA1-NT696) amino acids 1-696; (SPA1NT545) amino acids 1-545; (SPA1-CC) amino acids 521-696; (SPA1-CT509) amino acids 521-1029; (SPA1-CC) amino acids 546-646. The right side of the panel shows corresponding β-galactosidase activities. Error bars represent the standard deviation.
To examine whether direct protein interaction between SPA1 and HY5 is involved in the SPA1-mediated control of HY5 level, we used a yeast two-hybrid assay. Full-length SPA1 fused with the LexA DNA-binding domain interacts with the GAL4 activation-domain fusion of HY5 (Fig. 2E). This interaction is greatly reduced by point mutations in the COP1-binding domain of HY5 (HY5VP-AA; Holm et al. 2001). Truncated forms of SPA1 lacking either the WD40 domain or the coiled-coil region do not interact with HY5. This suggests that as found with the COP1-HY5 interaction, the SPA1-HY5 interaction is mediated through the WD40 domain, and presence of both WD40 domain and coiled-coil region are required. Furthermore, two-hybrid analyses using mutant versions of SPA1 and COP1 confirmed that, as previously reported (Hoecker and Quail 2001), the coiled-coil regions of both proteins are necessary and sufficient for the COP1-SPA1 interaction (Fig. 3F,G). Interestingly, the interaction between the coiled-coil regions and their full-length partners is considerably greater than that between the two full-length proteins (Fig. 3F,G). A small deletion or a single amino acid substitution within the WD40 domain of COP1 (cop1-8 and cop1-9, respectively) also inhibits the COP1-SPA1 interaction (Fig. 3G). Thus, it appears that presence of a misfolded WD40 domain in COP1 impairs its coiled-coil-mediated interaction with SPA1, whereas the complete loss of the WD40 domain does not.
COP1 interacts with SPA1 in vivo, which is negatively regulated by light
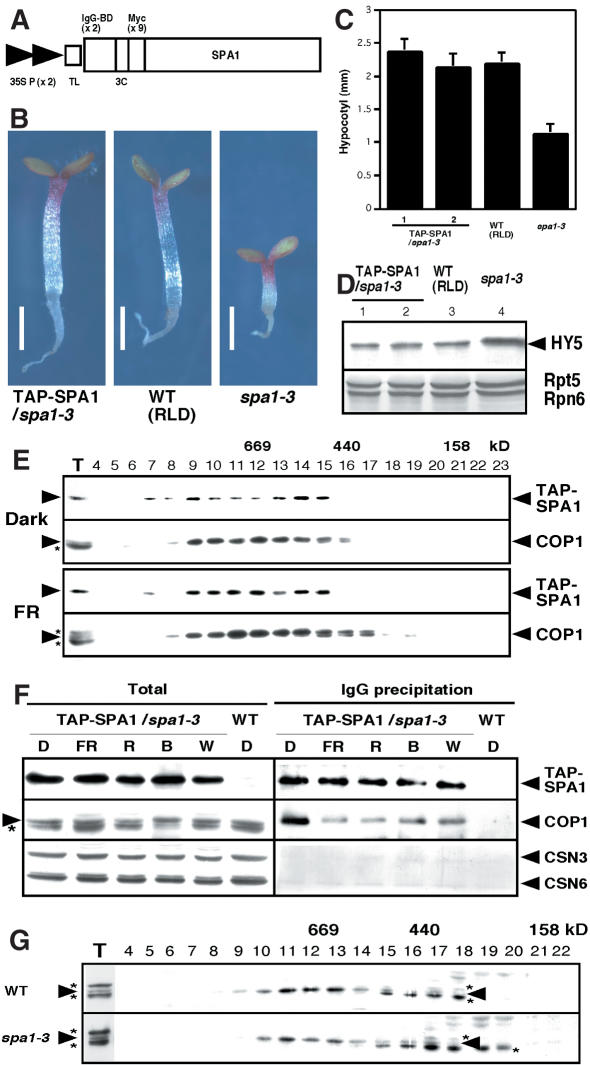
To further investigate the COP1-SPA1 interaction and its role in vivo, we generated transgenic Arabidopsis plants by introducing an N-terminal fusion protein of SPA1 with a modified tandem affinity purification (TAP) tag (Fig. 4A; Rigaut et al. 1999). As shown in Figure 4B and C, this construct complemented the spa1-3 mutation and restored HY5 protein abundance to its wild-type level (Fig. 4D). To address our hypothesis that SPA1 acts as a component of the COP1 complex(es), we first compared gel filtration profiles of both proteins from dark- and FRc-grown seedlings (Fig. 4E). Both the TAP-SPA1 fusion protein and COP1 largely cofractionate in the large-molecular-size fractions (∼700 kD) under both darkness and FRc.
Figure 4.
COP1 interacts with SPA1 in vivo, but light induces their dissociation. (A) The modified TAP-SPA1 construct used for plant transformation, consisting of two copies of the IgG-binding domain (IgG-BD), a human rhinovirus 3C protease cleavage site (3C), and nine copies of the myc epitope tag downstream of two tandem copies of the cauliflower mosaic virus 35S promoter (35S P) with a tobacco mosaic virus translational enhancer sequence (TL). (B-D) TAP-SPA1 complements spa1-3 under continuous far-red light at the level of seedling morphology (B), hypocotyl length (C), and HY5 abundance (D). Bars, 1 mm. Error bars represent the standard deviation of >20 plants each. An immunoblot probed with anti-Rpt5 and anti-Rpn6 antibodies is also shown as a loading control. (E) Gel filtration analyses of 4-day-old dark-grown and continuous far-red-grown TAP-SPA1 transgenic plants. (F) IgG precipitation using the TAP-SPA1 plants grown under various light conditions. (D) Continuous darkness; (FR) continuous far-red light; (R) continuous red light; (B) continuous blue light; (W) continuous white light. (G) Gel filtration profiles of COP1 under continuous far-red light (FRc). The wild-type (WT) ecotype used is RLD. The arrowheads and asterisks in panels D-G indicate protein positions and the anti-COP1 antibody cross-reacting bands, respectively.
To confirm the association of COP1 and SPA1 in vivo, we performed coprecipitation assays with immunoglobin G (IgG)-agarose beads, which specifically precipitate TAP-SPA1 from crude plant extracts. As shown in Figure 4F, COP1 is coprecipitated with TAP-SPA1 from the transgenic plants but not from the wild-type controls. Although almost equal amounts of TAP-SPA1 were precipitated, the yield of coprecipitated COP1 is higher from dark-grown seedlings than from any other light-grown seedlings (Fig. 4F, right). Because the amounts of COP1 and SPA1 as a proportion of total protein extracts are essentially the same under all growth conditions (Fig. 4F, left), this difference may reflect a decrease in the amount of SPA1 associated with COP1 in the light. Thus, it is possible that light-mediated inactivation of COP1 is achieved, at least in part, through this light-induced dissociation of SPA1 from COP1. Even though SPA1 function is most prominent under FRc, we detected no enhancement in the COP1-SPA1 interaction under FRc compared with the other light conditions. Apparently, direct interaction with COP1 alone does not account for the functional specificity of SPA1 toward the PHYA-mediated FRc responses. Rather, it suggests that a posttranslational modification on SPA1 and/or COP1 may occur predominantly under FR, possibly mediated by PHYA. Nevertheless, both cofractionation and coprecipitation suggest a tight physical association between COP1 and SPA1. In contrast, the signals for two subunits of the CSN (CSN3 and CSN6) in the IgG-precipitated samples are not substantially elevated over the mock-precipitated control.
It seems likely that SPA1 acts as a component of the COP1 complex(es). However, further gel filtration analyses did not reveal any significant difference in the profile of the COP1 complex(es) in the FRc-grown spa1-3 seedlings (Fig. 4G), in which a nonsense mutation precedes the coiled-coil region of SPA1 protein (Hoecker et al. 1999). This indicates that at least one form of the COP1 complex(es) maintains its integrity without SPA1. It is possible that different COP1 complexes exist and at least one of them contains SPA1 as an associated factor. In this regard, the Arabidopsis genome has at least two genes that encode proteins highly homologous in structure to SPA1 (GenBank accession nos. T08190 and AAF87859) that may partially compensate for SPA1 activity in its absence. Alternatively, the COP1-SPA1 interaction may be an intercomplex interaction. However, it should be noted that no such super-large complex was detected in our gel filtration analyses.
COP1 shows an E3 ubiquitin ligase activity on HY5 in vitro, which is altered by SPA1
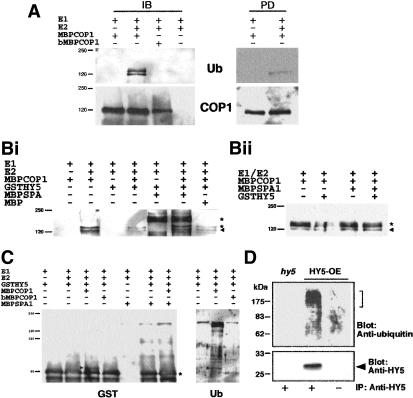
Because all the results suggest that the COP1-SPA1 complex(es) plays a role in regulating ubiquitination pathways, we investigated whether COP1 has intrinsic E3 ubiquitin ligase activity by using in vitro ubiquitination assays with maltose-binding protein (MBP) fusions of full-length COP1 and SPA1. In the presence of recombinant yeast E1 and Arabidopsis E2, the purified MBPCOP1 exhibits autoubiquitination activity (Fig. 5A). Two bands corresponding to ubiquinated proteins were seen in immunoblots only in the presence of E1, E2, and functional COP1. Using amylose resin to pull-down the MBP tag (Fig. 5A, right panels), it was determined that the lower band represents ubiquinated COP1, whereas the upper band is an unknown protein that is also ubiquinated by COP1. The signal for ubiquinated COP1 overlaps with that of unmodified protein, and we were unable to distinguish multiple forms of COP1 using either COP1 (Fig. 5A, lower panel) or MBP antibodies (data not shown). This suggests the addition of one or possibly two ubiquitin molecules to only a small proportion of the total amount of COP1 in the assay mixture.
Figure 5.
COP1-mediated ubiquitination of HY5. Recombinant COP1, SPA1, and HY5 fusions proteins were purified from E. coli, and in vitro ubiquination assays were performed. (A) Demonstration of COP1 E3 autoubiquination activity of active COP1 (MBPCOP1) or denatured COP1 (bMBPCOP1) in vitro after immunoblotting (IB) or precipitation with amylose resin and subsequent immonublotting (PD). For IB, MBPCOP1 was probed with antibodies against COP1, and ubiquitinated products were detected by specific binding of peroxidase-conjugated streptavidin to biotinylated ubiquitin followed by chemiluminescence detection. (B) The left and right panels show the alteration of COP1 autoubiquination activity by SPA1. Ubiquitinated products were detected as described above. The arrowhead indicates positions of autoubiquitinated COP1. The lower asterisk indicates an unidentified ubiquitinated protein (see text), the upper asterisk in the left panel represents nonspecific cross-reacting band associated with some MBPSPA1 preparations. (C) COP1-mediated ubiquination of HY5 and the effect of SPA1. GSTHY5 in the left panel was detected by using antibodies against GST; ubiquitinated GSTHY5 in the right panel was visualized as described above. Arrowheads indicate position of ubiquinated HY5, whereas the asterisk indicates position of unmodified GSTHY5. (D) Three-day-old white-light-grown HY5-overexpressing (HY5-OE) and hy5-ks50 seedlings were transferred to media containing proteasome inhibitors (MG115, MG132, and PSI h), and subjected to darkness for 20 h. Immunoprecipitated protein (IP) with anti-HY5 serum or preimmune serum was analyzed by immunoblotting with anti-ubiquitin and anti-HY5 antibodies.
To investigate the effects of SPA1 on the E3 ligase activity of COP1, we first examined the effects of MBPSPA1 on COP1 autoubiquitination activity (Fig. 5B, left and right panels). As expected, bands corresponding to ubiquinated COP1 and the unidentified protein described above were observed only in the presence of E1, E2, and COP1 (Fig. 5B, left panel). Interestingly, the addition of GSTHY5 (Fig. 5B, left panel), but not GST alone (Fig. 5B, right panel), caused a decrease in the amount of these two ubiquinated proteins. The addition of approximately equimolar (relative to COP1) amounts of MBPSPA1 returned amounts of ubiquinated COP1 to that observed in the absence of HY5 (Fig. 5B). However, in the absence of GSTHY5, the addition of SPA1 had no detectable effect on the level of COP1 autoubiquitination (Fig. 5B, right panel).
We next investigated the E3 ligase activity of COP1 on GSTHY5 by using GST antibodies. The addition of E1, E2, and active COP1 resulted in the formation of a larger form of GSTHY5 (Fig. 5C, left panel). This larger form of GSTHY5 was ubiquinated (Fig. 5C, right panel), with the size shift indicating the addition of one ubiquitin unit. Interestingly, the addition of SPA1 (but not MBP; data not shown) inhibits the formation of ubiquinated GSTHY5 (Fig. 5C).
Because proteins that are destined for proteasome-mediated degradation are generally conjugated to a polyubiquitin chain (Pickart 2001), we wanted to investigate the ubiquination status of HY5 in vivo. To this end, we took light-grown Arabidopsis seedlings and transferred them to the dark, a process that results in the rapid degradation of HY5 (Osterlund et al. 2000). Treatment of the seedlings with proteasome inhibitors enabled detection of polyubiquitinated forms of HY5 (Fig. 5D), demonstrating that in vivo HY5 is polyubiquitinated before its degradation via the proteasome pathway.
Recently, Seo et al. (2003) have also demonstrated the E3 ligase activity of COP1 on itself and the MYB transcription factor LAF1. However, in contrast to these results, they reported that SPA1 enhances the COP1-mediated ubiquination of LAF1. There are a number of possibilities to reconcile these two sets of data. Firstly, although Seo et al. (2003) used recombinant MBPCOP1, they used mammalian E1 and E2 in their in vitro assays, in contrast to the yeast E1 and Arabidopsis E2 used in this work. We have ourselves noticed variations in the efficiencies of different Arabidopsis E2s with COP1. For example, this work uses the E2 AtUbc9, whereas we were unable to detect any activity using AtUbc8, which was capable of functioning with the Arabidopsis E3 CIP8 (Hardtke et al. 2002). Second, in a previous study, Seo et al. (2003) use only the coiled-coiled domain of SPA1, whereas we use the full-length fusion protein in our assays. Although the coiled-coil domain of SPA1 is necessary and sufficient for interaction with COP1, the WD40 region also has an important role to play in both substrate interaction and interaction with COP1 (Fig. 3). One possible model to explain our in vitro data is that SPA1 inhibits COP1-mediated ubiquination of HY5 by sequestering the substrate away from the active site of COP1. If such sequestration of LAF1 involves the WD40 region, as is the case with HY5, this interaction would not be seen with a truncated protein that lacks the WD40 domain. Finally, the differences between these results and those previously reported may simply be due to the use of different COP1 substrates. Although both LAF1 and HY5 are transcription factors that positively regulate photomorphogenesis, LAF1 is specific to far-red light, whereas HY5 regulates red-, far-red-, and blue-light-mediated signaling (Oyama et al. 1997; Ballesteros et al. 2001).
Treatment of seedlings with proteasome inhibitors enabled detection in vivo of polyubiquitinated forms of HY5 (Fig. 5D), demonstrating that HY5 is polyubiquitinated before its degradation. Given this observation, there appears to be a discrepancy between the results from in vitro biochemical assays and our observations in vivo. The genetic data suggest that SPA1 and COP1 act together to reduce the abundance of HY5, ultimately leading to reduced photomorphogenesis. The reduction in the abundance of HY5 involves degradation by the proteasome, and we can detect polyubiquinated HY5 in vivo under conditions in which it is being actively degraded (Fig. 5D). Given these observations, we might have expected to see an enhancement in the COP1-mediated ubiquination of HY5 in the presence of SPA1, whereas we, in fact, see an inhibitory effect (Fig. 5C). One possibility to explain this discrepancy is that the SPA1 is acting as a switch, changing the ubiquination activity of COP1 on HY5 from monoubiquitination to polyubiquination. It is possible that presence of SPA1 changes the interaction between COP1 and HY5, producing a different type of ubiquination event. Clearly, our in vitro assay system lacks the other as-yet-unidentified components of the COP1 complex(es), and it may be that one of these is essential for the polyubiquination of HY5. This missing activity may reflect the action of another ubiquitin ligase, an E4 polyubiquitination ligase subtype (Pickart, 2001), and/or that further activation of COP1 may be required to generate the polyubiquitinated forms of HY5. Another RING-finger protein, CIP8, which is capable of ubiquitinating HY5 (Hardtke et al. 2002) and directly interacting with COP1 (Torii et al. 1999), might fulfill the missing link to polyubiquitination. It is also possible that further modification of HY5, which is absent from our in vitro experiments, is required in planta for polyubiquitination.
The E3 activity of COP1 observed in vitro and its SPA1-mediated alteration may represent only part of their enzymatic function. Further study is required to reveal the physiological significance of the monoubiquination activity of COP1 on HY5. Nevertheless, it is certain that SPA1 is inherently capable of modulating the E3 ubiquitin ligase activity of COP1. It is conceivable that distinct light signals are acting through different COP1 complex components, of which SPA1 is the component primarily responsible for mediating far-red light regulation of COP1 activity. The direct interaction between COP1 and SPA1 provides a molecular basis for the convergence of PHYA-mediated signaling and COP1, and how far red light regulates activity of photomorphogenesis promoting transcription factors such as HY5 and thus developmental pattern.
Materials and methods
Plant materials and growth conditions
The wild-type plants used were of the Wassilewskija ecotype unless otherwise stated. The cop, spa1-3, hy5-ks50 mutants and the HY5-overexpression lines used have been described previously (McNellis et al. 1994; Oyama et al. 1997; Hoecker et al. 1999; Hardtke et al. 2002; Suzuki et al. 2002). Seed sterilization and plant growth conditions were performed as previously described (Hardtke et al. 2002). Vernalized seeds were exposed to white light for 6-10 h, and then transferred to continuous light conditions (6 µM m-2 s-1 for far-red light, 10 µM m-2 s-1 for red light, 4 µM m-2 s-1 for blue light, and 30 µM m-2 s-1 for white light), unless otherwise stated. Anthocyanin content of seedlings was determined as described previously (McNellis et al. 1994). Total RNA extractions, Northern blot procedures, and microarray analysis and hierarchical clustering were performed as previously described (Osterlund et al. 2000; Wang et al. 2002). For raw data, see http://plantgenomics.biology.yale.edu.
Western blot analysis, gel filtration chromatography, and immunoprecipitation
Protein extraction, gel filtration, and Western blot analyses were essentially performed as described previously (Hardtke et al. 2002) except using buffer A (50 mM Tris at pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 25 mM β-glycerophosphate, 2 mM sodium orthovanadate, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM PMSF, and 1× complete protease inhibitor cocktail; Roche) for lysis and gel filtration. A Superose 6 (Amersham) column was used for gel filtration. For IgG-bead precipitation, after incubation of the extracts with IgG-agarose beads (Amersham) in buffer A without DTT (buffer B) for 3 h at 4°C, beads were washed twice in buffer B, and then twice more in buffer B containing 200 mM NaCl. The precipitated protein was eluted into buffer C (100 mM glycine at pH 2.5, 100 mM NaCl), and then concentrated by using Strataresin (Stratagene). Immunoprecipitation was performed essentially as IgG precipitation described above, except that protein A-Sepharose beads (Pierce) coupled to anti-HY5 serum or pre-immune serum were used, and all the processes were performed in presence of proteasome inhibitors (MG132, MG115, and PS1 at 25 µM each; Calbiochem). Anti-myc monoclonal antibody (the 9E10 clone) was used to detect TAP-SPA1 fusion protein.
Yeast two-hybrid analysis
The assay system, all the procedures, and all the constructs for COP1 have been described previously (Wang et al. 2001). The HY5VP-AA mutations were introduced via PCR as described previously (Holm et al. 2001).
TAP-SPA1 transgenic plants
The SPA1 cDNA was inserted into a modified N-terminal TAP tag (Rigaut et al. 1999) in pZP222 (Hajdukiewicz et al. 1994). Sequence analyses revealed that the resulted SPA1 sequence contains one amino acid substitution in the very C terminus (V1029C). At least two independent T3 transgenic lines were used for subsequent analyses.
In vitro ubiquitination assays
N-terminal fusions between MBP and Arabidopsis COP1 and SPA1 coding regions were expressed in BL21 codon plus (Stratagene) Escherichia coli cells, and purified by using Amylose resin (New England Biolabs). For experiments with denatured MBPCOP1, the protein was boiled for 10 min at 95°C before addition to the ubiquination reaction. The GST-fusion protein with Arabidopsis HY5 was prepared as described previously (Hardtke et al. 2002). An N-terminal 6xHis tag with Arabidopsis UBC9 was produced in the plasmid pQE30 (Qiagen) and purified by using Ni-NTA resin (Qiagen) according to manufacturer's protocol. In vitro ubiquitination assays were performed in a total volume of 30 µL consisting of 50 mM Tris (pH 7.5), 10 mM MgCl2, 10 mM ATP, 1 µg biotinylated ubiquitin (Affiniti), ∼25 ng yeast E1 (Sigma), 25 ng AtUBC9, 500 ng GSTHY5, 500 ng MBP-COP1, and 500 ng MBP-SPA1. Reactions were incubated for 2 h at 30°C before being terminated by the addition of SDS sample buffer. Products conjugated with biotinylated ubiquitin were detected by incubation of Western blots with streptavidin-conjugated horseradish peroxidase (Amersham), followed by chemiluminescence visualization.
Acknowledgments
We thank S.P. Dinesh-Kumar for the construction of the TAP-fusion plant binary vector, and T. Tsuge for the monoclonal myc antibodies. Our work was supported by grants from National Institutes of Health (GM47850) and National Science Foundation (MCB-0115870) to X.W.D. and fund from Boyce Thompson Institute to H.W. Y.S. is a Japanese Society for Promotion of Science postdoctoral fellow; J.A.S., V.R., and L.M. are Human Frontier Science Program Organization postdoctoral fellows.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1122903.
References
- Ballesteros M.L., Bolle, C., Lois, L.M., Moore, J.M., Vielle-Calzada, J.P., Grossniklaus, U., and Chua, N.H. 2001. LAF1: A MYB transcriptional activator for phytochrome A signaling. Genes & Dev. 15: 2613-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Minami, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. 1992. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71: 791-801. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab, Z., and Maliga, P. 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989-994. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Okamoto, H., Stoop-Myer, C., and Deng, X.W. 2002. Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30: 385-394. [DOI] [PubMed] [Google Scholar]
- Hoecker U. and Quail, P.H. 2001. The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276: 38173-38178. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Tepperman, J.M., and Quail, P.H. 1999. SPA1: A WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496-499. [DOI] [PubMed] [Google Scholar]
- Holm M., Hardtke, C.S., Gaudet, R., and Deng, X.W. 2001. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20: 118-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. 1994. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke, C.S., Wei, N., and Deng, X.W. 2000. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462-466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura, Y., and Okada, K. 1997. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Dev. 11, 2983-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503-533. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A genetic protein purification method for protein complex characterization and proteome exploration. Nat. Biotech. 17: 1030-1032. [DOI] [PubMed] [Google Scholar]
- Seo S.H., Yang, J., Ishikawa, M., Bolle, B., Ballesteros, M.L., and Chua, N.H. 2003. LAF1 ubiquination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995-999. [DOI] [PubMed] [Google Scholar]
- Suzuki G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. 2002. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes & Dev. 16: 554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., McNellis, W., and Deng, X.W. 1998. Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17: 5577-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., Stoop-Myer, C.D., Okamoto, H., Coleman, J.E., Matsui, M., and Deng, X.W. 1999. The RING finger motif of photomorphogenic repressor COP1 specifically interacts with the RING-H2 motif of a novel Arabidopsis protein. J. Biol. Chem. 274: 27674-27681. [DOI] [PubMed] [Google Scholar]
- Wang, H. and Deng, X.W. 2002. Phytochrome signaling mechanism. In The Arabidopsis book (eds. C.R. Somerville and E.M. Meyerowitz). American Society of Plant Biologists, Rockville, MD; doi/10.1199/tab.0074, http://www.aspb.org/publications/arabidopsis (published electronically).
- Wang H., Ma, L.-G., Li, J.-M., Zhao, H.-Y., and Deng, X.W. 2001. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154-158. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma, L.-G., Habashi, J., Li, J., Zhao, H., and Deng, X.W. 2002. Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32: 723-733. [DOI] [PubMed] [Google Scholar]