Abstract
It is proposed that ischemic preconditioning (PC) initiates signaling that converges on mitochondria and results in cardioprotection. The outcome of this signaling on mitochondrial enzyme complexes is yet to be understood. We therefore used proteomic methods to test the hypothesis that PC and pharmacological preconditioning similarly alter mitochondrial signaling complexes. Langendorff-perfused murine hearts were treated with the specific GSK-3 inhibitor AR-A014418 (GSK Inhib VIII) for 10 min or subjected to four cycles of 5-min ischemia-reperfusion (PC) before 20-min global ischemia and 120-min reperfusion. PC and GSK Inhib VIII both improved recovery of postischemic left ventricular developed pressure, decreased infarct size, and reduced lactate production during ischemia compared with their time-matched controls. We used proteomics to examine mitochondrial protein levels/posttranslational modifications that were common between PC and GSK Inhib VIII. Levels of cytochrome-c oxidase subunits Va and VIb, ATP synthase-coupling factor 6, and cytochrome b-c1 complex subunit 6 were increased while cytochrome c was decreased with PC and GSK Inhib VIII. Furthermore, the amount of cytochrome-c oxidase subunit VIb was found to be increased in PC and GSK Inhib VIII mitochondrial supercomplexes, which are comprised of complexes I, III, and IV. This result would suggest that changes in complex subunits associated with cardioprotection may affect supercomplex composition. Thus the ability of PC and GSK inhibition to alter the expression levels of electron transport complexes will have important implications for mitochondrial function.
Keywords: glycogen synthase kinase-3, preconditioning, electron transport chain, supercomplex
murry et al. (26) were the first to describe the preconditioning (PC) phenomenon in which brief episodes of ischemia and reperfusion protect the heart from a subsequent prolonged ischemic period. Preconditioned hearts have reduced infarct size, arrhythmias, and postischemic contractile dysfunction. Although the mechanism by which PC mediates cardioprotection has not been fully elucidated, the current paradigm is that PC releases agonists (e.g., adenosine, bradykinin, and opioids) into the cellular milieu, which bind to G protein-coupled receptors, and initiates a protein kinase signaling cascade leading to protection. Phosphatidylinositol 3-kinase (PI3-kinase) plays an essential role in PC signaling (40) and leads to downstream phosphorylation and inactivation of glycogen synthase kinase (GSK)-3β (41).
Earlier studies have suggested that GSK-3β Ser9 phosphorylation before ischemia reduces infarct size after ischemia-reperfusion (I/R) in the heart and short-term GSK-3β inhibition with pharmacological agents mimics PC cardioprotection (13, 18, 41). Gross et al. (13) further showed that addition of GSK inhibitors at the start of reperfusion reduced infarct size in rat heart. Juhaszova et al. (18) suggested that GSK-3β inhibition by Ser9 phosphorylation reduces mitochondrial permeability transition pore opening in cardiomyocytes. Although the signaling kinases activated by PC have been thoroughly described, the mechanisms by which these kinases result in protection are poorly understood. Recent studies have suggested that many of these kinase signals converge on the mitochondria to initiate cardioprotection. Thus the primary goal of this study was to identify the alterations in the mitochondrial proteome that are involved in cardioprotection by using a proteomics approach. We identified changes in the mitochondrial proteome that are consistent in two cardioprotective approaches, PC and treatment with a GSK-3 inhibitor. We hypothesize that both PC and GSK inhibition will lead to a number of modifications in the mitochondrial proteome, many of which will not be involved in cardioprotection. However, by focusing on the proteome alterations that occur in both PC and GSK inhibition, we can distinguish changes that are likely to be involved in cardioprotection. We report that specific GSK-3 inhibitor AR-A014418 (GSK Inhib VIII) treatment and PC both resulted in changes in several proteins in the electron transport chain and these alterations are associated with altered assembly of mitochondrial supercomplexes.
MATERIALS AND METHODS
Animals.
All animals were treated and cared for in accordance with the Guide for the Care and Use of Laboratory Animals [National Institutes of Health (NIH), revised 1996], and protocols were approved by the Institutional Animal Care and Use Committee. Adult male C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) (age 12–16 wk) were used for this study.
Experimental protocol.
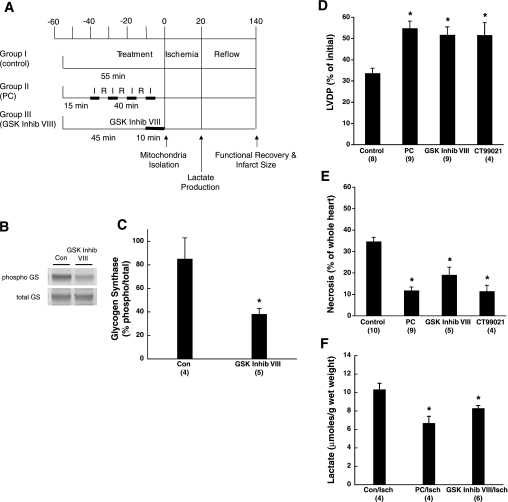
Hearts were perfused as described in Fig. 1A. Briefly, the hearts were isolated and Langendorff perfused in Krebs-Henseleit (KH) buffer for 55 min (control), preconditioned (PC; 4 cycles of 5-min ischemia and 5-min reperfusion) after a 15-min equilibration, or equilibrated for 45 min and given 1 μM AR-A014418 (GSK Inhib VIII) (Calbiochem, Gibbstown, NJ) for 10 min. Hearts were subjected to 20 min of no-flow ischemia. Recovery of left ventricular developed pressure (LVDP), expressed as a percentage of the initial LVDP before ischemia or preconditioning, was measured at 2 h of reflow.
Fig. 1.
Ischemic preconditioning (PC) and glycogen synthase kinase (GSK)-3 inhibitor AR-A014418 (GSK Inhib VIII) protect murine hearts against ischemia-reperfusion injury. A: experimental protocol showing duration and time course of ischemia (I) and reperfusion (R). Mitochondria were isolated after each 55-min protocol, and lactate measurements were made after 20 min of ischemia. Functional recovery and infarct size were assessed after 2 h of reflow. B: representative Western blot of phospho-glycogen synthase (GS) and total GS from control (Con) and GSK Inhib VIII-treated hearts. C: densitometry of % phosphorylated GS normalized to total GS of Con and GSK Inhib VIII-treated hearts. D: recovery of left ventricular developed pressure (LVDP) measured after 2 h of reperfusion. Data are normalized to preischemic LVDP values. E: infarct size (necrosis) as % of whole heart measured with 2,3,5-triphenyltetrazolium chloride (TTC) staining and representative heart cross sections. F: lactate production after 20 min of ischemia. Data are expressed as micromoles of lactate produced per gram of wet weight. Numbers of hearts are shown in parentheses. *P < 0.05 compared with Con.
Necrosis measurement.
At the end of the 2-h reperfusion, hearts were incubated in 1% (wt/vol) 2,3,5-triphenyltetrazolium chloride (TTC) in KH buffer. Samples were sliced into cross sections, and the area of infarct as a percentage of the whole heart was quantified.
Lactate production.
Hearts were snap-frozen after the 20-min ischemic period, minced, and extracted in 3.6% perchloric acid. The samples were centrifuged at 3,000 g, and the supernatant was neutralized with 1 M KOH to pH 5–6. Lactate assays were performed in phosphate buffer with 1 M glycine (pH 8.3), 1 M hydrazine, 20 mM NAD+, and 0.02 U l-lactate dehydrogenase. The assay plate was incubated at 37°C for 30 min, and NADH fluorescence was measured at 355-nm excitation and 460-nm emission.
Immunoblotting.
Hearts were snap-frozen after each 55-min treatment and lysed in 50 mM Tris·HCl (pH 7.4), 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100. The homogenates were centrifuged at 10,000 g, and the supernatants were used for electrophoresis. Samples (40 μg) were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes. After blocking with Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE), the membranes were incubated with the following primary antibodies at 1:1,000 dilutions: anti-phospho-glycogen synthase (Ser641 and Ser645) (Invitrogen) and anti-total-glycogen synthase (Chemicon, Billerica, MA). Appropriate secondary antibodies were used and visualized on the ODYSSEY Infrared Imaging System (LI-COR).
Mitochondria isolation.
Mitochondria were isolated by differential centrifugation according to standard procedures (36). Because we were interested in identifying modifications of known mitochondrial proteins, we utilized a rapid isolation of mitochondria (to preserve posttranslational modifications) rather than a prolonged purification of the crude mitochondrial pellet. Briefly, hearts were weighed; minced in (mM) 225 mannitol, 75 sucrose, 5 MOPS, 0.5 EGTA, and 2 taurine (pH 7.25) (buffer A); and homogenized by Polytron. To digest the contractile proteins, trypsin (0.001 g/0.1 g wet tissue) in buffer A was added to the homogenate for 5 min on ice. Digestion was stopped with buffer A containing protease and phosphatase inhibitors. The homogenate was centrifuged at 500 g, and the resulting supernatant was spun at 11,000 g to pellet the mitochondria. The final mitochondrial pellet was resuspended in buffer A with protease and phosphatase inhibitors. Protein content was determined with a Bradford assay.
Two-dimensional gel electrophoresis and gel staining.
CyDye two-dimensional (2D) fluorescence difference gel electrophoresis (DIGE) and Pro-Q staining were performed as described previously (15, 16). Briefly, mitochondrial proteins were solubilized in lysis buffer [30 mM Tris·HCl, 7 M urea, 2 M thiourea, and 4% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)]. Homogenates were centrifuged at 12,000 g, and the pellets were discarded. Individual samples (50 μg) were labeled on Lys residues with Cy3 and Cy5 (GE Healthcare, Piscataway, NJ). A 50-μg internal standard consisting of equal protein amounts of all samples was labeled with Cy2. The labeled samples and internal standard were combined with 175 μg of each sample (unlabeled) to bring the total protein content for each gel to 500 μg. Unlabeled samples (500 μg) were utilized for Pro-Q gels. First-dimension isoelectric focusing was carried out on 24-cm Immobiline DryStrip gels pH 3–10 nonlinear (GE Healthcare) in an Ettan IPG Phor electrophoresis unit (GE Healthcare), rehydrating at 30 V for 10–12 h followed by focusing for ∼66,667 Vh. The strips were loaded into precast 10–15% Optigel polyacrylamide gels (NextGen Sciences, Ann Arbor, MI), and the proteins were separated on an Ettan DALT-12 electrophoresis unit (GE Healthcare) at constant voltage (∼2,350 Vh). All gels were scanned on a Typhoon 9400 variable imager (GE Healthcare) at a resolution of 100 μm. Image analysis was performed by using the cross-stain analysis function with Progenesis Discovery software (NonLinear Dynamics, Durham, NC).
Protein identification.
For all protein identifications from 2D gels, protein spots were picked with the Ettan Spot Handling Workstation (GE Healthcare). Protein identification was carried out with a matrix-assisted laser desorption/ionization (MALDI)-time of flight (TOF)/TOF instrument (4700 Proteomics Analyzer, Applied Biosystems, Foster City, CA) with reflector positive ion mode. For mass spectrometry (MS) analysis, an 800–4,000 mass-to-charge ratio (m/z) mass range was used with 1,500 shots per spectrum. Result-dependent analysis (RDA) was used for MS/MS selection. A maximum of six precursors per protein were selected, with a confidence interval (CI) percentage of 50 or higher and a minimum signal-to-noise ratio of 50. In addition, a low-confidence investigation (peptides not matched to top proteins) was used to allow a maximum of five precursors per spot with minimum signal-to-noise ratio of 50 and selected for data-dependent MS/MS analysis. A 1-kV collision energy was used for collision-induced dissociation (CID), and 1,500 acquisitions were accumulated for each MS/MS spectrum. For both MS and MS/MS analysis, the default calibration was performed with 4700 mass standard peptide mix (Applied Biosystems) achieving a mass accuracy within 50 ppm. Internal calibration was used for all MS runs with trypsin autolysis peaks of 842.51 m/z, 1,045.56 m/z, and 2,211.11 m/z. When one or more of the trypsin peaks were not found within the mass tolerance of 0.1 m/z, default processing was used.
The peak lists were generated with GPS Explorer software using default parameters (version 3.0, Applied Biosystems). A Mascot search engine was used (version 2.2, Matrix Science, Boston, MA) for peptide and protein identifications with the following search criteria: enzyme, trypsin; miscleavages, one; fixed modifications, cysteine carbamidomethylation; variable modifications, methionine oxidation; mass tolerance for precursor ions, 100 ppm; and mass tolerance for fragment ions, 0.5 Da. The SwissProt protein knowledgebase database (Sprot, release 57.4, 16 June 2009; 497,293 sequences) was searched against, and MS peak filtering was set for all trypsin autolysis peaks. The species selected was Mus musculus (mouse), and the number of sequence entries searched in the M. musculus database was 16,183 of 497,293 total sequence entries. The acceptance criteria for individual MS/MS spectra had a significance threshold set to P < 0.05 with expectation values (E values) < 0.05 (number of different peptides with scores equivalent to or better than the result reported that are expected to occur in the database search by chance). The P value was chosen to reflect a 95% probability that the protein identification is correct. The identification of two peptides or more had to meet the following criteria: the protein was characterized as a mitochondrial protein, E value < 0.05, and molecular weight had to match the position where the spot was picked on the 2D gel. Calculated protein isoelectric points (pI) were obtained from the Expert Protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (SIB) and are used in Tables 3–5. Mitochondrial proteins often have a mitochondrial targeting sequence that is processed/cleaved when entering the mitochondria, and these cleaved proteins are referred to as “clipped proteins.” The sequence of the clipped proteins is a truncated sequence of the total protein without the mitochondrial targeting sequence. The calculated pIs shown in Tables 3–5 are for the processed clipped proteins.
Table 3.
Con/PC 2D-DIGE protein differences
| Spot No. | Name | Accession No. | Calc MW | Obs MW | Calc pI | Obs pI | Function | Gels IDed | Effect in PC | P |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (Q91VD9) NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial precursor | NDUS1_MOUSE | 80,724 | 80,000 | 5.24 | 5.30 | Ox Phos | 3 | 21.45% lower | 0.017 |
| 2 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 5.83 | β-Oxidation | 1 | 33.91% lower | 0.052 |
| 3 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 6.00 | β-Oxidation | 3 | 29.63% lower | 0.051 |
| 4 | (Q99NB1) Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor | ACS2L_MOUSE | 75,317 | 75,000 | 5.98 | 6.00 | Metabolite metabolism/biosyn | 2 | 35.36% lower | 0.001 |
| 5 | (P38647) Stress-70 protein, mitochondrial precursor | GRP75_MOUSE | 73,768 | 75,000 | 5.50 | 5.30 | Protein transport/folding | 3 | 30.65% lower | 0.052 |
| 6 | (P38647) Stress-70 protein, mitochondrial precursor | GRP75_MOUSE | 73,768 | 75,000 | 5.50 | 5.35 | Protein transport/folding | 3 | 31.46% lower | 0.015 |
| 7 | (P38647) Stress-70 protein, mitochondrial precursor | GRP75_MOUSE | 73,768 | 75,000 | 5.50 | 5.40 | Protein transport/folding | 2 | 32.84% lower | 0.011 |
| 8 | (Q91ZA3) Propionyl-CoA carboxylase α chain, mitochondrial precursor | PCCA_MOUSE | 80,498 | 75,000 | 6.13 | 5.91 | Metabolite metabolism/biosyn | 3 | 12.59% lower | 0.041 |
| 9 | (Q8R1S0) Ubiquinone biosynthesis monooxygenase COQ6 (EC1.14.13) | COQ6_MOUSE | 50,959 | 60,000 | 6.55 | 6.70 | Metabolite metabolism/biosyn | 2 | 15.90% lower | 0.002 |
| 10 | (P50544) Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADV_MOUSE | 71,230 | 71,000 | 7.72 | 7.40 | β-Oxidation | 1 | 24.87% lower | 0.032 |
| 11 | (P50544) Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADV_MOUSE | 71,230 | 71,000 | 7.72 | 7.60 | β-Oxidation | 2 | 15.18% lower | 0.051 |
| 12 | (Q8CHT0) Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor | AL4A1_MOUSE | 62,228 | 70,000 | 7.70 | 7.50 | Amino acid degradation | 2 | 7.66% lower | 0.030 |
| 13 | (Q9CZ13) Cytochrome b-c1 complex subunit 1, mitochondrial precursor | QCR1_MOUSE | 53,420 | 53,000 | 5.28 | 5.35 | Ox Phos | 3 | 14.24% lower | 0.017 |
| 14 | (P29758) Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) | OAT_MOUSE | 48,400 | 50,000 | 5.73 | 5.75 | Metabolite metabolism/biosyn | 3 | 14.97% lower | 0.005 |
| 15 | (Q91WD5) NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial | NDUS2_MOUSE | 52,991 | 50,000 | 5.86 | 5.87 | Ox Phos | 2 | 18.83% lower | 0.007 |
| 16 | (Q9JK42) [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, mitochondrial | PDK2_MOUSE | 46,267 | 50,000 | 5.60 | 5.88 | Glucose metabolism | 2 | 13.64% lower | 0.053 |
| 17 | (Q9QYR9) Acyl-coenzyme A thioesterase 2, mitochondrial precursor (EC 3.1.2.2) | ACOT2_MOUSE | 49,849 | 48,000 | 6.14 | 6.10 | Metabolite metabolism/biosyn | 2 | 10.71% lower | 0.037 |
| 18 | (P51174) Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | ACADL_MOUSE | 48,277 | 47,000 | 6.50 | 6.35 | β-Oxidation | 3 | 14.46% lower | 0.008 |
| 19 | (P51174) Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | ACADL_MOUSE | 48,277 | 47,000 | 6.50 | 6.55 | β-Oxidation | 3 | 11.66% lower | 0.008 |
| 20 | (P51174) Long-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | ACADL_MOUSE | 48,277 | 47,000 | 6.50 | 6.80 | β-Oxidation | 3 | 15.90% lower | 0.010 |
| 21 | (Q6P8J7) Creatine kinase, sarcomeric mitochondrial precursor (EC 2.7.3.2) | KCRS_MOUSE | 47,811 | 49,000 | 7.72 | 7.90 | ATP regeneration | 3 | 8.42% lower | 0.011 |
| 22 | (Q99LC5) Electron transfer flavoprotein subunit α, mitochondrial precursor | ETFA_MOUSE | 35,272 | 35,000 | 8.44 | 7.00 | Electron transport | 3 | 10.10% lower | 0.003 |
| 23 | (Q60930) Voltage-dependent anion-selective channel protein 2 (VDAC-2) | VDAC2_MOUSE | 32,353 | 35,000 | 7.44 | 7.40 | Ion transport | 3 | 4.76% lower | 0.050 |
| 24 | (Q9D6J6) NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | NDUV2_MOUSE | 27,610 | 28,000 | 5.31 | 5.40 | Electron transport | 3 | 15.90% lower | 0.013 |
| 25 | (P11240) Cytochrome-c oxidase subunit 5A, mitochondrial precursor (cytochrome c) | COX5A_MOUSE | 16,347 | 15,000 | 5.01 | 5.15 | Ox Phos | 3 | 13.20% higher | 0.006 |
| 26 | (P99028) Cytochrome b-c1 complex subunit 6, mitochondrial precursor | QCR6_MOUSE | 10,713 | 11,000 | 4.84 | 5.15 | Ox Phos | 2 | 15.50% higher | 0.026 |
| 27 | (P97450) ATP synthase-coupling factor 6, mitochondrial precursor | ATP5J_MOUSE | 12,489 | 10,000 | 5.40 | 5.15 | Ox Phos | 3 | 14.20% higher | 0.044 |
| 28 | (P97450) ATP synthase-coupling factor 6, mitochondrial precursor | ATP5J_MOUSE | 12,489 | 10,000 | 5.40 | 5.47 | Ox Phos | 3 | 16.90% higher | 0.007 |
| 29 | (P56391) Cytochrome-c oxidase subunit Vlb isoform 1 (COX Vlb-1) | CX6B1_MOUSE | 10,293 | 10,000 | 8.91 | 8.50 | Ox Phos | 2 | 110.50% higher | 0.038 |
| 30 | (P62898) Cytochrome c, somatic | NDUA2_MOUSE | 11,023 | 11,000 | 9.61 | 9.15 | Ox Phos | 2 | 10.37% lower | 0.051 |
| 31 | (P56391) Cytochrome-c oxidase subunit Vlb isoform 1 (COX Vlb-1) | CX6B1_MOUSE | 10,293 | 10,000 | 8.91 | 9.20 | Ox Phos | 3 | 105.70% higher | 0.042 |
Spots were picked based on P < 0.05 from 3 replicates. Identifications were confirmed by dual mass spectrometry (MS/MS) in at least 2 separate gels. A spot identified in only 1 gel was confirmed if a spot in its near vicinity of the same protein was found in at least 2 separate gels. Data are expressed as % change in PC compared with control. Accession numbers are from the SwissProt/Uniprot database. Proteins for spots numbered 25–31 have the same trend as Control (Con)/GSK Inhib VIII gels. 2D-DIGE, 2-dimensional difference gel electrophoresis; biosyn, biosynthesis; Calc MW, calculated protein molecular weight; Calc pI, calculated protein isoelectric point for clipped protein [Expert Protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (SIB)]; Obs MW, observed protein molecular weight; Obs pI, observed protein isoelectric point; Ox Phos, oxidative phosphorylation. Mitochondrial proteins often have a mitochondrial targeting sequence that is processed/cleaved when entering the mitochondria, and these cleaved proteins are referred to as “clipped proteins.” The calculated pIs shown are for the processed clipped proteins.
Table 5.
Proteins with similar changes in PC and GSK Inhib VIII hearts
| Spot No. | Name | Accession No. | Calc MW | Obs MW | Calc pI | Obs pI | Function | Effect in PC | Effect in GSK Inhib VIII |
|---|---|---|---|---|---|---|---|---|---|
| 29, 67 | (P56391) Cytochrome-c oxidase subunit VIb isoform 1 (COX VIb-1). | CX6B1_MOUSE | 10,293 | 10,000 | 8.91 | 8.50, 8.15 | Ox Phos | 110.50% higher | 146.10% higher |
| 31, 69 | (P56391) Cytochrome-c oxidase subunit VIb isoform 1 (COX VIb-1) | CX6B1_MOUSE | 10,293 | 10,000 | 8.91 | 9.20, 9.10 | Ox Phos | 105.70% higher | 266.91% higher |
| 30, 68 | (P62898) Cytochrome c, somatic | NDUA2_MOUSE | 11,023 | 11,000 | 9.61 | 9.15, 9.00 | Ox Phos | 10.37% lower | 58.09% lower |
| 27, 65 | (P97450) ATP synthase-coupling factor 6, mitochondrial precursor | ATP5J_MOUSE | 12,489 | 10,000 | 5.40 | 5.15, 5.15 | Ox Phos | 14.20% higher | 25.90% higher |
| 28, 66 | (P97450) ATP synthase-coupling factor 6, mitochondrial precursor | ATP5J_MOUSE | 12,489 | 10,000 | 5.40 | 5.47, 5.40 | Ox Phos | 16.90% higher | 35.00% higher |
| 25, 64 | (P11240) Cytochrome-c oxidase subunit 5A, mitochondrial precursor | COX5A_MOUSE | 16347 | 15,000 | 5.01 | 5.15, 5.05 | Ox Phos | 13.20% higher | 20.40% higher |
| 26, 63 | (P99028) Cytochrome b-c1 complex subunit 6, mitochondrial precursor | QCR6_MOUSE | 10713 | 11,000 | 4.84 | 5.15, 5.00 | Ox Phos | 15.50% higher | 24.70% higher |
Spot numbers (1st no. PC, 2nd no. GSK Inhib VIII), accession numbers (SwissProt/Uniprot database), calculated and observed MW, calculated and observed pI (1st no. PC, 2nd no. GSK Inhib VIII), and protein function are shown. The calculated pIs shown are for the processed clipped proteins. Data are expressed as % change in PC or GSK Inhib VIII relative to Con.
Cytochrome c content.
Levels of cytochrome c were measured spectrophotometrically as previously described (2).
Blue Native-PAGE electrophoresis and immunoblotting.
Mitochondrial fractions were solubilized in Native-PAGE sample buffer with 1 mM phenylmethylsulfonyl fluoride, 2.1 mM leupeptin, and 1% digitonin. The lysate was centrifuged at 18,000 g, and protein concentrations were determined on the resulting supernatant. Solubilized mitochondria (75 μg) were loaded onto NativePAGE 3–12% gels with Dark Blue Cathode Buffer per manufacturer instructions (Invitrogen) and run until the dye front reached one-third down the gel. The buffer was switched to Light Blue Cathode Buffer until the dye front could no longer be seen. Gels were visualized on the ODYSSEY Infrared Imaging System (LI-COR) before transfer to polyvinylidene difluoride membranes. Membranes were blocked with Odyssey Blocking Buffer (LI-COR) and incubated with the primary antibody cytochrome-c oxidase subunit VIb (Mitosciences, Eugene, OR) at a 1:1,000 dilution. Appropriate secondary antibodies were used and visualized on the ODYSSEY Infrared Imaging System (LI-COR).
Data analysis.
All data are presented as means ± SE. Statistic analyses were performed with a one-way ANOVA analysis followed by a Tukey post hoc test. A value of P < 0.05 was considered significant.
RESULTS
Hemodynamic parameters.
In this study, we were interested in determining the changes in the mitochondrial proteome that are involved with cardioprotection. We chose the concentration of 1 μM GSK Inhib VIII because it was 10-fold higher than the IC50 value (104 nM) (4) and the lowest concentration we found to protect in our preliminary dosing study. There were no differences in LVDP, heart rate, or rate-pressure product between GSK Inhib VIII treatment and time-matched controls at the end of either the equilibration (Table 1) or treatment (Table 2) period. To test whether the concentration of GSK Inhib VIII used blocked GSK-3 activity, the amount of phosphorylated glycogen synthase was measured in control and GSK Inhib VIII-treated hearts (Fig. 1B). Because GSK-3 is a kinase that phosphorylates glycogen synthase, it would be expected that an inhibitor of GSK-3 activity would decrease the amount of phosphorylated glycogen synthase. As shown in Fig. 1, B and C, the 1 μM dose of GSK Inhib VIII used in these studies significantly decreased the percentage of phospho-glycogen synthase from ∼85% to ∼35% (P < 0.05), indicating that GSK Inhib VIII can inhibit GSK-3 activity.
Table 1.
Hemodynamic parameters
| End of Equilibration |
||||||
|---|---|---|---|---|---|---|
| n | Body wt, g | HR, bpm | LVDP, cmH2O | RPP (LVDP × HR) | Flow, ml/min | |
| Control | 8 | 27.0±0.8 | 378.8±10.9 | 122.4±7.5 | 46,301±2,938 | 2.6±0.3 |
| PC | 9 | 29.6±0.6 | 367.4±12.1 | 142.7±10.0 | 52,232±3,821 | 2.9±0.2 |
| GSK Inhib VIII | 9 | 26.1±0.9 | 349.3±10.5 | 127.1±10.5 | 44,633±3,972 | 2.6±0.3 |
Values are means ± SE for n mice. HR, heart rate; bpm, beats per minute; LVDP, left ventricular developed pressure; RPP, rate-pressure product; PC, ischemic preconditioning; GSK Inhib VIII, glycogen synthase kinase (GSK)-3 inhibitor AR-A014418.
Table 2.
Hemodynamic parameters
| End of Treatment |
End of 120-min Reperfusion |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| n | HR, bpm | LVDP, cm H2O | RPP (LVDP × HR) | Flow, ml/min | HR, bpm | LVDP, cm H2O | RPP (LVDP × HR) | Flow, ml/min | |
| Control | 8 | 358.3±12.7 | 40.8±3.0* | 14,447±872* | 1.8±0.3 | ||||
| PC | 9 | 369.3±24.6 | 105.7±13.2* | 37,699±3,119* | 2.5±0.3 | 326.6±8.4 | 72.2±4.4*‡ | 23,629±1,598*‡ | 1.8±0.3 |
| GSK Inhib VIII | 9 | 332.7±13.8 | 115.9±6.8 | 38,559±1,953 | 2.9±0.3 | 326.6±20.1 | 64.8±6.1*† | 21,176±2,302*† | 2.0±0.3 |
Values are means ± SE for n mice.
P < 0.05 vs. parameter of group at end of equilibration.
P < 0.05 vs. parameter of group at end of treatment;
P < 0.05 vs. parameter of Control at end of 120-min reperfusion.
GSK-3 inhibition is cardioprotective.
To test the hypothesis that GSK-3 inhibition is cardioprotective in mice, we treated hearts with 1 μM GSK Inhib VIII for 10 min before 20 min of ischemia and 120 min of reperfusion (Table 2). As shown in Fig. 1D, control hearts recovered 33.6 ± 2.4% of their preischemic LVDP. In contrast, PC- and GSK Inhib VIII-treated hearts had improved postischemic LVDP recovery (57.0 ± 2.5 and 51.7 ± 3.8%; P < 0.05). To evaluate whether GSK-3 inhibition reduced infarct size, we measured TTC staining after 2 h of reperfusion. Consistent with improved functional recovery, we observed decreased infarct size in PC- and GSK Inhib VIII-treated hearts compared with control (11.9 ± 1.6 and 19.2 ± 3.5 vs. 34.8 ± 1.9%; P < 0.05) (Fig. 1E). In addition, we also treated hearts with another GSK-3 inhibitor, CT99021 (0.4 μM), for 10 min before ischemia and found similar improvement in LVDP recovery and infarct size (51.6 ± 4.6% LVDP and 11.5 ± 2.7% infarct; P < 0.05; Fig. 1, D and E).
Murry and colleagues (26) first observed that PC delayed cell injury while reducing lactate production during sustained ischemia. We have also found that a number of cardioprotective agents, such as diazoxide (9), Bcl-2 overexpression (17), and adenosine (10), also result in a decrease in ischemic acidosis or lactate production. We measured lactate levels in control and PC- and GSK Inhib VIII-treated hearts after 20 min of ischemia to evaluate whether GSK Inhib VIII treatment slowed anaerobic glycolysis during ischemia. As shown in Fig. 1F, lactate levels were significantly lower in PC- and GSK Inhib VIII-treated hearts compared with control (6.69 ± 0.73 and 8.30 ± 0.29 vs. 10.34 ± 0.67 μmol lactate/g wet wt; P < 0.05). Together, these results demonstrate that GSK-3 inhibition protects mice from I/R injury to a similar extent as PC by increasing functional recovery, reducing infarct size, and decreasing lactate production during ischemia.
PC and GSK-3 inhibitor treatment alter proteins involved in mitochondrial energetics.
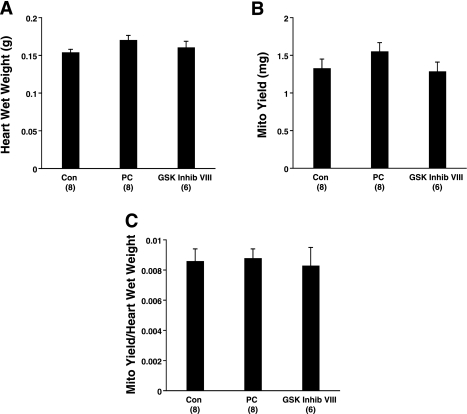
Because cardioprotection is thought to involve alterations in the mitochondria, we were interested in determining whether consistent changes in the mitochondrial proteome occur in PC- and GSK Inhib VIII-treated hearts at the end of the treatment period. We reasoned that since these different treatments are both cardioprotective, common proteomic changes are likely to be involved in protection. Mitochondrial fractions were isolated at the end of the treatment period for all sample groups. In our mitochondrial isolation, the wet weight of the hearts (Fig. 2A), mitochondrial content (Fig. 2B), and mitochondrial yield normalized to heart wet weight (Fig. 2C) were comparable between all three treatment groups. To ensure that there was similar integrity of the outer mitochondrial membrane between the treatment groups, cytochrome c-dependent oxygen consumption was measured, and we did not find a significant difference in state 3 consumption on the addition of cytochrome c, suggesting similar outer membrane integrity between all treatment groups.
Fig. 2.
Mitochondrial yield is similar between all treatment groups. A: wet weights (g) of hearts from Con, PC, and GSK Inhib VIII treatment groups. B: mitochondrial (Mito) content (mg protein) from Con, PC, and GSK Inhib VII hearts. C: mitochondrial yield normalized to heart wet weight for all treatment groups.
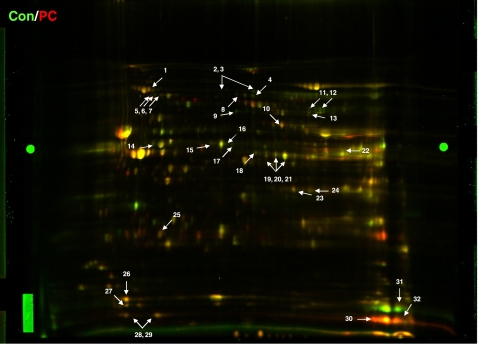
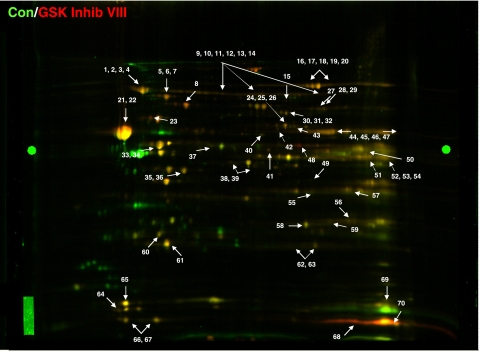
The isolated mitochondrial fractions were resolved by 2D-DIGE, and we performed a comparative proteomic analysis of control and PC mitochondria (Fig. 3) as well as control and GSK Inhib VIII mitochondria (Fig. 4). We used Progenesis software to identify spots from either PC- or GSK Inhib VIII-treated hearts that had significant changes (P < 0.05) relative to control. Protein identifications for each spot were confirmed in at least two gels with a minimum of two unique peptides. Table 3 summarizes the proteins with significant differences between control and PC hearts. A total of 23 unique protein spots were significantly altered between control and PC. The majority of the identified changes occurred in proteins involved in mitochondrial energetics, such as oxidative phosphorylation, β-oxidation, metabolism, and ATP regeneration. GSK Inhib VIII treatment led to the modification of 38 distinct proteins that spanned a wide range of cellular functions (Table 4).
Fig. 3.
Mitochondrial proteome comparing Control and PC. Representative 2-dimensional difference gel electrophoresis (2D-DIGE) image of proteins from Con (labeled green, Cy3) and PC (labeled red, Cy5) mitochondria. All experiments were performed in triplicate, and arrows indicate proteins of interest. Bright green circles midway in the gel on far left and far right are anchor markers used for spot picking.
Fig. 4.
Mitochondrial proteome comparing control and GSK Inhib VIII. Representative 2D-DIGE image of mitochondrial pellets from Con (labeled green, Cy3) and GSK Inhib VIII (labeled red, Cy5) hearts. All experiments were performed in triplicate, and arrows indicate proteins of interest. Bright green circles midway in the gel on far left and far right are anchor markers used for spot picking.
Table 4.
Con/GSK Inhib VIII 2D-DIGE protein differences
| Spot No. | Name | Accession No. | Calc MW | Obs MW | Calc pI | Obs pI | Function | Gels IDed | Effect in GSK Inhib VIII | P |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | (Q91VD9) NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial precursor | NDUS1_MOUSE | 80,724 | 80,000 | 5.24 | 5.12 | Protein transport/folding | 3 | 41.90% higher | 0.011 |
| 2 | (Q91VD9) NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial precursor | NDUS1_MOUSE | 80,724 | 80,000 | 5.24 | 5.15 | Ox Phos | 3 | 30.30% higher | 0.013 |
| 3 | (Q91VD9) NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial precursor | NDUS1_MOUSE | 80,724 | 80,000 | 5.24 | 5.17 | Ox Phos | 3 | 36.70% higher | 0.010 |
| 4 | (Q91VD9) NADH-ubiquinone oxidoreductase 75-kDa subunit, mitochondrial precursor | NDUS1_MOUSE | 80,724 | 80,000 | 5.24 | 5.22 | Ox Phos | 3 | 36.20% higher | 0.007 |
| 5 | (P38647) Stress-70 protein, mitochondrial precursor | GRP75_MOUSE | 73,768 | 75,000 | 5.24 | 5.42 | Protein transport/folding | 3 | 44.20% higher | 0.015 |
| 6 | (P38647) Stress-70 protein, mitochondrial precursor | GRP75_MOUSE | 73,768 | 75,000 | 5.50 | 5.50 | Protein transport/folding | 3 | 43.70% higher | 0.003 |
| 7 | (P38647) Stress-70 protein, mitochondrial precursor | GRP75_MOUSE | 73,768 | 75,000 | 5.50 | 5.55 | Protein transport/folding | 3 | 45.20% higher | 0.039 |
| 8 | (Q8BMF4) Dihydrolipoyllysine-residue acetyltransferase component of pyruvate | ODP2_MOUSE | 68,469 | 70,000 | 5.70 | 5.62 | Glucose metabolism | 3 | 39.50% higher | 0.025 |
| 9 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 5.82 | β-Oxidation | 2 | 46.80% higher | 0.020 |
| 10 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 5.92 | β-Oxidation | 1 | 48.30% higher | 0.012 |
| 11 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 6.05 | β-Oxidation | 3 | 48.30% higher | 0.011 |
| 12 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 6.25 | β-Oxidation | 2 | 34.00% higher | 0.007 |
| 13 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 7.15 | β-Oxidation | 2 | 45.20% higher | 0.002 |
| 14 | (Q8BMS1) Trifunctional enzyme subunit α, mitochondrial precursor (TP-α) | ECHA_MOUSE | 83,302 | 80,000 | 9.06 | 7.60 | β-Oxidation | 2 | 31.80% higher | 0.001 |
| 15 | (Q8CAQ8) Mitochondrial inner membrane protein (Mitofilin). | IMMT_MOUSE | 84,247 | 75,000 | 6.18 | 6.90 | Mitochondria morphology | 3 | 111.60% higher | 0.006 |
| 16 | (Q99KI0) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) | ACON_MOUSE | 86,121 | 85,000 | 7.40 | 7.25 | TCA cycle | 3 | 34.50% higher | 0.011 |
| 17 | (Q99KI0) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) | ACON_MOUSE | 86,121 | 85,000 | 7.40 | 7.40 | TCA cycle | 3 | 31.30% higher | 0.010 |
| 18 | (Q99KI0) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) | ACON_MOUSE | 86,121 | 85,000 | 7.40 | 7.50 | TCA cycle | 3 | 32.80% higher | 0.026 |
| 19 | (Q99KI0) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) | ACON_MOUSE | 86,151 | 85,000 | 7.40 | 7.55 | TCA cycle | 3 | 32.00% higher | 0.022 |
| 20 | (Q99KI0) Aconitate hydratase, mitochondrial precursor (EC 4.2.1.3) | ACON_MOUSE | 86,121 | 85,000 | 7.40 | 7.77 | TCA cycle | 2 | 27.90% higher | 0.018 |
| 21 | (P56480) ATP synthase subunit β, mitochondrial precursor (EC 3.6.3.14) | ATPB_MOUSE | 56,265 | 55,000 | 4.99 | 5.00 | Ox Phos | 3 | 62.50% higher | 0.016 |
| 22 | (P56480) ATP synthase subunit β, mitochondrial precursor (EC 3.6.3.14) | ATPB_MOUSE | 56,265 | 55,000 | 4.99 | 5.05 | Ox Phos | 3 | 48.70% higher | 0.005 |
| 23 | (P63038) 60-kDa heat shock protein, mitochondrial precursor (heat shock protein) | CH60_MOUSE | 61,088 | 65,000 | 5.35 | 5.32 | Protein transport/folding | 3 | 47.80% higher | 0.017 |
| 24 | (Q9D2G2) Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | ODO2_MOUSE | 49,236 | 55,000 | 5.98 | 5.35 | Glucose metabolism | 3 | 23.20% higher | 0.032 |
| 25 | (O08749) Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) | DLDH_MOUSE | 54,751 | 55,000 | 6.43 | 6.60 | Glucose metabolism | 3 | 29.20% higher | 0.012 |
| 26 | (O08749) Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) | DLDH_MOUSE | 54,751 | 55,000 | 6.43 | 6.85 | Glucose metabolism | 3 | 27.70% higher | 0.026 |
| 27 | (P50544) Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADV_MOUSE | 71,230 | 72,000 | 7.72 | 7.60 | β-Oxidation | 3 | 17.80% higher | 0.045 |
| 28 | (P47934) Carnitine O-acetyltransferase (EC 2.3.1.7) (carnitine acetylase) (CAT) | CACP_MOUSE | 71,335 | 72,000 | 8.52 | 7.65 | β-Oxidation | 1 | 23.90% higher | 0.011 |
| 29 | (P47934) Carnitine O-acetyltransferase (EC 2.3.1.7) (carnitine acetylase) (CAT) | CACP_MOUSE | 71,335 | 72,000 | 8.52 | 7.70 | β-Oxidation | 2 | 20.30% higher | 0.033 |
| 30 | (Q921G7) Electron transfer flavoprotein-ubiquinone oxidoreductase | ETFD_MOUSE | 68,903 | 70,000 | 6.47 | 6.75 | Electron transport | 3 | 26.70% higher | 0.002 |
| 31 | (Q921G7) Electron transfer flavoprotein-ubiquinone oxidoreductase | ETFD_MOUSE | 68,903 | 70,000 | 6.47 | 6.80 | Electron transport | 3 | 21.90% higher | 0.003 |
| 32 | (Q921G7) Electron transfer flavoprotein-ubiquinone oxidoreductase | ETFD_MOUSE | 68,903 | 70,000 | 6.47 | 6.85 | Electron transport | 1 | 21.20% higher | 0.006 |
| 33 | (Q9CZ13) Cytochrome b-c1 complex subunit 1, mitochondrial precursor | QCR1_MOUSE | 53,420 | 50,000 | 5.28 | 5.42 | Ox Phos | 3 | 20.90% higher | <0.001 |
| 34 | (Q9CZ13) Cytochrome b-c1 complex subunit 1, mitochondrial precursor | QCR1_MOUSE | 53,420 | 50,000 | 5.28 | 5.52 | Ox Phos | 3 | 35.10% higher | 0.011 |
| 35 | (Q9D6R2) Isocitrate dehydrogenase [NAD] subunit α, mitochondrial precursor | IDH3A_MOUSE | 40,044 | 40,000 | 5.60 | 5.60 | TCA cycle | 3 | 15.20% higher | 0.014 |
| 36 | (P29758) Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) | OAT_MOUSE | 48,400 | 50,000 | 5.73 | 5.75 | Metabolite metabolism/biosyn | 3 | 13.30% higher | 0.006 |
| 37 | (Q99LC3) NADH dehydrogenase [ubiquinone] 1 α subcomplex subunit 10 | NDUAA_MOUSE | 40,863 | 40,000 | 5.96 | 5.88 | Ox Phos | 3 | 19.50% higher | 0.005 |
| 38 | (Q99LC3) NADH dehydrogenase [ubiquinone] 1 α subcomplex subunit 10 | NDUAA_MOUSE | 40,863 | 40,000 | 5.96 | 5.98 | Ox Phos | 2 | 36.00% higher | <0.001 |
| 39 | (P47738) Aldehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.3) | ALDH2_MOUSE | 57,015 | 55,000 | 5.96 | 6.25 | Metabolite metabolism/biosyn | 3 | 11.80% higher | 0.017 |
| 40 | (Q8BFR5) Elongation factor Tu, mitochondrial precursor | EFTU_MOUSE | 49,876 | 50,000 | 6.20 | 6.45 | Protein biosynthesis | 3 | 29.00% higher | 0.037 |
| 41 | (Q8BGH2) Sorting and assembly machinery component 50 homolog | SAM50_MOUSE | 52,230 | 57,000 | 6.34 | 6.70 | Protein transport/folding | 3 | 25.30% higher | 0.044 |
| 42 | (Q60936) Chaperone activity of bc1 complex-like, mitochondrial precursor | ADCK3_MOUSE | 72,750 | 65,000 | 6.03 | 7.05 | Protein transport/folding | 2 | 16.00% higher | 0.009 |
| 43 | (Q03265) ATP synthase subunit α, mitochondrial precursor | ATPA_MOUSE | 59,830 | 60,000 | 8.28 | 7.80 | Ox Phos | 3 | 32.60% higher | 0.014 |
| 44 | (Q03265) ATP synthase subunit α, mitochondrial precursor\ | ATPA_MOUSE | 59,830 | 60,000 | 8.28 | 8.00 | Ox Phos | 3 | 36.60% higher | 0.033 |
| 45 | (Q03265) ATP synthase subunit α, mitochondrial precursor | ATPA_MOUSE | 59,830 | 60,000 | 8.28 | 8.75 | Ox Phos | 3 | 37.50% higher | 0.015 |
| 46 | (Q03265) ATP synthase subunit α, mitochondrial precursor | ATPA_MOUSE | 59,830 | 60,000 | 8.28 | 9.05 | Ox Phos | 1 | 31.70% higher | 0.007 |
| 47 | (P35486) Pyruvate dehydrogenase E1 component subunit α, somatic form | ODPA_MOUSE | 43,888 | 50,000 | 6.78 | 7.25 | Glucose metabolism | 3 | 34.50% higher | 0.049 |
| 48 | (P54071) Isocitrate dehydrogenase [NADP], mitochondrial precursor (EC 1.1.1.42) | IDHP_MOUSE | 51,391 | 45,000 | 8.49 | 8.75 | TCA cycle | 3 | 14.40% higher | 0.008 |
| 49 | (Q6P8J7) Creatine kinase, sarcomeric mitochondrial precursor (EC 2.7.3.2) | KCRS_MOUSE | 47,811 | 48,000 | 7.72 | 8.50 | ATP regeneration | 2 | 14.90% higher | 0.003 |
| 50 | (Q8QZT1) Acetyl-CoA acetyltransferase, mitochondrial precursor (EC 2.3.1.9) | THIL_MOUSE | 45,129 | 47,000 | 8.18 | 8.50 | Metabolite metabolism/biosyn | 2 | 15.50% higher | <0.001 |
| 51 | (Q8BWT1) 3-Ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) | THIM_MOUSE | 42,288 | 47,000 | 8.33 | 8.75 | β-Oxidation | 1 | 16.40% higher | <0.001 |
| 52 | (Q8BWT1) 3-Ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) | THIM_MOUSE | 42,288 | 47,000 | 8.33 | 8.90 | β-Oxidation | 1 | 18.60% higher | 0.018 |
| 53 | (Q8BWT1) 3-Ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) | THIM_MOUSE | 42,288 | 47,000 | 8.33 | 9.05 | β-Oxidation | 3 | 14.60% higher | 0.006 |
| 54 | (Q60930) Voltage-dependent anion-selective channel protein 2 (VDAC-2) | VDAC2_MOUSE | 32,353 | 35,000 | 7.44 | 7.50 | Ion transport | 2 | 7.00% higher | 0.054 |
| 55 | (Q9DCW4) Electron transfer flavoprotein subunit β (β-ETF) | ETFB_MOUSE | 27,834 | 30,000 | 8.24 | 8.00 | Electron transport | 2 | 16.00% higher | 0.019 |
| 56 | (Q61425) Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial precursor | HCDH_MOUSE | 34,613 | 35,000 | 8.26 | 8.25 | β-Oxidation | 2 | 14.10% higher | 0.051 |
| 57 | (Q9CR68) Cytochrome b-c1 complex subunit Rieske, mitochondrial precursor | UCRI_MOUSE | 29,634 | 30,000 | 8.91 | 7.35 | Ox Phos | 3 | 16.80% higher | 0.007 |
| 58 | (Q8BH95) Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) | ECHM_MOUSE | 31,853 | 30,000 | 6.98 | 7.80 | β-Oxidation | 3 | 21.90% higher | 0.001 |
| 59 | (Q9D6J6) NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | NDUV2_MOUSE | 27,610 | 28,000 | 7.78 | 5.42 | Ox Phos | 3 | 18.60% higher | 0.003 |
| 60 | (Q9DCX2) ATP synthase subunit d, mitochondrial | ATP5H_MOUSE | 18,795 | 27,000 | 5.31 | 5.50 | Ox Phos | 3 | 21.30% higher | 0.003 |
| 61 | (P09671) Superoxide dismutase [Mn], mitochondrial precursor (EC 1.15.1.1) | SODM_MOUSE | 24,816 | 25,000 | 5.52 | 7.10 | ROS maintenance | 2 | 19.40% higher | 0.025 |
| 62 | (P09671) Superoxide dismutase [Mn], mitochondrial precursor (EC 1.15.1.1) | SODM_MOUSE | 24,816 | 25,000 | 7.30 | 7.55 | ROS maintenance | 3 | 9.50% higher | 0.058 |
| 63 | (P99028) Cytochrome b-c1 complex subunit 6, mitochondrial precursor | QCR6_MOUSE | 10,713 | 11,000 | 4.84 | 5.00 | Ox Phos | 2 | 24.70% higher | 0.017 |
| 64 | (P11240) Cytochrome-c oxidase subunit 5A, mitochondrial precursor | COX5A_MOUSE | 16,347 | 15,000 | 5.01 | 5.05 | Ox Phos | 3 | 20.40% higher | 0.016 |
| 65 | (P97450) ATP synthase-coupling factor 6, mitochondrial precursor | ATP5J_MOUSE | 12,489 | 10,000 | 5.40 | 5.15 | Ox Phos | 3 | 25.90% higher | <0.001 |
| 66 | (P97450) ATP synthase-coupling factor 6, mitochondrial precursor | ATP5J_MOUSE | 12,489 | 10,000 | 5.40 | 5.40 | Ox Phos | 2 | 35.00% higher | 0.002 |
| 67 | (P56391) Cytochrome-c oxidase subunit VIb isoform 1 (COX VIb-1) | CX6B1_MOUSE | 10,293 | 10,000 | 8.91 | 8.15 | Ox Phos | 2 | 146.10% higher | 0.054 |
| 68 | (P62898) Cytochrome c, somatic | NDUA2_MOUSE | 11,023 | 11,000 | 9.61 | 9.00 | Ox Phos | 2 | 58.09% lower | 0.038 |
| 69 | (P56391) Cytochrome-c oxidase subunit VIb isoform 1 (COX VIb-1) | CX6B1_MOUSE | 10,293 | 10,000 | 8.91 | 9.10 | Ox Phos | 2 | 266.91% higher | 0.053 |
Spots were picked based on P < 0.05 from 3 replicates. Identifications were confirmed by MS/MS in at least 2 separate gels. A spot identified in only 1 gel was confirmed if a spot in its near vicinity of the same protein was found in at least 2 separate gels. Data are expressed as %change in GSK Inhib VIII compared with Con. Accession numbers are from the SwissProt/Uniprot database. Proteins for spots numbered 63–69 have the same trend as Con/PC gels. The calculated pIs shown are for the processed clipped proteins.
We were particularly interested in changes that were consistent between PC and GSK-3 inhibition. We examined Tables 3 and 4 for protein spots that were higher or lower (i.e., the same trend) in both PC and GSK Inhib VIII treatment because these changes could be responsible for cardioprotection. Although seven protein spots were found to have the same trends in PC- and GSK Inhib VIII-treated hearts (Table 5), these spots were identified as only five distinct proteins because of multiple spots with the same identification (i.e., cytochrome-c oxidase subunit VIb and ATP synthase-coupling factor 6). PC and GSK Inhib VIII treatment both increased the protein abundance of cytochrome-c oxidase subunits Va and VIb, ATP synthase-coupling factor 6, and cytochrome b-c1 complex subunit 6, while cytochrome c levels were decreased.
Five spots at different pI values were identified for NADH-ubiquinone 75 kDa; these five spots likely represent differences in posttranslational modifications (Figs. 3 and 4). Interestingly, we found that treatment with GSK Inhib VIII increased the levels of NADH-ubiquinone 75 kDa at four spots located at more acidic pIs (Fig. 4, Table 4), whereas PC decreased the level of NADH-ubiquinone 75 kDa at a spot with a more basic pI (Fig. 3, Table 3). Together, these data would be consistent with cardioprotection (GSK-3 inhibitor treatment and PC) resulting in an increase in posttranslational modification (such as phosphorylation) of NADH-ubiquinone 75 kDa; this would be apparent as an increase in a spot at the more acidic pI as observed with GSK-3 inhibition and a decrease in a spot at the more basic pI as observed in PC.
To better illustrate protein levels in the DIGE data, we magnified DIGE overlay images of the five proteins for control/PC and control/GSK Inhib VIII (Fig. 5, A–C, left) and compared them to DIGE single (nonoverlay) images for each treatment group (Fig. 5, A–C, middle). With the exception of cytochrome c, the DIGE overlays show an increase of the proteins in Table 5 as evident by the darker protein densities in the PC and GSK Inhib VIII single DIGE images. In summary, PC and GSK-3 inhibition both lead to consistent changes in protein abundance of several electron transport chain components, namely complexes III, IV, and V.
Fig. 5.
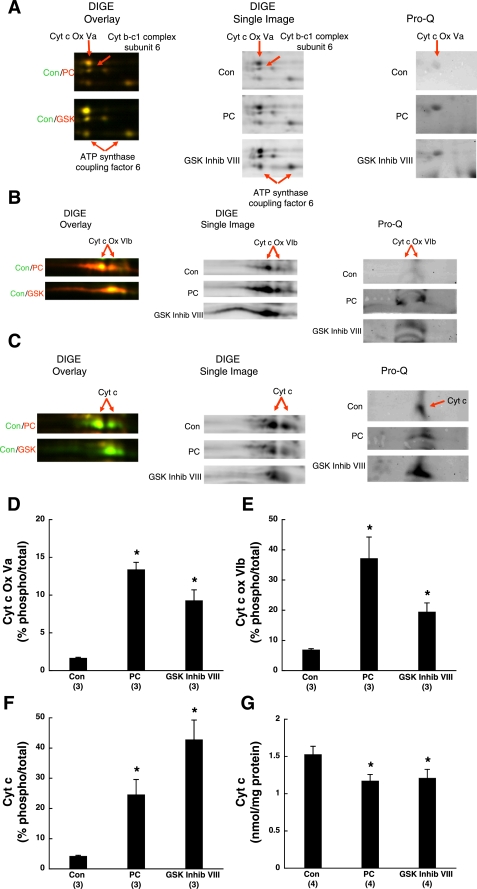
Expression levels and phosphorylation changes for proteins of interest. A–C: magnified DIGE overlay (Con/PC or Con/GSK Inhib VIII) (left), DIGE single images (center), and Pro-Q images (right) of cytochrome-c oxidase (Cyt c Ox) subunit Va (A), cytochrome-c oxidase subunit VIb (B), and cytochrome c (Cyt c; C) from Con, PC, and GSK Inhib VIII hearts. Red arrows indicate proteins of interest. D–F: densitometry of Pro-Q images for cytochrome-c oxidase subunit Va (D); cytochrome-c oxidase subunit VIb (E), and cytochrome c (F) from control, PC, and GSK Inhib VIII hearts. Pro-Q phosphoprotein peak volumes were normalized to Sypro Ruby total protein peak volumes, and % changes are shown. G: Cyt c content of Con, PC, and GSK Inhib VIII hearts. *P < 0.05 compared with Con.
Alterations of protein amount in PC and GSK-3 inhibitor treatment likely due to changes in protein degradation.
The identified differences in protein subunits of complexes III–V in PC- and GSK Inhib VIII-treated hearts could be attributed to several factors, such as protein synthesis, protein degradation, or posttranslational modification that caused a pI shift. Protein synthesis during the 10-min GSK-3 inhibitor treatment or the 40-min PC protocol would have been minimal; therefore, the changes observed would be more likely due to degradation or posttranslational modifications, such as phosphorylation. To determine whether PC and GSK Inhib VIII were associated with phosphorylation changes, we imaged 2D gels with Pro-Q Diamond phosphoprotein stain. Pro-Q has been used extensively to directly detect phosphate groups attached to Tyr, Ser, or Thr residues and will tend to select for proteins with high stable phosphorylation levels (37, 38). The dye has been shown to accurately quantify protein loads ranging from 1 ng to 1 μg on an electrophoresis gel. Relative to control, PC- and GSK Inhib VIII-treated hearts had increased Pro-Q staining for cytochrome-c oxidase subunits Va (Fig. 5A, right) and VIb (Fig. 5B, right) and cytochrome c (Fig. 5C, right), suggesting enhanced phosphorylation of these proteins.
To investigate whether these increases in phosphorylation were associated with alterations in protein expression, the volume of each Pro-Q spot was normalized to the volume of the Sypro Ruby total protein spot as would be performed in Western blot densitometry. The percentage of phosphorylated cytochrome-c oxidase subunit Va to total protein was elevated from 1.68 ± 0.08% to 13.40 ± 0.94% in PC (P < 0.05; ∼8-fold change) and 9.30 ± 1.40% in GSK Inhib VIII (P < 0.05; ∼5.5-fold change) (Fig. 5D). In a similar fashion, the percentage of phospho-cytochrome-c oxidase subunit VIb increased from 6.93 ± 0.35% in control to 37.20 ± 7.02% in PC (P < 0.05; ∼5-fold change) and 19.48 ± 2.92% in GSK Inhib VIII (P < 0.05; ∼3-fold change) (Fig. 5E). This would suggest that cardioprotection through PC and GSK-3 inhibition increased the phosphorylation levels of these complex IV subunits, implicating a possible role of phosphorylation in protein stability since these protein levels were increased. Interestingly, we also observed an increase in the percentage of phosphorylated cytochrome c from 4.27 ± 0.21% in control to 24.61 ± 5.05% in PC (P < 0.05; ∼5.5-fold change) and 42.82 ± 6.42% in GSK Inhib VIII (P < 0.05; ∼10-fold change) (Fig. 5F). This would be consistent with a role for increased phosphorylation of cytochrome c resulting in an increase in its degradation since cytochrome c levels were decreased. To confirm the decrease in cytochrome c in PC- and GSK Inhib VIII-treated hearts observed in the proteomics data, we measured cytochrome c content by spectrophotometric methods and found a ∼20–25% decrease in cytochrome c in PC and GSK Inhib VIII treatment (Fig. 5G; P < 0.05). This result would be consistent with cytochrome c phosphorylation playing a role in its stability. In contrast, we did not observe Pro-Q staining for cytochrome b-c1 complex subunit 6 or ATP synthase-coupling factor 6, which would suggest that these proteins were not modified by phosphorylation, but there could be other posttranslational modifications or a change in protein amount.
PC and GSK-3 inhibitor cardioprotection leads to change in complex subunit assembly into mitochondrial supercomplexes.
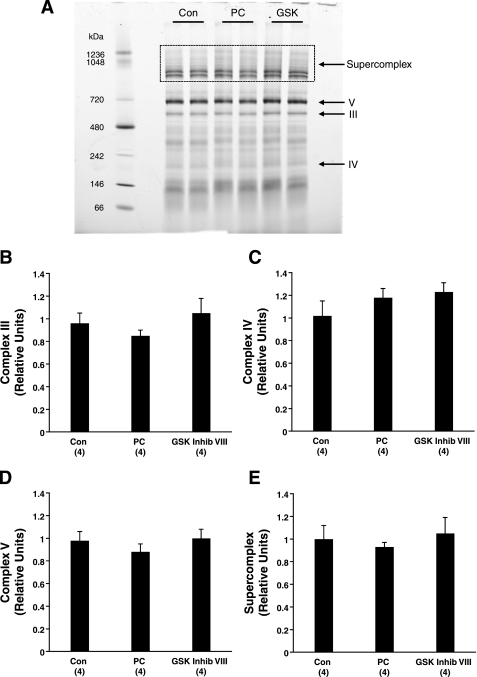
We were interested in the functional consequences of the increased levels of specific subunits of complexes III–V (Table 5). To distinguish whether the changes in subunit amount (and/or phosphorylation) increased the complex formation or supercomplex levels/composition, we used Blue Native (BN)-PAGE gels to separate complexes in their native form and allow for quantification of the total complex amount (Fig. 6A). We found that the complex amounts were comparable between the treatment groups (Fig. 6, B–D). Therefore, the increases detected in the subunits of Table 5 did not alter total complex amount.
Fig. 6.
Content of complexes III, IV, V, and supercomplex are similar between treatment groups. A: representative Blue Native (BN)-PAGE gel of mitochondria isolated from Con, PC, and GSK Inhib VIII (GSK) hearts. III, IV, V, complex III, IV, and V bands; box, mitochondrial supercomplex. B–E: densitometry of the amounts of complex III (B), complex IV (C), complex V (D), and supercomplex (E) in relative units.
We also evaluated the BN-PAGE gels for any changes in the amount of mitochondrial supercomplexes, which are formed between complexes I, III, and IV and often referred to as respirasomes (Fig. 6A) (34, 35). Typically, supercomplexes contain complex I, a dimer of complex III, and zero to four units of complex IV, where higher-molecular-weight supercomplexes contain more units of complex IV. Similar to the individual complexes, we did not observe a change in supercomplex amount in PC- and GSK Inhib VIII-treated hearts (Fig. 6E).
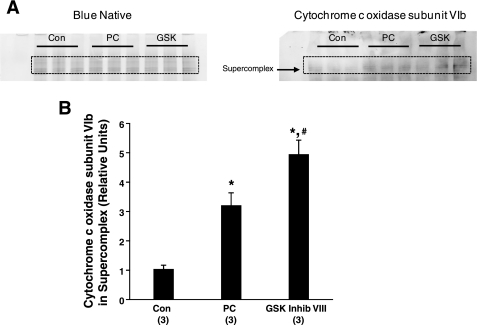
Although supercomplex levels were unaltered, we were interested in whether PC and GSK-3 inhibition and the resultant increase in subunit levels may lead to differences in supercomplex composition. We measured levels of cytochrome-c oxidase subunit VIb, which was increased by PC and GSK-3 inhibition, in the supercomplex and found approximately threefold and approximately fivefold increases in the levels of the subunit in PC and GSK Inhib VIII mitochondrial supercomplexes, respectively (Fig. 7; P < 0.05). This would suggest that the increase in subunit levels by these cardioprotective treatments may lead to changes in supercomplex assembly.
Fig. 7.
Levels of cytochrome-c oxidase subunit VIb are elevated in PC and GSK Inhib VIII mitochondrial supercomplexes. A: BN-PAGE gel of mitochondrial fractions from Con, PC, and GSK Inhib VIII-treated hearts and BN-Western blot for cytochrome-c oxidase (complex IV) subunit VIb. The supercomplex is indicated by the box. B: densitometry of cytochrome-c oxidase subunit VIb content in the mitochondrial supercomplex. *P < 0.05 compared with Con; #P < 0.05 compared with PC.
DISCUSSION
The primary goal of this study was to identify the alterations in the mitochondrial proteome that are consistent in two approaches, PC and GSK-3 inhibition. Preischemic treatment with GSK-3 inhibitors mimics PC cardioprotection (13, 41). In addition, various cardioprotective drugs result in GSK-3 inhibition (13, 27), and GSK-3 seems to serve as a convergence point for many signaling pathways (18). Previous work by our group (41) and others (12, 13, 18) demonstrated that PC and other forms of cardioprotection lead to increased GSK-3 phosphorylation and inactivation through a PI3-kinase-Akt-dependent pathway. However, in contrast to these studies, a recent study found that GSK-3 inactivation was not required for cardioprotection (28). Possible reasons for this discrepancy have been discussed elsewhere (25).
PC can also result in GSK-3 phosphorylation through pathways other than Akt because GSK-3 serves as the convergence point for several protein kinases such as Erk, p70S6 kinase, and protein kinase C (PKC) (8, 19). GSK-3 is also known to be phosphorylated in signaling mechanisms other than PC, such as in insulin receptor activation, which mainly acts through Akt to phosphorylate GSK-3 (24). However, it is unclear whether the localization of GSK-3 following PC and insulin stimulation are the same. Also, PC and insulin likely result in activation of different signaling pathways that could alter the context in which GSK-3 is phosphorylated. Additional studies will be needed to directly compare the effect of insulin to PC.
Consistent with earlier studies revealing the importance of GSK-3 inhibition in preconditioned rats (13, 41), we find in mice that GSK-3 inhibitors GSK Inhib VIII and CT99021 provide protection similar to PC with increased functional recovery and reduced infarct size. The protection afforded by PC and pharmacological preconditioning, such as GSK-3 inhibition, has been suggested to involve the mitochondria. We therefore used a proteomics approach to determine whether cardioprotection with PC and a GSK-3 inhibitor results in consistent changes in the mitochondrial proteome. We were interested in identifying changes in posttranslational modifications of mitochondrial proteins. We were concerned that these modifications would be lost during the extensive isolation needed to purify mitochondria, and thus we used rapid isolation techniques to obtain crude mitochondrial extracts for our DIGE proteomics. A proteomics approach has the advantage of providing an unbiased assessment of changes in protein levels and posttranslational modifications. However, a limitation of these proteomic studies is that hydrophobic and membrane proteins often have difficulty entering 2D gels. Thus there could be other changes in hydrophobic and membrane proteins that are not detected.
Overall, there was little overlap in protein changes between PC and GSK-3 inhibition because we identified only five proteins with consistent changes between PC and GSK-3 inhibition (Table 5). Similarly, Arrell et al. (1) also found only six similar protein changes between cardioprotection with adenosine and diazoxide in rabbit ventricular myocytes. Interestingly, PC primarily leads to a reduction in mitochondrial protein expression (Table 3) and GSK-3 inhibition leads to an increase in expression in most proteins (Table 4). PC includes multiple ischemic episodes, and ischemia might be expected to increase degradation. Furthermore, modifications of protein stability have been associated with PC, specifically in the case of the PTEN phosphatase. PTEN negatively regulates the serine-threonine kinase Akt that is a key player in PC signaling, and this phosphatase is rapidly degraded in ischemic hearts. Cai and Semenza (5) showed that pretreatment of perfused rat hearts with a proteasome inhibitor blocks ischemia-induced PTEN degradation and PC. Thus these results demonstrate that degradation and the resulting loss of activity of PTEN is necessary for the induction of PC.
In contrast to the decrease in protein abundance observed with PC, treatment with the GSK inhibitor resulted primarily in an increase in protein abundance. This finding might be explained if GSK-3 phosphorylation resulted in reduced protein stability; thus inhibition of GSK-3 would increase protein stability. In fact, GSK-3 has been implicated in the modulation of protein stability such as in the Wnt pathway and its downstream target β-catenin (3, 7).
When comparing the protein abundance changes that were the result of PC and GSK-3 inhibition, we identified seven spots representing five proteins with consistent alterations between the cardioprotective treatments. Four of the five proteins showed an increased abundance with PC and GSK inhibitor treatment; these include cytochrome-c oxidase subunits Va and VIb, ATP synthase-coupling factor 6, and cytochrome b-c1 complex. We also found one protein (cytochrome c) that was reduced in PC- and GSK inhibitor-treated hearts. On the basis of Pro-Q Diamond staining, it appears that several of these proteins (cytochrome-c oxidase subunits Va and VIb and cytochrome c) showed an increase in phosphorylation or some other posttranslational modification. Thus the changes in protein abundance occur with concomitant changes in phosphorylation. It is interesting to speculate that the changes in phosphorylation of these proteins might alter their stability. Alterations in mitochondrial protein abundance in PC- and GSK Inhib VIII-treated hearts could result from changes in protein synthesis and/or degradation. Protein synthesis during the 10-min GSK inhibitor treatment or the 40-min PC protocol would have been minimal. In addition, Thornton et al. (39) demonstrated that PC cardioprotection is not blocked by inhibitors of protein synthesis, and thus it is unlikely that synthesis of a protective protein is the mechanism of PC protection. Therefore, the protein abundance changes induced by PC and GSK-3 inhibition would be more likely due to alterations in protein degradation or stability, and changes in phosphorylation are well known to influence protein stability.
An example of how phosphorylation may alter protein stability is our observed decrease in cytochrome c with PC and GSK-3 inhibition (Table 5), which was also confirmed by spectrophotometric methods (Fig. 5G). Although a reduction in mitochondrial cytochrome c would usually be interpreted as a detrimental event coinciding with an increase of cytosolic cytochrome c and other apoptotic-inducing factors, it is likely that cytochrome c levels in the intermembrane space can also be regulated, perhaps by phosphorylation. The decrease we observe in cytochrome c in PC- and GSK inhibitor-treated hearts is not associated with increased injury, because these treatments protect against I/R injury (Fig. 1). We measured cytochrome c content in the corresponding cytosolic fractions and found no differences in cytochrome c levels between the treatment groups (data not shown), which demonstrates that the decrease in mitochondrial cytochrome c does not coincide with an increase in cytosolic cytochrome c content. We also found similar outer membrane integrity between the treatment groups; therefore, the decrease in cytochrome c could not be attributed to increased outer membrane permeability. We speculate that the increase in cytochrome c phosphorylation might alter its stability. The site of phosphorylation has not been identified, but cytochrome c has been shown to be tyrosine phosphorylated in vivo at Tyr48 in bovine liver and Tyr97 in bovine heart, which are conserved residues in eukaryotes (21, 43). Furthermore, Yu and coworkers (43) find that cytochrome c Tyr48 phosphorylation inhibits mitochondrial respiration by slowing down the reaction kinetics between cytochrome c and cytochrome-c oxidase. It has also been suggested that the posttranslational modification may alter the role of cytochrome c in apoptosis or its cardiolipin peroxidase activity (20, 21).
The effect of increased phosphorylation of cytochrome-c oxidase subunits Va and VIb will need further study. It has been shown that complex IV activity is elevated during PC in a PKC-ϵ-dependent manner and that PKC-ϵ can coimmunoprecipitate with cytochrome-c oxidase subunit IV (14, 29). Yu et al. (44) showed that I/R injury decreases levels of complex IV subunits I and Va. However, PC administered before I/R reduced the loss of subunit I and inhibited the Va subunit losses completely. Thus these data suggest that PC can modulate levels of specific complex subunits as observed in the present study.
We considered that a possible consequence of the increase of specific complex subunits might be an alteration in the assembly of mitochondrial supercomplexes because they typically consist of complexes I, III, and IV. We found that PC and GSK-3 inhibition lead to increased levels of cytochrome-c oxidase subunit VIb in the supercomplex (Fig. 7), which might suggest that cardioprotection can modulate the assembly of supercomplexes. The precise relationship between supercomplex composition and function is still being elucidated, but it appears that the higher-molecular-weight complexes are associated with structural stabilization (34), increased electron transport rate (11, 32), sequestration of reactive intermediates (35), and prevention of excess reactive oxygen species production (31).
Supercomplex formation or stabilization may be a mechanism for cardioprotection and preservation of mitochondrial function. In fact, a recent study by Rosca et al. (33) showed that oxidative phosphorylation was significantly decreased in mitochondria from a canine heart failure model because of a reduced amount of electron transport chain supercomplexes. This work shows that supercomplexes are not only important in terms of assembly and mitochondrial structure but may be relevant as a mechanism in mitochondria-related dysfunction.
In conclusion, the results of this study demonstrate that PC and GSK-3 inhibition alter the protein abundance of subunits of complexes III, IV, and V. It is likely that the expression changes observed were caused by alterations in protein degradation or stability due to the short time frame of our treatments, and these differences could contribute to the cardioprotection provided by PC and GSK-3 inhibition by altering the assembly of complexes into mitochondrial supercomplexes.
GRANTS
This work was supported by the National Heart, Lung, and Blood Institute, NIH intramural program.
DISCLOSURES
No conflicts of interest are declared by the author(s).
REFERENCES
- 1.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marban E, Van Eyk JE. Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res 99: 706–714, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Balaban RS, Mootha VK, Arai A. Spectroscopic determination of cytochrome c oxidase content in tissues containing myoglobin or hemoglobin. Anal Biochem 237: 274–278, 1996 [DOI] [PubMed] [Google Scholar]
- 3.Barandon L, Dufourcq P, Costet P, Moreau C, Allieres C, Daret D, Dos Santos P, Daniel Lamaziere JM, Couffinhal T, Duplaa C. Involvement of FrzA/sFRP-1 and the Wnt/frizzled pathway in ischemic preconditioning. Circ Res 96: 1299–1306, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Bhat R, Xue Y, Berg S, Hellberg S, Ormo M, Nilsson Y, Radesater AC, Jerning E, Markgren PO, Borgegard T, Nylof M, Gimenez-Cassina A, Hernandez F, Lucas JJ, Diaz-Nido J, Avila J. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J Biol Chem 278: 45937–45945, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Cai Z, Semenza GL. PTEN activity is modulated during ischemia and reperfusion: involvement in the induction and decay of preconditioning. Circ Res 97: 1351–1359, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Chen Q, Moghaddas S, Hoppel CL, Lesnefsky EJ. Ischemic defects in the electron transport chain increase the production of reactive oxygen species from isolated rat heart mitochondria. Am J Physiol Cell Physiol 294: C460–C466, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Ding VW, Chen RH, McCormick F. Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J Biol Chem 275: 32475–32481, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Downey JM, Krieg T, Cohen MV. Mapping preconditioning's signaling pathways: an engineering approach. Ann NY Acad Sci 1123: 187–196, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Forbes RA, Steenbergen C, Murphy E. Diazoxide-induced cardioprotection requires signaling through a redox-sensitive mechanism. Circ Res 88: 802–809, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Fralix TA, Murphy E, London RE, Steenbergen C. Protective effects of adenosine in the perfused rat heart: changes in metabolism and intracellular ion homeostasis. Am J Physiol Cell Physiol 264: C986–C994, 1993 [DOI] [PubMed] [Google Scholar]
- 11.Genova ML, Bianchi C, Lenaz G. Supercomplex organization of the mitochondrial respiratory chain and the role of the Coenzyme Q pool: pathophysiological implications. Biofactors 25: 5–20, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Gomez L, Paillard M, Thibault H, Derumeaux G, Ovize M. Inhibition of GSK3beta by postconditioning is required to prevent opening of the mitochondrial permeability transition pore during reperfusion. Circulation 117: 2761–2768, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Gross ER, Hsu AK, Gross GJ. Opioid-induced cardioprotection occurs via glycogen synthase kinase beta inhibition during reperfusion in intact rat hearts. Circ Res 94: 960–966, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Guo D, Nguyen T, Ogbi M, Tawfik H, Ma G, Yu Q, Caldwell RW, Johnson JA. Protein kinase C-epsilon coimmunoprecipitates with cytochrome oxidase subunit IV and is associated with improved cytochrome-c oxidase activity and cardioprotection. Am J Physiol Heart Circ Physiol 293: H2219–H2230, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Hoffert JD, van Balkom BW, Chou CL, Knepper MA. Application of difference gel electrophoresis to the identification of inner medullary collecting duct proteins. Am J Physiol Renal Physiol 286: F170–F179, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry 45: 2524–2536, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imahashi K, Schneider MD, Steenbergen C, Murphy E. Transgenic expression of Bcl-2 modulates energy metabolism, prevents cytosolic acidification during ischemia, and reduces ischemia/reperfusion injury. Circ Res 95: 734–741, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Juhaszova M, Zorov DB, Kim SH, Pepe S, Fu Q, Fishbein KW, Ziman BD, Wang S, Ytrehus K, Antos CL, Olson EN, Sollott SJ. Glycogen synthase kinase-3beta mediates convergence of protection signaling to inhibit the mitochondrial permeability transition pore. J Clin Invest 113: 1535–1549, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhaszova M, Zorov DB, Yaniv Y, Nuss HB, Wang S, Sollott SJ. Role of glycogen synthase kinase-3beta in cardioprotection. Circ Res 104: 1240–1252, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 1: 223–232, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Lee I, Salomon AR, Yu K, Doan JW, Grossman LI, Huttemann M. New prospects for an old enzyme: mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry 45: 9121–9128, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Lesnefsky EJ, Gudz TI, Migita CT, Ikeda-Saito M, Hassan MO, Turkaly PJ, Hoppel CL. Ischemic injury to mitochondrial electron transport in the aging heart: damage to the iron-sulfur protein subunit of electron transport complex III. Arch Biochem Biophys 385: 117–128, 2001 [DOI] [PubMed] [Google Scholar]
- 23.Lesnefsky EJ, Tandler B, Ye J, Slabe TJ, Turkaly J, Hoppel CL. Myocardial ischemia decreases oxidative phosphorylation through cytochrome oxidase in subsarcolemmal mitochondria. Am J Physiol Heart Circ Physiol 273: H1544–H1554, 1997 [DOI] [PubMed] [Google Scholar]
- 24.MacAulay K, Woodgett JR. Targeting glycogen synthase kinase-3 (GSK-3) in the treatment of Type 2 diabetes. Expert Opin Ther Targets 12: 1265–1274, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy E, Steenbergen C. Does inhibition of glycogen synthase kinase protect in mice? Circ Res 103: 226–228, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation 74: 1124–1136, 1986 [DOI] [PubMed] [Google Scholar]
- 27.Nishihara M, Miura T, Miki T, Sakamoto J, Tanno M, Kobayashi H, Ikeda Y, Ohori K, Takahashi A, Shimamoto K. Erythropoietin affords additional cardioprotection to preconditioned hearts by enhanced phosphorylation of glycogen synthase kinase-3 beta. Am J Physiol Heart Circ Physiol 291: H748–H755, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Nishino Y, Webb IG, Davidson SM, Ahmed AI, Clark JE, Jacquet S, Shah AM, Miura T, Yellon DM, Avkiran M, Marber MS. Glycogen synthase kinase-3 inactivation is not required for ischemic preconditioning or postconditioning in the mouse. Circ Res 103: 307–314, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Ogbi M, Johnson JA. Protein kinase Cepsilon interacts with cytochrome c oxidase subunit IV and enhances cytochrome c oxidase activity in neonatal cardiac myocyte preconditioning. Biochem J 393: 191–199, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem 272: 24735–24738, 1997 [DOI] [PubMed] [Google Scholar]
- 31.Panov A, Dikalov S, Shalbuyeva N, Hemendinger R, Greenamyre JT, Rosenfeld J. Species- and tissue-specific relationships between mitochondrial permeability transition and generation of ROS in brain and liver mitochondria of rats and mice. Am J Physiol Cell Physiol 292: C708–C718, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Ragan CI, Heron C. The interaction between mitochondrial NADH-ubiquinone oxidoreductase and ubiquinol-cytochrome c oxidoreductase. Evidence for stoichiometric association. Biochem J 174: 783–790, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res 80: 30–39, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schagger H. Respiratory chain supercomplexes. IUBMB Life 52: 119–128, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholz TD, Balaban RS. Mitochondrial F1-ATPase activity of canine myocardium: effects of hypoxia and stimulation. Am J Physiol Heart Circ Physiol 266: H2396–H2403, 1994 [DOI] [PubMed] [Google Scholar]
- 37.Schulenberg B, Goodman TN, Aggeler R, Capaldi RA, Patton WF. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis 25: 2526–2532, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Steinberg TH, Agnew BJ, Gee KR, Leung WY, Goodman T, Schulenberg B, Hendrickson J, Beechem JM, Haugland RP, Patton WF. Global quantitative phosphoprotein analysis using Multiplexed Proteomics technology. Proteomics 3: 1128–1144, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Thornton J, Striplin S, Liu GS, Swafford A, Stanley AW, Van Winkle DM, Downey JM. Inhibition of protein synthesis does not block myocardial protection afforded by preconditioning. Am J Physiol Heart Circ Physiol 259: H1822–H1825, 1990 [DOI] [PubMed] [Google Scholar]
- 40.Tong H, Chen W, Steenbergen C, Murphy E. Ischemic preconditioning activates phosphatidylinositol-3-kinase upstream of protein kinase C. Circ Res 87: 309–315, 2000 [DOI] [PubMed] [Google Scholar]
- 41.Tong H, Imahashi K, Steenbergen C, Murphy E. Phosphorylation of glycogen synthase kinase-3beta during preconditioning through a phosphatidylinositol-3-kinase-dependent pathway is cardioprotective. Circ Res 90: 377–379, 2002 [DOI] [PubMed] [Google Scholar]
- 42.van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. J Biol Chem 277: 17901–17905, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Yu H, Lee I, Salomon AR, Yu K, Huttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta 1777: 1066–1071, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Q, Nguyen T, Ogbi M, Caldwell RW, Johnson JA. Differential loss of cytochrome-c oxidase subunits in ischemia-reperfusion injury: exacerbation of COI subunit loss by PKC-epsilon inhibition. Am J Physiol Heart Circ Physiol 294: H2637–H2645, 2008 [DOI] [PubMed] [Google Scholar]