Abstract
Gene activation and repression regulated by acetylation and deacetylation represent a paradigm for the function of histone modifications. We provide evidence that, in contrast, histone H2B monoubiquitylation and its deubiquitylation are both involved in gene activation. Substitution of the H2B ubiquitylation site at Lys 123 (K123) lowered transcription of certain genes regulated by the acetylation complex SAGA. Gene-associated H2B ubiquitylation was transient, increasing early during activation, and then decreasing coincident with significant RNA accumulation. We show that Ubp8, a component of the SAGA acetylation complex, is required for SAGA-mediated deubiquitylation of histone H2B in vitro. Loss of Ubp8 in vivo increased both gene-associated and overall cellular levels of ubiquitylated H2B. Deletion of Ubp8 lowered transcription of SAGA-regulated genes, and the severity of this defect was exacerbated by codeletion of the Gcn5 acetyltransferase within SAGA. In addition, disruption of either ubiquitylation or Ubp8-mediated deubiquitylation of H2B resulted in altered levels of gene-associated H3 Lys 4 methylation and Lys 36 methylation, which have both been linked to transcription. These results suggest that the histone H2B ubiquitylation state is dynamic during transcription, and that the sequence of histone modifications helps to control transcription.
Keywords: Histone, modifications, ubiquitylation, methylation, transcription, SAGA
The posttranslational modification of core histones within chromatin, including acetylation, methylation, and phosphorylation, is involved in transcriptional regulation (Bradbury 1992; Strahl and Allis 2000). Characterization of the histone-modifying complexes as well as their antagonistic counterparts in the yeast Saccharomyces cerevisiae and other model systems led to a paradigm wherein modifications contribute to “on/off” switches for gene regulation. Thus, for example, typically lysine acetylation is activating and its deacetylation is repressing.
The SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex is composed of more than 15 subunits and acetylates lysine residues on the N-terminal tails of histones H3 and H2B (Grant et al. 1997; Sanders et al. 2002). SAGA is modular in structure, and has distinct functional units including an acetylation module (Gcn5, Ada2, and Ada3), a TBP-regulatory module (Spt3, Spt7, and Spt8), an activator interaction subunit (Tra1), a Taf histone-fold structure (TAFs and Ada1), and subunits required for assembly of these modules (Ada1, Spt20; Grant et al. 1998a,b; Dudley et al. 1999; Sterner et al. 1999; Belotserkovskaya et al. 2000; Gangloff et al. 2000, 2001). Gcn5 is required for the proper expression of ∼5% of yeast genes in rich media (Holstege et al. 1998). Histone deacetylases, such as Rpd3 and Hda1, act as antagonists to Gcn5 and other histone acetyltransferases (HATs) by removing acetyl groups from histones and turning off transcription (Peterson 2002; Robyr et al. 2002).
Histone ubiquitylation in higher eukaryotes has long been associated with active transcription (Nickel et al. 1989; Davie and Murphy 1990). In yeast, Rad6-mediated monoubiquitylation of histone H2B on Lys 123 (K123) is required for meiotic progression (Robzyk et al. 2000), and yeast Rad6 can ubiquitylate H2B in vitro (Sung et al. 1988; Sharon et al. 1991). Rad6 ubiquitin conjugating activity has been implicated in gene repression in yeast (Turner et al. 2002). Localization of Rad6 to constitutively active gene promoters requires the histone ubiquitin ligase Bre1 (Hwang et al. 2003; Wood et al. 2003a). Thus, although the localization of relevant enzymes suggests that H2B ubiquitylation may be involved in constitutive transcription, as yet there is neither direct evidence for a role of H2B ubiquitylation in gene induction nor has it been demonstrated that H2B ubiquitylation levels correlate with transcription.
Analysis of bulk histone modifications on yeast nucleosomes indicates that, in a “trans-tail” process, histone H2B K123 ubiquitylation leads to methylation of histone H3 K4 and K79, but not K36 (mediated by Set1, Dot1, and Set2, respectively; Briggs et al. 2002; Dover et al. 2002; Sun and Allis 2002). The precise mechanism underlying this dependency is not known, but several functional possibilities have emerged concerning the connection between H2B ubiquitylation and H3 methylation. First, Rad6/ubH2B (ubiquitylated histone H2B) and H3 K4/K79 methylation are involved in telomeric silencing (Briggs et al. 2001; Bryk et al. 2002; Sun and Allis 2002; van Leeuwen et al. 2002). However, methylation appears to be localized preferentially to euchromatic regions compared with heterochromatic regions (Ng et al. 2003b), which therefore has been suggested to be a mechanism to help sequester Sir silencing proteins to heterochromatin (van Leeuwen et al. 2002).
Second, methylation has been correlated with transcription itself. K4 methylation occurs in open reading frames (ORFs; Bernstein et al. 2002), and correlates with gene up-regulation (Litt et al. 2001; Noma et al. 2001; Santos-Rosa et al. 2002). Recent evidence indicates that histone methylation functions during transcriptional elongation (Dover et al. 2002; Li et al. 2002, 2003; Krogan et al. 2003a,b; Ng et al. 2003a; Schaft et al. 2003; Wood et al. 2003b; Xiao et al. 2003). Both Set1 and Set2, the K4 and K36 methyltransferases, respectively, associate with the Paf1 elongation complex (Shi et al. 1996), which associates with RNA polymerase II as it traverses the transcribed gene. Rad6 is also associated with the Paf1 complex, and components of the Paf1 complex are required for H2B ubiquitylation when examined in whole-cell extracts. Finally, the interaction of Rad6 with Paf1 is required for K4 methylation, but not K36 methylation. Thus, Rad6 is required to establish Paf1-dependent, elongation-specific K4 methylation. However, it is unclear whether H2B is a key target of Rad6 at promoters and transcribed regions. In summary, the interrelationship between Rad6/H2B ubiquitylation and H3 methylation is complex, but appears to divide into a global function in helping to establish euchromatin identity and a local function in regulating transcriptional initiation/elongation.
Many questions persist concerning the role of histone ubiquitylation in gene regulation. As mentioned above, it is not clearly established whether histone ubiquitylation is involved in gene induction, and if so, whether the bulky ubiquitin moiety is removed once present. Are there specific activities that remove ubiquitin, and if so, what is their role? Here we provide direct evidence for a sequence of histone H2B K123 ubiquitylation and deubiquitylation during gene activation and identify an H2B deubiquitylating enzyme in the SAGA complex. In addition, we find that disruption of normal H2B ubiquitylation/deubiquitylation alters levels of H3 methylation.
Results
Ubiquitylation of H2B during transcriptional activation
We tested a possible role of histone ubiquitylation in transcription of the GAL1 and SUC2 genes because these genes are well-characterized, inducible, and dependent on chromatin regulation, making them candidates for dependence on additional histone modifications, such as ubiquitylation (Pollard and Peterson 1997; Dudley et al. 1999; Bhaumik and Green 2001). Quantitative S1 nuclease RNA protection analysis was performed on RNA isolated from cells grown in noninducing or inducing conditions for each gene. The strains were either wild type for histone H2B, or bore a single substitution of the ubiquitylation target residue in H2B (htb1-K123R, hereafter called htb1-KR; Robzyk et al. 2000). The htb1-KR substitution reduced H2B GAL1 RNA levels fourfold, whereas SUC2 levels were reduced to 40% of wild-type levels (Fig. 1A). These data, along with the localization of the Rad6/Bre1 complex at a constitutively transcribed gene (Wood et al. 2003a) and recent evidence of an elongation role (Ng et al. 2003c; Wood et al. 2003b), suggest that H2B ubiquitylation may be involved in gene activation, in addition to its reported role in telomeric silencing (Sun and Allis 2002).
Figure 1.
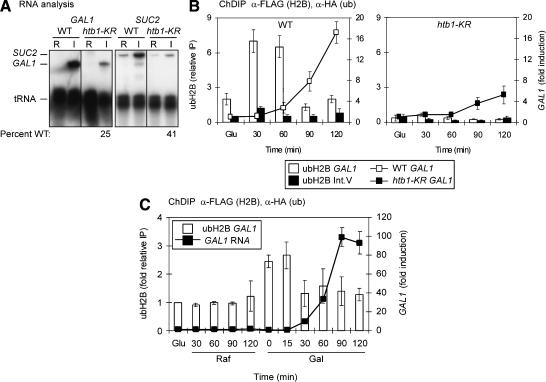
Role of ubiquitylated H2B in the expression of SAGA-dependent genes. (A) GAL1 and SUC2 transcription in HTB1 and htb1-KR cells. RNA was analyzed by S1 nuclease protection assay. Fold induction was calculated as the level of expression under inducing conditions (I) compared with repressing conditions (R) and is presented as the percentage of wild-type induction (set at 100%). tRNA was used as a loading control for the gene-specific RNA. The repressing and inducing conditions were, respectively, GAL1, 2% glucose and 2% galactose for 2.5 h at OD600 nm = 0.8; SUC2, 2% glucose and 0.05% glucose for 2.5 h at OD600 nm < 0.5. The strains were wild type (HTB1+) and htb1-KR, bearing a lysine-to-arginine substitution at residue 123, the known H2B ubiquitylation site (Robzyk et al. 2000). Quantitation was done by PhosphorImager analysis. (B) Double chromatin immunoprecipitation (ChDIP) of ubH2B and RNA analysis during galactose induction. The histogram represents ChDIP analysis, and the line graph represents RNA analysis. Formaldehyde cross-linked chromatin was obtained from wild-type (left) and htb1-KR (right) strains bearing Flag-tagged H2B and HA-tagged ubiquitin in glucose medium (Glu) or during the indicated time course in galactose-containing medium. Sonicated chromatin was immunoprecipitated first with anti-Flag antibody (M2, Sigma) and eluted with 3× Flag peptide (Sigma). Eluates were then immunoprecipitated with anti-HA antibody (12CA5, Roche) before elution. Quantitative PCR analysis of eluted DNA was done in real time. The level of immunoprecipitated chromatin from the GAL1 promoter (white bars) and Int. V region (black bars) is shown as relative immunoprecipitation, a ratio of immunoprecipitated material to input chromatin (left Y-axis). GAL1 RNA levels were assayed in the same samples. RNA was isolated from wild-type (left) or htb1-KR (right) samples and analyzed by S1 analysis. GAL1 expression was normalized to tRNA levels and is presented as fold change in expression relative to expression in glucose (right Y-axis). (C) ubH2B ChDIP and GAL1 RNA analysis in wild-type cells during derepression and activation. Treatments were the same as in B except that cells were incubated in raffinose (GAL1 derepressing) for 2 h prior to the addition of galactose. Samples were taken in glucose and then at indicated times in raffinose and in galactose. Data are normalized to input and Int. V levels and are presented as fold relative IP with the glucose input-normalized immunoprecipitated values set at 1.0 and all others compared with these samples. RNA was treated as in B.
To determine whether ubiquitylation at K123 of histone H2B is, indeed, linked to gene activation, we performed chromatin double immunoprecipitation (ChDIP) and quantitative PCR in real-time. Wild-type or htb1-KR strains containing Flag-tagged H2B and HA-tagged ubiquitin were sampled during growth in glucose (GAL1-repressing) and galactose (GAL1-inducing). Chromatin was immunoprecipitated with Flag antibody and eluted with Flag peptide, and supernatants were subjected to HA immunoprecipitation. DNA was eluted from anti-HA immunoprecipitation and evaluated by PCR in real time.
ubH2B levels at the GAL1 TATA region were low in glucose, increased at 30-60 min after galactose induction, and then decreased at 90-120 min (Fig. 1B, left). PCR of samples from the htb1-KR strain exhibited a greatly reduced signal (Fig. 1B, right), identifying histone H2B K123 as the major ubiquitylated species detected in the ChDIP of wild-type HTB1 samples. The PCR signal of a nontranscribed intergenic region of Chromosome V (Int. V) was very low in all immunoprecipitated samples (Fig. 1B). RNA collected from cells grown under the ChDIP conditions was analyzed by S1 nuclease assay. Increased levels of GAL1 RNA were observed at 60 min and continued to increase to 120 min (Fig. 1B, left); the GAL1 RNA level was lower at each time point in the htb1-KR strain (Fig. 1B, right). Thus, histone H2B ubiquitylation decreased prior to significant accumulation of GAL1 RNA, suggesting that ubiquitylation has a transient role.
Recent data indicate that nucleosomes are evicted from the PHO5 promoter during activation (Boeger et al. 2003; Reinke and Hörz 2003). We tested whether H2B at GAL1 is targeted for removal, by amplification of both the initial anti-Flag elution (containing both ubH2B and H2B) and the second anti-HA elution (containing only ubH2B). The relative levels of H2B at GAL1 compared with Int. V were constant over the time course (data not shown). Thus, during GAL1 induction, histone H2B levels were not lowered at the time that there is a drop in ubH2B levels and a rise in RNA levels indicating that nucleosomes are not evicted. However, these results do not rule out possible replacement of ubH2B.
During the switch from growth in glucose to galactose, the GAL1 promoter is first derepressed to remove DNA-bound repressors and then activated via Gal4 recruitment of coactivator complexes as well as the transcriptional machinery (Schuller 2002). Events of derepression can be experimentally distinguished from activation using raffinose as a carbon source, which derepresses, but does not activate, the promoter. Thus, to further dissect the point at which H2B ubiquitylation occurs, cells were grown in media containing raffinose prior to addition of galactose. The level of ubH2B did not change significantly in raffinose but exhibited an increase when the cells were exposed to galactose that remained high at 15 min. The signal then decreased at 30 min and was low through 120 min in galactose (Fig. 1C). These observations indicate that H2B ubiquitylation increases early during activation, and then decreases. As before, RNA levels [as determined by reverse transcriptase PCR (RT-PCR) in real time] increased following the decline of ubH2B levels (Fig. 1C). Together, these data imply that ubiquitylated H2B is either targeted for turnover (but not depletion), or is deubiquitylated prior to high-level RNA accumulation.
The putative ubiquitin protease, Ubp8, is present in the SAGA and SALSA/SLIK complexes and recruited to promoters simultaneously with Gcn5
Many histone-modifying enzymes (e.g., HATs and histone kinases) are opposed by enzymes that remove the modification (e.g., HDACs or phosphatases). A putative ubiquitin protease, Ubp8, was recently reported to associate with components of the SAGA complex (Gavin et al. 2002; Ho et al. 2002; Sanders et al. 2002). SAGA, and the closely related SALSA/SLIK (SAGA altered, Spt8 absent; SAGA-like) complexes are both involved in transcription, whereas the smaller ADA (Adaptor) complex, which retains Gcn5, does not function in transcription (Eberharter et al. 1999; Pray-Grant et al. 2002; Sterner et al. 2002; Wu and Winston 2002). In particular, SAGA was shown to have a crucial role in GAL1 gene activation (Bhaumik and Green 2001; Larschan and Winston 2001). Because levels of SAGA increase at GAL1 as the gene is induced, during the timeframe in which we observed decreased levels of ubH2B, we investigated the possibility that SAGA-associated Ubp8 may target H2B for deubiquitylation.
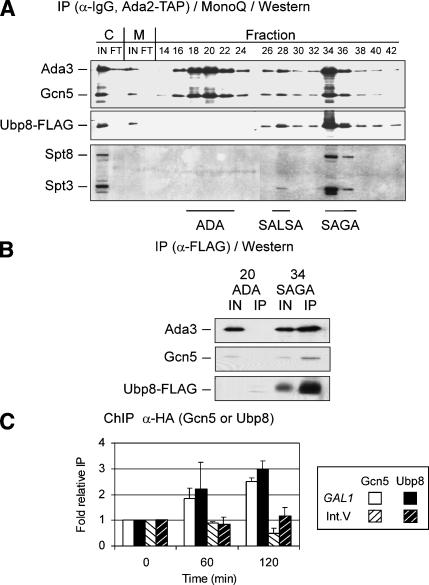
We first examined the association of Ubp8 with three Gcn5 complexes, using multistep purification from a strain bearing an Ada2-TAP affinity tag, because Ada2 is a component of each complex. Cell extracts were fractionated first via the TAP affinity tag followed by MonoQ ion exchange chromatography, resulting in near homogeneous purification of these Gcn5/Ada2 complexes (Grant et al. 1997; Puig et al. 2001; L. Duggan, unpubl.). Flag-tagged Ubp8 was found to cofractionate with SAGA (fractions 34-36) and SALSA (fraction 28) but not with the ADA complex (fractions 18-22; Fig. 2A). Although the Western blot analysis does not reveal whether Ubp8 is present in similar amounts relative to other components of the complexes, it is clear that the stoichiometry of Ubp8 is similar within SAGA compared with SAGA/SLIK. To confirm that Ubp8 is stably and specifically associated with SAGA, Flag immunoprecipitation of Ubp8-Flag was performed using the SAGA or ADA fractions. Ubp8-Flag was detected in fractions containing SAGA but not ADA, and the relative levels of coimmunoprecipitated Gcn5 and Ada3 remained similar compared with the inputs from the SAGA MonoQ column fractions (Fig. 2B). Thus, Ubp8 is a stable component of the transcriptionally relevant SAGA and SALSA/SLIK complexes, and not a component of ADA.
Figure 2.
Analysis of Ubp8 association within Ada2-containing complexes. (A) Chromatographic fractionation of Ubp8 from whole-cell extracts. Cell extracts containing Ada2-TAP and Ubp8-Flag were fractionated first via the TAP purification method (Puig et al. 2001) followed by MonoQ ion exchange chromatography. Even-numbered fractions (14-42) from the MonoQ column were subjected to Western blotting to detect Ubp8-Flag, which was compared with ADA/SALSA/SAGA-associated (as indicated) Ada3 and Gcn5, SALSA/SAGA-associated Spt3, and SAGA-specific Spt8. Samples from inputs (IN) and flowthroughs (FT) for calmodulin-bead binding (C) and MonoQ column (M) purification steps were included. (B) Association of Ubp8 in ADA or SAGA complexes. ADA and SAGA fractions eluted from the MonoQ column were immunoprecipitated with anti-Flag affinity resin (Sigma), and subjected to SDS-PAGE and Western blotting to detect Ubp8-Flag, Ada3, and Gcn5. Input represents 30% of material in the immunoprecipitation. (C) ChIP analysis of Gcn5 and Ubp8 at the GAL1 promoter. Gcn5-3HA (open bars) and Ubp8-3HA (closed bars) binding in a wild-type background were analyzed by ChIP at the GAL1 promoter in glucose (0 time point) and in galactose (60 and 120 min time points). This association was compared with the Int. V region [Gcn5-3HA sample (open bars, black stripes) or Ubp8-3HA (closed bars, white stripes)]. Data are presented as fold relative IP with glucose input-normalized immunoprecipitated values set to 1.0 and all others compared with these samples.
The SAGA complex is required for GAL1 expression and is necessary for TBP recruitment (Bhaumik and Green 2001; Larschan and Winston 2001). As a component of SAGA, Ubp8 is predicted to increase at GAL1 in a galactose-dependent manner. ChIP analysis was performed on triple HA-tagged Gcn5 and Ubp8 during galactose induction. The levels of both proteins at the GAL1 TATA region increased after the shift to galactose medium from glucose and exhibited similar timing of increased association (Fig. 2C). In contrast, the levels of Gcn5-3HA and Ubp8-3HA at the untranscribed Int. V region did not increase during GAL1 induction.
Ubp8 lowers H2B ubiquitylation in vivo and in vitro
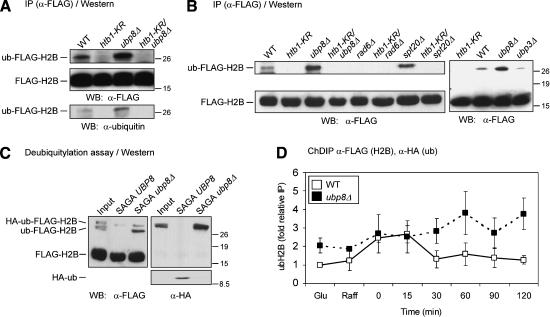
The correlation between increased recruitment of Ubp8 to the GAL1 TATA region and reduction in ubiquitylation of H2B led us to test Ubp8 for its ability to deubiquitylate H2B. First, the level of H2B ubiquitylation was examined in cell lysates from wild-type or ubp8Δ strains expressing H2B-Flag. The levels of H2B-Flag were monitored by anti-Flag immunoprecipitation and Western blotting (Fig. 3A). Because the ubiquitin moiety is relatively large (76 amino acids), it reduces the electrophoretic mobility of H2B (Robzyk et al. 2000). Flag detection showed a slow-migrating H2B species in wild-type extracts, which, as expected (Robzyk et al. 2000), was not present in cell extracts from strains containing either the htb1-KR mutation (Fig. 3A) or the rad6 deletion (Fig. 3B). The level of ubH2B was increased in extracts prepared from the ubp8Δ strain, and this elevated level was eliminated in the htb1-KR/ubp8Δ strain (Fig. 3A). To confirm that this altered species was ubH2B, Western analysis was performed on a separate blot using ubiquitin antibody. Western blotting with anti-Flag and anti-ubiquitin antibodies detected the same molecular weight species, as shown by electrophoretic migration relative to size standards (Fig. 3A, lower panel). These results confirmed that ubH2B increased in ubp8Δ extracts compared with UBP8 extracts.
Figure 3.
H2B ubiquitylation in UBP8 and ubp8Δ strains. (A) Modified H2B in wild-type and ubp8Δ strains. Anti-Flag immunoprecipitates from N-terminally Flag-tagged H2B in wild-type and indicated mutant strains were evaluated by Western blot analysis to determine the relative levels of ubiquitin-modified and unmodified Flag-H2B as described previously (Robzyk et al. 2000). The antibodies used for Western blot analysis are listed in Table 3. Detection of ub-Flag-H2B with anti-Flag antibody and anti-ubiquitin antibody was done in separate Western blots, and their migration was comparable as indicated by molecular weight standards (shown in kilodaltons). (B) Ubiquitylation of H2B in rad6Δ, spt20Δ, and ubp3Δ strains. Western blotting was performed as in A with anti-Flag immunoprecipitated lysates from the indicated strains. The location of molecular weight standards is indicated. (C) In vitro deubiquitylation of ubH2B by Ubp8 within SAGA. SAGA was purified from UBP8 and ubp8Δ strains, and equivalent amounts (see Fig. 5B) were incubated for 30 min with ubH2B and H2B, which bore either single or double tags as indicated. ubH2B and H2B were obtained by anti-Flag immunoprecipitation from ubp8Δ cell lysates. The input was a mock-treated ubH2B sample. Western blotting was performed with anti-Flag (left; to detect H2B and ubH2B containing both tagged and untagged ubiquitin) or with anti-HA (right; to detect ubiquitin-modified H2B and free ubiquitin). (Lower panel, right) Intact HA-ubiquitin (judged from molecular weight standards) that was hydrolyzed from ubH2B by the SAGA UBP8 sample, but not from SAGA ubp8Δ. (D) ChDIP analysis of ubH2B at the GAL1 promoter in ubp8Δ. Data represent ChDIP analysis for ubH2B in both wild-type (open boxes, solid line) and ubp8Δ (closed boxes, broken line) backgrounds as performed in Figure 1C.
The specificity of in vivo deubiquitylation of H2B by Ubp8 was explored in several ways. First, disruption of Spt20, which is required for SAGA integrity (Sterner et al. 1999; Wu and Winston 2002), also resulted in increased levels of ubH2B (Fig. 3B), although the ubH2B level in spt20Δ was slightly lower than in ubp8Δ. This suggests that Ubp8 functions to target ubH2B for hydrolysis primarily as a component of SAGA (or SALSA/SLIK). In addition, we tested the specificity of Ubp8 for H2B deubiquitylation. In particular, Ubp3 displays genetic interactions with SAGA (Martens et al. 1996; Saleh et al. 1998), and therefore could regulate ubH2B levels. However, in contrast to the increased level of ubH2B in the ubp8Δ strain, the level of ubH2B was not increased in ubp3Δ (Fig. 3B).
The ability of Ubp8 to directly deubiquitylate ubH2B was tested in vitro. SAGA complex was purified from UBP8 or ubp8Δ strains using Ada2-TAP-affinity followed by MonoQ fractionation (see Figs. 2A, 5A, below), and equivalent amounts of each complex were incubated with ubH2B. SAGA-UBP8 deubiquitylated ubH2B, using both HA-ub-Flag-H2B and ub-Flag-H2B as a substrate, and released HA-ub (Fig. 3C). In contrast, SAGA prepared from the ubp8Δ strain was unable to hydrolyze ubH2B (Fig. 3C). The slightly higher levels of ub H2B apparent in the SAGA-ubp8Δ sample were not seen in repetitions of the experiment and thus were likely to be a loading effect.
Figure 5.
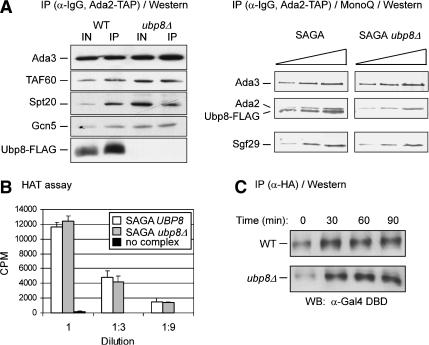
Effect of loss of Ubp8 on SAGA function and Gal4 stability. (A) SAGA stability in wild-type and ubp8Δ strains. (Left) Immunoblots of IgG-immunoprecipitated samples obtained from Ada2-TAP-tagged strains in which Ubp8 was double-Flag-tagged (WT) or deleted (ubp8Δ). Equivalent amounts of protein were used for each immunoprecipitation. (Right) SAGA was purified via standard TAP procedure followed by MonoQ ion exchange chromatography (as in Fig. 2A). Complexes from wild-type and ubp8Δ strains were analyzed by Western blotting using twofold serially diluted samples of each purified SAGA complex. (B) HAT activity of SAGA derived from wild-type or ubp8Δ strains. Threefold serial dilutions of equivalent amounts of wild-type (white bars) or ubp8Δ (gray bars) SAGA complex were assayed for HAT activity on core histones (Sigma) as described previously (John et al. 2000). Background (black bars) indicates incorporation of 3H-acetate without added SAGA complex. (C) Gal4 stability in wild-type and ubp8Δ strains. The Gal4 activator was triple HA-tagged in the wild-type or ubp8Δ backgrounds. Equivalent amounts of protein from lysates collected in glucose (0 min) or galactose-induced cultures (30, 60, and 90 min) were immunoprecipitated with anti-HA antibody and analyzed by Western blotting with an anti-Gal4 DNA-binding domain antibody (Table 3).
Finally, we tested whether Ubp8 has a role in H2B deubiquitylation at the GAL1 TATA region in vivo. The ChDIP assay was used to compare ubH2B levels at GAL1 in UBP8+ and ubp8Δ strains. The ubH2B level was examined in glucose, raffinose, and then during a time course in galactose medium. As previously observed (Fig. 1C), in the wild-type strain there was no increase in ubH2B in raffinose, but it increased transiently after shifting to galactose and then declined. In contrast, higher ubH2B levels were detected in ubp8Δ relative to the wild-type strain (Fig. 3D), both in glucose and raffinose medium. After the addition of galactose, at the peak of ubH2B in UBP8+, higher levels were not detected in ubp8Δ. However, when ubH2B levels decreased in the wild-type strain in galactose, the levels in ubp8Δ remained high. These data suggest that ubH2B is a target of Ubp8 at the GAL1 TATA region.
Ubp8 is required for optimal gene activation
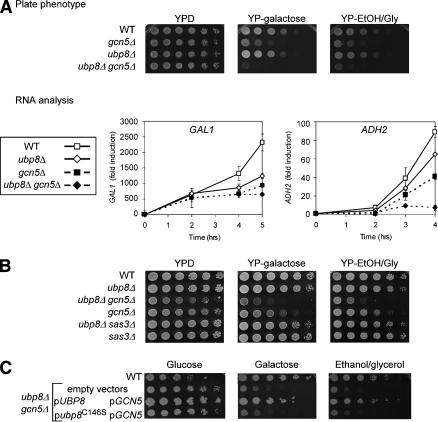
We then examined the role of Ubp8 in growth and gene expression. Semiquantitative growth assays in galactose and in ethanol/glycerol medium showed slight impairment by the loss of Ubp8 (Fig. 4A). Transcription of GAL1 and ADH2 (another SAGA-dependent gene, induced in ethanol/glycerol medium; Chiang et al. 1996) were both lowered in the ubp8Δ strain relative to UBP8 (Fig. 4A). These effects were most apparent late in induction when RNA levels are very high (i.e., at 3-4 h of galactose induction), which followed H2B deubiquitylation (Fig. 1B). Because Ubp8 is present in SAGA, a Gcn5-dependent HAT complex, the role of Gcn5 relative to Ubp8 was also tested. In the growth assay, the gcn5Δ strain showed more severe effects in both media than ubp8Δ, but the combined double ubp8Δ gcn5Δ mutant grewmore poorly than either single mutant (Fig. 4A). Transcriptional effects were consistent with the growth defects, because GAL1 and ADH2 levels were reduced more strongly in the ubp8Δ gcn5Δ double-mutant strain than in either single-mutant strain.
Figure 4.
Role of Ubp8 in SAGA-dependent expression. (A) Cell growth and expression in ubp8Δ. (Upper panels) Fivefold serial dilutions of exponentially growing yeast were spotted on plates containing rich medium, with glucose (YPD), galactose (YP-galactose; GAL1-inducing), or ethanol/glycerol (YP-EtOH/Gly; ADH2-inducing). Wild-type or indicated mutant strains were plated and incubated at 30°C for 48 h. (Lower panels) RNA was extracted from cell pellets collected at the indicated times during incubation in galactose (inducing for GAL1) or ethanol/glycerol (inducing for ADH2) in the indicated strains. Expression was analyzed by RT-PCR in real time and normalized to ACT1 levels. Data are presented as fold induction with wild-type expression in glucose medium set at 1.0. (B) Genetic interaction of ubp8Δ is specific to gcn5Δ and not sas3Δ. Fivefold serial dilutions of the indicated strains were treated as in A. (C) Analysis of Ubp8 catalytic mutant. The ubp8Δ gcn5Δ mutant was transformed with plasmids that contained GCN5 (promoter and open reading frame; pGCN5) and either a wild-type UBP8 gene (pUBP8) or an allele bearing a substitution mutation in the putative catalytic site of Ubp8 (pubp8C146S). Wild type and ubp8Δ gcn5Δ were also transformed with vectors alone. Fivefold serial dilutions of exponentially growing cells were spotted onto selective plates as indicated and grown at 30°C for 48 h.
Thus, combined loss of both Ubp8 and Gcn5 lowered SAGA's function in regulating transcription. Gcn5 is a histone H3 HAT, and displays a similar pattern of acetylation with the Sas3 HAT (John et al. 2000; Howe et al. 2001). We tested whether disruption of Ubp8 combined with disruption of Sas3 exhibited synthetic effects on growth in galactose or ethanol/glycerol medium. The ubp8Δ sas3Δ double mutant grewcomparably to ubp8Δ alone, in contrast to the severe effect of the ubp8Δ gcn5Δ double mutant (Fig. 4B). Thus, we conclude that loss of Ubp8 combined uniquely with loss of the Gcn5 HAT is detrimental to transcription.
A putative catalytic substitution was generated within Ubp8 (C146S), based on its conserved sequence with previously characterized ubiquitin proteases (Hu et al. 2002). This mutation did not alter association of Ubp8 with the SAGA complex, but it did impair catalytic function, similar to ubp8Δ (K. Ingvarsdottir, N.C. Tolga Emre, and S.L. Berger, in prep.). The catalytic Ubp8 mutant was compared with wild-type Ubp8 in growth complementation of Ubp8 loss. Plasmids bearing either wild-type UBP8 or the ubp8-C146S mutant were co-transformed with GCN5 into the double disrupted strain ubp8Δ gcn5Δ. Whereas growth complementation with the unmutated genes was comparable to the wild-type strain, ubp8-C146S displayed poor complementation (Fig. 4C). These data indicate that catalytic activity of Ubp8 is required for its role in these functional pathways.
Ubp8 does not disrupt SAGA complex or alter its HAT activity
We then examined several explanations for the reduction in GAL1 transcription caused by loss of Ubp8. We considered several potential effects of deleting Ubp8, which would not involve H2B. First, deletion of Ubp8 could disrupt the SAGA complex. To examine this possibility, Ada2-TAP complexes were purified from wild-type and ubp8Δ cell extracts, followed by Western blotting to detect components of SAGA. The levels of Ada3, Gcn5, Spt20, and Taf60 were comparable in Ada2-TAP-containing complexes and those immunoprecipitated from a strain lacking Ubp8 (Fig. 5A). Further purification of these complexes by MonoQ fractionation showed stability of both complexes, because the SAGA components were still present with or without Ubp8 (Fig. 5B).
Previous data indicate that Spt7 in SAGA is ubiquitylated (Saleh et al. 1998). It is possible that Spt7 or other ubiquitylated SAGA components could be targets for deubiquitylation by Ubp8, and that Ubp8 absence could manifest as altered HAT activity. We established a linear range of HAT activity with wild-type SAGA and then found that comparable amounts of SAGA lacking Ubp8 exhibited similar HAT activity using either core histones (Fig. 5B) or nucleosomal histones (data not shown) as substrates. Thus, reduced GAL1 transcription in the ubp8Δ strain was not caused by SAGA destabilization or altered HAT activity within the isolated complex.
Another possible explanation for defective GAL1 transcription is that the absence of Ubp8 could alter Gal4 activator levels. Activators have been seen to be monoubiquitylated during gene activation, as well as polyubiquitylated during proteolysis (Salghetti et al. 2001; Conaway et al. 2002). To test this, Western blotting of Gal4-3HA was performed on immunoprecipitated extracts prepared from wild-type and ubp8Δ strains during galactose induction. The levels of Gal4 increased following galactose induction and remained constant in both strains over the time course in galactose (Fig. 5D), indicating that Ubp8 does not regulate Gal4 levels under inducing conditions.
Loss of ubiquitylation or deubiquitylation reciprocally alters the levels of Lys 4 and Lys 36 methylation in histone H3
Histone H3 methylation has been implicated in transcriptional elongation. As described above, both K4 trimethylation and K36 methylation occur during transcription, and Set1 and Set2 are both associated with RNA polymerase II by way of interactions with the Paf1 elongation complex. Furthermore, analysis of bulk nucleosomal histones revealed that the absence H2B K123 ubiquitylation strongly lowers H3 K4 methylation, but not K36 methylation. Taken together, these observations suggest that TATA-directed H2B K123 ubiquitylation may stimulate H3 K4 methylation. Moreover, because both Set1 and Set2 associate with the Paf1 complex, but only K4 methylation is stimulated by H2B ubiquitylation, it may be that alteration of ubiquitylation levels may affect relative levels of K4 and K36 methylation at specific genes.
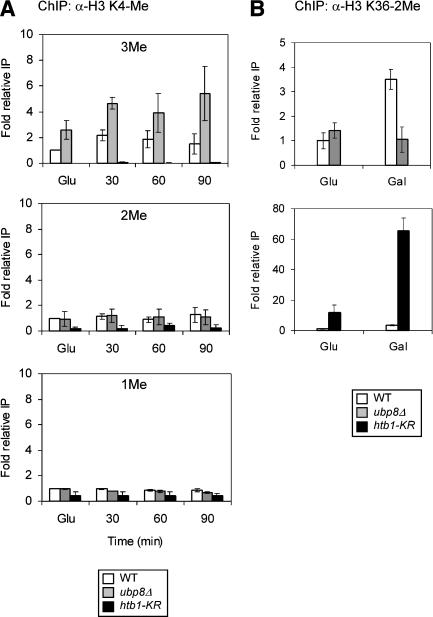
We tested this hypothesis using ChIP analysis, examining the level of K4 and K36 methylation at GAL1 during galactose induction and comparing the levels in wild-type, htb1-KR, and ubp8Δ strains. First, we examined K4 methylation and detected an increase in trimethylation from glucose to galactose (Fig. 6A, top), as observed previously for other inducible genes (Santos-Rosa et al. 2002). K4 trimethylation was very low in the htb1-KR strain relative to wild type (Fig. 6A, top), as expected from previous observations in bulk histones that H2B K123 ubiquitylation strongly stimulates K4 methylation (Sun and Allis 2002). In contrast, the level of K4 trimethylation was increased in the ubp8Δ strain (Fig. 6A, top). This increase in K4 trimethylation in ubp8Δ was detected in glucose (twofold), but was increased further in galactose (threefold at 90 min). Neither K4 dimethylation nor monomethylation levels at GAL1 were altered in the ubp8Δ strain (Fig. 6A, middle and bottom). Thus, the absence of Ubp8 (i.e., higher H2B ubiquitylation) resulted in higher levels specifically of K4 trimethylation.
Figure 6.
Effect of altered histone H2B ubiquitylation on histone H3 methylation status. (A) Analysis of H3 K4 methylation in htb1-KR or ubp8Δ strains. ChIP analysis of the GAL1 promoter in wild-type and ubp8Δ strains. Antibodies specific for different methylation states of histone H3 K4 (3Me, top; 2Me, middle; and 1Me, bottom). Samples were taken from wild-type (open bars), ubp8Δ (gray bars), and htb1-KR (black bars) strains grown in glucose (Glu) and at the times indicated after transfer to galactose medium. The data represent the amplification product from immunoprecipitated material normalized to input material and compared with wild-type glucose IP levels (set as 1.0). (B) Histone H3 methylation of K36. ChIP analysis of the strains used in A was performed using antibodies to H3 K36 dimethylated as in A. The upper panel shows wild type and ubp8Δ; the lower panel shows wild type and htb1-KR. Cells were grown in glucose-containing medium (Glu), washed, and transferred to raffinose medium for 2 h prior to the addition of galactose (Gal).
We then tested the second part of the hypothesis, that manipulation of ubiquitylation levels may alter K36 methylation. Because ubiquitylation promotes K4, but not K36 methylation, and because both Set1 and Set2 are associated with Paf1, it is possible that K36 methylation may be altered in an opposite direction to that of K4 methylation. That is, K36 methylation may be increased in the absence of H2B K123 ubiquitylation and decreased in the absence of Ubp8. First, in the wild-type strain, K36 methylation at the TATA region of GAL1 increased in the switch from glucose to galactose growth (Fig. 6B, top), as also observed for K4 trimethylation (Fig. 6A, top). Second, the level of K36 methylation was increased in the htb1-KR strain, showing a large increase in glucose, but increasing much more in galactose (Fig. 6B, bottom). Thus, the absence of K123 ubiquitylation resulted in high levels of K36 methylation at the GAL1 TATA region. Finally, the level of K36 methylation was similar in the wild-type and ubp8Δ strains in glucose, but, in contrast to wild type, did not increase in the ubp8Δ strain in galactose (three- to fourfold lower compared with wild type; Fig. 6B). Overall, it appears that loss of H2B ubiquitylation compared with loss of Ubp8-mediated deubiquitylation results in opposite effects on K4 versus K36 methylation. This is consistent with the idea that defective H2B ubiquitylation/deubiquitylation alters levels of K4 methylation relative to K36 methylation.
Discussion
Histone ubiquitylation and sequential deubiquitylation in gene activation
In this report, we investigated histone H2B ubiquitylation in gene activation. Our experiments showthat H2B ubiquitylation levels increase at the proximal promoter during gene activation, and persist for a short time before declining. Our results suggest that a sequence of H2B ubiquitylation, followed by deubiquitylation, occurs during gene activation, and that both the addition and subtraction of ubiquitin is required for optimal induced transcription. Notably, addition and removal of ubiquitin occurs rapidly after switching to inducing conditions (Fig. 1B,C), and occurs well before the reduction in RNA levels caused by failure to remove ubiquitin (in the ubp8Δ strain) becomes apparent (Fig. 4A). Thus, ubiquitylation is highly transient, and likely results in further alterations of chromatin that are persistent, such as histone methylation (see below).
Present paradigms hold that the presence of a covalent histone modification has an opposite effect compared with its removal. For example, acetylation and phosphorylation are typically activating modifications, and their absence is repressing. In contrast, our observations suggest that ubiquitylation and deubiquitylation of H2B are both required for optimal induced transcription. First, H2B ubiquitylation levels rise early during gene activation, and then fall as stable RNA is detected. Second, Ubp8, a stable component of the SAGA complex, is capable of deubiquitylating H2B both in vivo and in vitro. Significantly, substitution of the histone H2B ubiquitylation site or deletion of Ubp8 decreases transcription, and to a similar extent.
Ubp8 within SAGA deubiquitylates H2B and collaborates with Gcn5 in gene activation
Ubp8 is a stable component of the SAGA (and SALSA/SLIK) complex. Several lines of evidence from our study suggest that Ubp8 deubiquitylates H2B, and does so as a component of the SAGA complex. First, the absence of Ubp8 correlates with an increase in intracellular levels of ubH2B, and ubH2B is also increased in spt20Δ (Fig. 3), which significantly disrupts SAGA (Wu and Winston 2002). Second, Ubp8 is recruited to the GAL1 TATA region, where levels of ubiquitylation rise and fall (Fig. 1D), and the kinetics of Ubp8 recruitment is similar to that of Gcn5 (Fig. 2C). Third, SAGA, but not SAGA lacking Ubp8, deubiquitylates ubH2B in vitro (Fig. 3C). Fourth, the combined loss of Ubp8 and Gcn5 causes a more severe effect on transcription than loss of either single enzyme (Fig. 4A).
However, two observations suggest that Ubp8 may deubiquitylate H2B also as a monomer or as a component of alternate complexes. The levels of ubH2B increase in the absence of Ubp8 even in rich media (Fig. 3A), where the number and activity of SAGA-dependent genes expressed is low, compared with SAGA-dependent gene activity in stress and nutrient-limiting conditions. Second, the increase in intracellular ubH2B levels in spt20Δ, required for SAGA integrity, is actually somewhat lower than the ubH2B increase in ubp8Δ (Fig. 2B). This suggests that a portion of Ubp8's total H2B-deubiquitylating activity is outside of SAGA or related complexes, and may have alternate functions.
The SAGA complex contains numerous subunits involved in transcription, including the HAT Gcn5. Previous examination of the role of Gcn5 and histone acetylation at GAL1 showed slight effects (Dudley et al. 1999; Bhaumik and Green 2001), and as seen for other components of SAGA (Sterner et al. 1999), we detected synthetic defects in double disruption of Ubp8 and Gcn5 (Fig. 4A). This synergism was specific for Gcn5, because no synthetic defect was observed in the double disruption of Ubp8 and Sas3 (Fig. 4B), a HAT with similar acetylation specificity to Gcn5, but present in a distinct HAT complex (John et al. 2000; Howe et al. 2001). These results showthat the absence of both Ubp8 and Gcn5 is more detrimental than either deletion alone, suggesting that acetylation and ubiquitylation may work together to activate transcription and implying that loss of either function is tolerated better than loss of both functions. Other double disruptions within the SAGA complex cause synthetic phenotypes, such as loss of both Spt3 (which recruits TBP to GAL1) and Gcn5 (Sterner et al. 1999). These synthetic effects likely reflect SAGA's modularity in structure and function, as described above, and are caused by loss of separate, partial transcriptional activating mechanisms—in this case, loss of histone deubiquitylation and loss of histone acetylation. In addition, loss of ubiquitylation (i.e., the histone H2B K123R substitution) combined with loss of Gcn5 also exhibits synthetic defects (C.-F. Kao and M.A. Osley, unpubl.), again supporting the model that both ubiquitylation and deubiquitylation are required for activation. The timing of histone acetylation compared with ubiquitylation/deubiquitylation at GAL1 is not yet known.
Catalytic activity of Ubp8 appears to be required for its function in the pathways we have investigated (Fig. 4C). Thus, the effects we detect in the ubp8Δ strain are likely caused by Ubp8's hydrolysis of ubiquitin and not some other, yet unknown function. However, our experiments do not exclude the possibility that Ubp8 deubiquitylates other proteins, either within SAGA or other complexes.
A second potential ubiquitylation/deubiquitylation pathway has been implicated in SAGA regulation through the HECT domain ubiquitin ligase Tom1 (Saleh et al. 1998) and Ubp3 (Martens et al. 1996). We found that Ubp3 does not target ubH2B for hydrolysis (Fig. 3B), suggesting an alternate target within SAGA, such as Spt7, as previously suggested (Saleh et al. 1998). In addition, ubH2B may be the target of other ubiquitin proteases, given that there are 17 potential deubiquitylating genes in S. cerevisiae (Amerik et al. 2000). The substitution of K123 in H2B reduces telomeric silencing (Sun and Allis 2002), and this effect is likely exerted through H2B ubiqitylation/H3 K4 and K79 methylation occurring in euchromatin (van Leeuwen et al. 2002; Ng et al. 2003b). Ubiquitin proteolysis may play a role in maintaining low H2B ubiquitylation, and hence lowH3 methylation levels at telomeres.
Dynamic ubiquitylation/deubiquitylation may regulate transitions during transcription
The finding that both H2B ubiquitylation and Ubp8-mediated H2B deubiquitylation are involved in transcriptional activation is surprising, because removal of histone modifications typically opposes the effect of their addition, such as acetylation/deacetylation. However, recent observations indicate that the deacetylase Hos2 is involved in transcriptional activation through histone targets, possibly to regenerate a permissive, low-acetylated chromatin ground state during multiple rounds of transcription (Wang et al. 2002). Deubiquitylation may serve a similar role in “resetting” the promoter region to a lowered level of ubiquitylated H2B, during induction of highly transcribed genes.
Methylation is another histone modification that correlates with gene activation. We observed opposite effects on K4 and K36 methylation of hypo- or hyperubiquitylation. Thus, when there was low ubiquitylation (htb1-KR), there was low K4 trimethylation and high K36 methylation; in contrast, when there was excess ubiquitylation (ubp8Δ), there was high K4 trimethylation and lowK36 methylation (Fig. 6). This was observed at GAL1 even in the uninduced state (i.e., in glucose), although the effects were increased during activation (i.e., in galactose). Why is there an inverse relationship between levels of K4 and K36 methylation when ubiquitylation levels are altered? Both Set1 and Set2 bind to the Paf1 complex associated with RNA polymerase II (Krogan et al. 2003a,b; Ng et al. 2003a; Wood et al. 2003b; Xiao et al. 2003). Methylation of K4 via Set1 occurs during early elongation steps (Krogan et al. 2003a; Ng et al. 2003a), whereas K36 methylation by Set2 occurs during later elongation steps (Krogan et al. 2003b; Schaft et al. 2003). Finally, as analyzed in bulk histones, ubiquitylation stimulates Set1 methylation and not Set2 methylation (Briggs et al. 2002). Thus, we speculate that aberrant levels of ubiquitylation/deubiquitylation may alter the relative association of Set1 and Set2 with the Paf1 complex. Increased K4 methylation may be caused by increased association of Set1 signaled by higher ubiquitylation levels and may decrease the amount of Set2 that can associate with Paf1 (as in ubp8Δ). Similarly, decreased amounts of Set1 caused by reduced/absent ubiquitylation may conversely increase the amount of Set2 (the situation in htb1-KR). An alternative, but related idea is that the activities of Set1 and Set2 may be stimulated sequentially, and overstimulation of the first Set1 step via extra/persistence of ubiquitin may inhibit the second Set2 step. Either hypothesis predicts that the amounts of K4/K36 methylation will be altered relative to each other when the levels of ubiquitylation are altered in positive or negative fashion, as we have observed. Further investigation will reveal whether the relative association or activity of Set1 and Set2 is altered by the level of ubiquitylation.
As mentioned above, the effects of the histone H2B K123R substitution on H3 methylation at the GAL1 TATA region were more dramatic than the absence of Ubp8. There are several possible explanations. There may be other Ubp enzymes functioning at GAL1 that target histone H2B for deubiquitylation. Alternatively, K123 may be modified by other covalent marks that influence lysine methylation. In addition, the finding that ubiquitylation and methylation effects occur even in glucose may reflect their role in marking euchromatin and transcribed regions relative to heterochromatin (Bernstein et al. 2002; van Leeuwen et al. 2002).
Previous reports have shown constant K36 methylation levels in RAD6 deletion or K123R substitution strains, when assayed by Western analysis of bulk histones (Briggs et al. 2002). However, by ChIP assay we detected an increase in K36 methylation at the TATA region in the K123R substitution strain in glucose (10-fold) that was even higher in galactose (70-fold). These differences might be attributable to lowsensitivity of Western analysis of bulk histones, and that the signal may have been “saturated.” In addition, the increase in K36 methylation may be both gene-specific and most strongly manifested under specific induction conditions.
A model for the role of sequential ubiquitylation/deubiquitylation in gene activation
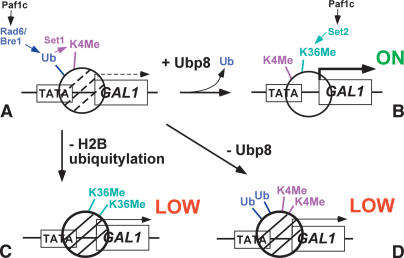
Our data indicate that ubiquitylation/deubiquitylation of H2B has a role in gene regulation. The level of ubiquitylation at the TATA region increases and then decreases during gene activation, through the targeted action of a deubiquitylating enzyme. A model to explain these effects is shown in Figure 7. In normal gene induction (Fig. 7A,B), ubiquitylation is transient, increasing (Fig. 7A) and then decreasing (Fig. 7B) to promote activation. Normal induction is accompanied by sequential K4 and K36 methylation (Fig. 7B). Ubiquitylation helps to establish H3 K4 methylation at the TATA region (Fig. 7A), which is revealed by low K4 methylation in the histone H2B K123R substitution (Fig. 7C). The absence of K4 methylation results in high K36 methylation (Fig. 7C). Deubiquitylation helps to establish the normal balance between K4 and K36 methylation (Fig. 7B), which is revealed by hyper-K4 methylation and lowK36 methylation in the ubp8Δ mutant (Fig. 7D). The result of either lowubiquitylation or lowdeubiquitylation is decreased transcription (Fig. 7C,D).
Figure 7.
Model of the role of ubiquitylation and deubiquitylation at GAL1. Schematic representation of the GAL1 promoter and a nucleosome over the TATA/5′ end of the ORF under poised (A; Ub + K4Me) and activated (B; K4Me + K36Me) conditions, showing the relationship between ubiquitylation/deubiquitylation and methylation in the wild-type strain. Paf1 complex loads both Set1 (K4Me) and Set2 (K36Me). The lower cartoons represent altered ubiquitylation and methylation in ubp8Δ and htb1-KR backgrounds. (C) In the absence of H2B ubiquitylation, there is no K4 methylation and high K36 methylation. (D) In the absence of Ubp8, there is high K4 methylation and lowK36 methylation. These altered ubiquitylation/methylation patterns lead to lowtranscription.
In summary, we have shown that ubiquitylation of H2B has a significant role in regulating transcriptional activation. In particular, a sequence of ubiquitylation followed by deubiquitylation is involved in gene induction, and the transient modification may help to establish certain levels of the more stable histone methylation. Future studies will reveal the nature of the link between ubiquitylation and methylation, and how deubiquitylation participates in transcriptional activation.
Materials and methods
S. cerevisiae strains
The genotypes of strains used in this study are listed in Table 1. Gene deletions were performed as described previously (Longtine et al. 1998) or by transformation with a PCR product obtained by amplification of genomic DNA from a 200-bp region surrounding the target gene, which had already been disrupted with the Escherishia coli KanMX gene (strains courtesy of Thomas Edlind, Drexel University College of Medicine, Philadelphia, PA). Except for ubiquitin, epitope tagging was performed as described previously (Longtine et al. 1998; Puig et al. 2001). HA-tagged ubiquitin under the constitutive GAPDH promoter was a kind gift from Dan Gottschling and Richard Gardner (Fred Hutchinson Cancer Research Center, Seattle, WA). The plasmid was digested with StuI and used for integration into the ura3-1 locus. Following appropriate selection, correct integration and tagging were tested by PCR amplification of genomic DNA with flanking and internal primers as well as Western blot analysis.
Table 1.
S. cerevisiae strains
| Name | Genotype | Reference |
|---|---|---|
| JR5-2A HTB1 | MATa ura-3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [HTB1-CEN-TRP1] | Robzyk et al. 2000 |
| JR5-2A htb1-KR | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [htb1-K123R-CEN-TRP1] | Robzyk et al. 2000 |
| JR5-2A FHTB1 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] | Robzyk et al. 2000 |
| JR5-2A Fhtb1-KR | MATa ura3-1, leu2-3,-112 his3-11,15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-htb1-K123R-CEN-TRP1] | Robzyk et al. 2000 |
| YKH002 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG ADA2-TAP:URA3 UBP8-2Flag:KanMX | This study |
| YKH007 | MATa ura3-1 leu2,3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] ubp8::KanMX | This study |
| YKH008 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-htb1-K123R-CEN-TRP1] ubp8::KanMX | This study |
| T29 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] ubp3::KanMX | This study |
| YKH009 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [HTB1-CEN-TRP1] GAL4-3HA:his5+ | This study |
| YKH012 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [HTB1-CEN-TRP1] GCN5-3HA:his5+ | This study |
| YKH013 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [HTB1-CEN-TRP1] UBP8-3HA:his5+ | This study |
| YKH023 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [HTB1-CEN-TRP1] ubp8::KanMX GAL4-3HA:his5+ | This study |
| YKH030 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] rad6::his5+ | This study |
| YKH032 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-htb1-K123R-CEN-TRP1] rad6::his5+ | This study |
| YKH039 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG ADA2-TAP:URA3 ubp8::KanMX | This study |
| YKH042 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] spt20::URA3 | This study |
| YKH043 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-htb1-K123R-CEN-TRP1] spt20::URA3 | This study |
| YKH045 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] pRG145 [GAPDHprom-3HA-UBI4-URA3 Integrative] | This study |
| YKH046 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-htb1-CEN-TRP1] pRG145 [GAPDHprom-3HA-UBI4-UBA3 Integrative] | This study |
| YKH047 | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [Flag-HTB1-CEN-TRP1] ubp8::KanMX pRG145 [GAPDHprom-3HA-UBI4-URA3 Integrative] | This study |
| Y131 | MATa his3-11,-15 leu2-3,112 trp1-1 ura3-1 ade2-1 can1-100 ssd1 hta-1htb1::LEU2 hta2-htb2:: pRS246-HTA1-HTB1 p RS314-Flag-HTB1 | Robzyk et al. 2000 |
| Y133 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 can1-100 ssd1 hta1-htb1::LEU2 hta2-htb2:: pRS426-HTA1-htb1-K123R pRS314-Flag-htb1-K123RTB1 | Robzyk et al. 2000 |
| YKH063 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 can1-100 ssd1 hta1-htb1::LEU2 hta2-htb2:: pRS426-HTA1-HTB1 pRS314-Flag-HTB1 ubp8::KanMX | This study |
| SB301 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG | Sterner et al. 2002 |
| SB303 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG gcn5::HIS3 | Sterner et al. 2002 |
| YKH067 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG ubp8::KanMX | This study |
| YKH068 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG gcn5::HIS3 ubp8::KanMX | This study |
| LPY8267 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG ubp8::KanMX sas3::HIS3 | This study |
| LPY8270 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG sas3::HIS3 | This study |
| YKH077 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG pRS314 (Cen-TRP1) pRS416 (Cen-URA3) | This study |
| YKH083 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG gcn5::HIS3 ubp8::KanMX pRS314 (Cen-TRP1) pRS416 (Cen-URA3) | This study |
| YKH084 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG gcn5::HIS ubp8::KanMX pC87 (GCN5-Cen-TRP1) pTE101 (UBP8-Cen-URA3) | This study |
| YKH085 | MATa his3Δ200 leu2Δ1 ura3-52 trp1ΔhisG gcn5::HIS3 ubp8::KanMX pC87 (GCN5-Cen-TRP1) pTE102 (ubp8C1465-Cen-URA3) | This study |
In vivo expression analysis
S1 nuclease protection assays were performed as described previously (Lo et al. 2000) using RNA extracted under GAL1- and SUC2-inducing conditions (2% galactose and 0.5% glucose medium, respectively). The oligonucleotides used for S1 analysis are listed in Table 2.
Table 2.
ChIP and TR-PCR amplification primers; S1 oligonucleodies
| Gene | Name | Primer sequence (5′ to 3′) | Location | Use |
|---|---|---|---|---|
| ChIP analysis | ||||
| GAL1 | GAL1 core QChIP F | AACAACCATAGGATGATAATGCGA | -238 to -215 | ChIP |
| GAL1 core QChIP R | ACCCCAGAAATAAGGCTAAAAAACTA | -213 to -187 | ChIP | |
| Intergenich Ch. V. | IntV QChIP F | TAAGAGGTGATGGTGATAGGCGT | 9762 to 9812 | ChIP |
| IntV QChIP R | CCCTCGGGTCAAACACTACAC | 9814 to 9834 | ChIP | |
| RNA expression | ||||
| ACT1 | ACT1 QRT-PCR F | TCGTTCCAATTTACGCTGGTT | 793 to 813 | RT-PCR |
| ACT1 QRT-PCR R | CGATTCTCAAAATGGCGTGA | 824 to 843 | RT-PCR | |
| GAL1 | GAL1 QRT-PCR F | CAGCGAGCTTTACTGCCGAC | 1238 to 1257 | RT-PCR |
| GAL1 QRT-PCR R | TCAAGGCACCAAATTGCTTG | 1269 to 1288 | RT-PCR | |
| ADH2 | ADH2 QRT-PCR F | AGGTATTGATGGTGGTCCAGGA | 597 to 618 | RT-PCR |
| ADH2 QRT-PCR R | CCACCGAGCGAGGTAAACAA | 628 to 648 | RT-PCR | |
| SUC2 | SUC2 S1 | GGCATCTTTTTCATCGTACCACAACCCATTTGG | 29 to 99 | S1 analysis |
| GTCATTCATCCAGCCCTTGTTGGGTGTGAAG | ||||
| TGGACC | ||||
| GAL1 | GAL1 S1 | TTTTCGGCCAATGGTCTTGGTAATTCCTTTGCG | 24 to 83 | S1 analysis |
| CTAGAATTGAACTCAGGTACAATCACT | ||||
| tRNA | tRNA S1 | GGAATTTCCAAGATTTAATTGGAGTCGAAAGCT | S1 analysis | |
| CGCCTTA |
RT-PCR was performed using RNA extracted according to the manufacturer's protocol (YeaStar RNA Kit, Zymo Research) from cells under GAL1- and ADH2-inducing conditions (2% galactose- and 2% ethanol/3% glycerol-containing medium, respectively). RNA was DNase-treated (DNA-free, Ambion) and used as a template for cDNA synthesis (TaqMan Reverse Transcriptase, Roche) according to the manufacturer's recommendations. cDNA was amplified in real time with an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) using ACT1 for normalization of expression. The primers used for RT-PCR are listed in Table 2.
Purification methods
Purification of Ada2-TAP-containing complexes was performed as described (Puig et al. 2001), followed by MonoQ ion exchange chromatography from strains in which Ubp8 was double Flag-tagged (YKH002) or deleted (YKH039). Then 1% of each even-numbered MonoQ fraction was run on a 10% SDS-PAGE gel, transferred to nitrocellulose, and subjected to Western blot analysis as described belowto determine Ubp8-containing fractions. Anti-Flag immunoprecipitation of fractions was performed using 4% of each fraction and anti-Flag affinity resin (Sigma; Table 3) according to the manufacturer's protocol and eluted with 200 ng/µL 3× Flag peptide (Sigma). Approximately 75% of the eluted material (equivalent to 3% of each fraction) was run on a 10% SDS-PAGE gel along with 1% from the appropriate fraction as an input control and transferred to nitrocellulose before Western analysis using the antibodies listed in Table 3.
Table 3.
Antibodies used in this study
| Antibody | Epitope | Source | Use |
|---|---|---|---|
| M2 | Flag | Sigma | Western blot |
| M2 Anti-Flag affinity resin | Flag | Sigma | IP, ChIP, ChDIP |
| 12CA5 | HA | Roche | Western blot, IP, ChIP, ChDIP |
| P4DA | Ubiquitin | Santa Cruz Biotechnology | Western blot |
| anti-GAL4 | Gal4 DBD | Upstate | Western blot |
| anti-histone H3 dimethyl K4 | H3 K4-2Me | Upstate | ChIP |
| ab8580 | H3 K4-3Me | Abcam | ChIP |
| ab8895 | H3 K4-1Me | Abcam | ChIP |
| anti-histone H3 dimethyl K36 | H3 K36-2Me | A. Shilatifard, St. Louis University | ChIP |
| anti-Gcn5 | Gcn5 | This laboratory | Western blot |
| anti-Ada2 | Ada2 | This laboratory | Western blot |
| anti-Ada3 | Ada3 | Leo Guarente, M.I.T. | Western blot |
| anti-Spt3 | Spt3 | F. Winston, Harvard Medical School | Western blot |
| anti-Spt8 | Spt8 | J. Workman, Penn State University | Western blot |
| anti-Spt20 | Spt20 | F. Winston, Harvard Medical School | Western blot |
| anti-Sgf29 | Sgf29 | This laboratory | Western blot |
| anti-TAF60 | TAF60 | M. Green, University of Massachusetts Medical | Western blot |
| School |
Coimmunoprecipitation of SAGA components with Ada2-TAP from lysates of either YKH002 or YKH039 was done with IgG Sepharose (Amersham). Equivalent amounts of protein were used for each IP with 10% of the elution and 5% of the input run on a 10% SDS-PAGE gel prior to Western analysis.
The relative levels of ubH2B in different strain backgrounds (JR5-2A Flag-HTB1, JR5-2A Flag-htb1-KR, YKH007, YKH008, YKH030, YKH032, YKH042, YKH043, and T29) were tested using an N-terminally Flag-tagged histone H2B to aid in the purification and detection using an anti-Flag antibody (Sigma) as described previously (Robzyk et al. 2000). Purification of ubH2B substrate from strain YKH047 for the deubiquitylation assay described belowwas also performed in this manner.
Anti-HA immunoprecipitation of Gal4 was performed with the 12CA5 antibody (Roche) using cell extracts from wild-type (YKH009) and ubp8Δ (YKH023) strains grown under repressing (2% glucose) and GAL1-inducing (2% galactose) conditions for the indicated times. Proteins were eluted by boiling in one volume of 2× SDS Sample buffer, and 20% of the eluted material was run on a 10% SDS-PAGE before transferring to nitrocellulose and Western blot analysis with a Gal4-specific antibody (Table 3).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as described previously (Burke et al. 2000) using 250 mL (ChDIP) or 50 mL (ChIP) of early-log-phase cultures grown in YPD (repressing), YP-2% raffinose (derepressing), or YP-2% galactose (inducing). For ChDIP immunoprecipitation, 20-30 mg of cross-linked protein was used at 4°C overnight, while 5-10 mg was used in normal ChIP applications.
To pull down antibody-bound protein:DNA complexes, Protein A agarose (Upstate) or Protein G Sepharose (Roche) in IP buffer containing 200 µg/mL sheared salmon sperm DNA and 100 µg/mL BSA was added to immunoprecipitations and incubated for 1 h. Agarose/Sepharose beads were washed, eluted, and prepared for PCR as described (Burke et al. 2000). The ChDIP procedure differed from the standard ChIP assay in that following elution of anti-Flag IP with Flag peptide, 90% of the eluted sample was immunoprecipitated with anti-HA antibody at 4°C overnight, and eluted under standard conditions (Burke et al. 2000). Several dilutions of resuspended DNA were tested to determine optimal amplification in real time. Typically, immunoprecipitated samples were initially diluted 1:250, and inputs were diluted 1:20,000 with ultrapure water.
In vitro deubiquitylation assay
The Flag-tagged H2B substrate (containing ubH2B and unmodified H2B) was obtained as described above. Then 500 ng of this substrate was incubated at 37°C for 30 min in DUB buffer (100 mM Tris-HCl at pH 8.0, 1 mM EDTA, 1 mM DTT, 5% glycerol, 1 µM PMSF, 1 µg/mL aprotinin and pepstatin A) with 1% of the MonoQ column fractions containing ADA or SAGA (+/- Ubp8) complex. As a control, substrate was also incubated in DUB buffer to which only MonoQ buffer was added. The reaction was stopped by freezing on dry ice, followed by boiling in one volume of 2× SDS Sample Buffer for 2 min, and running on a 15% SDS-PAGE gel. Gels were transferred to nitrocellulose, and Western blot analysis was performed using anti-Flag (to detect ubH2B and H2B) and anti-HA (to detect ubiquitin) antibodies.
HAT assay
Equivalent amounts of SAGA complex from MonoQ column fractions (+/- Ubp8) were incubated with either 4 µg of calf thymus histones (Sigma) or 2 µg of nucleosomes (a kind gift from Dan Bouchar and Ramin Sheikhatter, The Wistar Institute) in buffer containing 10 mM Tris-HCl (pH 7.5), 1% glycerol, 20 µM EDTA, 10 mM KCl, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate, and 3H-acetyl CoA (4.5 Ci/mmole). Samples were incubated at 30°C for 30 min, and placed on ice to stop the reaction. Then 15 µL of each reaction was spotted onto Whatman P-81 paper, allowed to dry, and washed three times for 15 min each wash in 50 mM sodium bicarbonate (pH 9.3), and one time for 30 min in acetone. After drying, samples were placed in scintillation tubes and 4 mL of scintillation fluid was added before radiolabel detection in a scintillation counter.
Acknowledgments
We thank D. Gottschling and R. Gartner for the generous gift of the constitutive yeast expression plasmid for HA-ubiquitin; K. Ingvarsdottir for the Ubp8 catalytic mutant plasmid; T. Edlind for the gift of deletion strains; M. Schwartz, G. Moore, Y. Henry, and R. Shiekhattar for critical reading of the manuscript; and members of the Berger laboratory for helpful discussions. This work was supported by research grants from the NIH (GM 55360 to S.L.B. and GM 40118 to M.A.O.) and the NSF (MCB-0078940 to S.L.B.). NIH training grants supported K.W.H. [Training Program in Basic Cancer Research to The Wistar Institute (CA09171) and National Research Service Award (F32 GM069207-01)] and A.W. (GM 007229).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1144003.
References
- Amerik A.Y., Li, S.-J., and Hochstrasser, M. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381: 981-992. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Sterner, D.E., Deng, M., Sayre, M.H., Lieberman, P.M., and Berger, S.L. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20: 634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Humphrey, E.L., Erlich, R.L., Schneider, R., Bouman, P., Liu, J.S., Kouzarides, T., and Schreiber, S.L. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. 99: 8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green, M.R. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 15: 1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H., Griesenbeck, J., Strattan, J.S., and Kornberg, R.D. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11: 1587-1598. [DOI] [PubMed] [Google Scholar]
- Bradbury E.M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays 14: 9-16. [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Bryk, M., Strahl, B.D., Cheung, W.L., Davie, J.K., Dent, S.Y., Winston, F., and Allis, C.D. 2001. Histone H2 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 15: 3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S.D., Xiao, T., Sun, Z.W., Caldwell, J.A., Shabanowitz, J., Hunt, D.F., Allis, C.D., and Strahl, B.D. 2002. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Bryk M., Briggs, S.D., Strahl, B.D., Curcio, M.J., Allis, C.D., and Winston, F. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12: 165-170. [DOI] [PubMed] [Google Scholar]
- Burke D., Dawson, D., and Stearns, T. 2000. Chromatin immunoprecipitation. In Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual, pp. 149-154. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chiang Y.C., Komarnitsky, P., Chase, D., and Denis, C.L. 1996. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J. Biol. Chem. 271: 32359-32365. [DOI] [PubMed] [Google Scholar]
- Conaway R.C., Brower, C.S., and Conaway, J.W. 2002. Emerging roles of ubiquitin in transcription regulation. Science 296: 1254-1258. [DOI] [PubMed] [Google Scholar]
- Davie J.R. and Murphy, L.C. 1990. Level of ubiquitinated histone H2B in chromatin is coupled to ongoing transcription. Biochemistry 29: 4752-4757. [DOI] [PubMed] [Google Scholar]
- Dover J., Schneider, J., Tawiah-Boatent, M.A., Wood, A., Dean, K., Johnston, M., and Shilatifard, A. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368-28371. [DOI] [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle, C., and Winston, F. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 13: 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A., Sterner, D.E., Schieltz, D., Hassan, A., Yates III, J.R., Berger, S.L., and Workman, J.L. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y.G., Werten, S., Romier, C., Carre, L., Poch, O., Moras, D., and Davidson, I. 2000. The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol. 20: 340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y.G., Sanders, S.L., Romier, C., Kirschner, D., Weil, P.A., Tora, L., and Davidson, I. 2001. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol. Cell. Biol. 21: 1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan, L., Cote, J., Roberts, S.M., Brownell, J.E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C.D., Winston, F., et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylated nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 11: 1640-1650. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz, D., Pray-Grant, M.G., Yates III, J.R., and Workman, J.L. 1998a. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2: 863-867. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Schieltz, D., Pray-Grant, M.G., Steger, D.J., Reese, J.C., Yates III, J.R., and Workman, J.L. 1998b. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45-53. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green M.R., Golub, T.R., Lander, E.S., and Young, R.A. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717-728. [DOI] [PubMed] [Google Scholar]
- Howe L., Auston, D., Grant, P., John, S., Cook, R.G., Workman, J.L., and Pillus, L. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes & Dev. 15: 3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li, P., Li, M., Li, W., Yao, T., Wu, J.W., Gu, W., Cohen, R.E., and Shi, Y. 2002. Crystal structure of a UBP-family deubiquitylating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111: 1041-1054. [DOI] [PubMed] [Google Scholar]
- Hwang W.W., Venkatasubrahmanyam, S., Ianculescu, A.G., Tong, A., Boone, C., and Madhani, H.D. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11: 261-266. [DOI] [PubMed] [Google Scholar]
- John S., Howe, L., Tafrov, S.T., Grant, P.A., Sternglanz, R., and Workman, J.L. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes & Dev. 14: 1196-1208. [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Ryan, O.W., Golshani, A., Johnston, M., et al. 2003a. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11: 721-729. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim, M., Tong, A., Galshani, A., Cagney, G., Canadien, V., Richards, D.P., Beattie, B.K., Emili, A., Boone, C., et al. 2003b. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 23: 4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E. and Winston, F. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 15: 1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Moazed, D., and Gygi, S.P. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcriptional elongation. J. Biol. Chem. 277: 49383-49388. [DOI] [PubMed] [Google Scholar]
- Li B., Howe, L., Anderson, S., Yates III, J.R., and Workman, J.R. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278: 8897-8903. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson, M., Gaszner, M., Allis, C.D., and Felsenfeld, G. 2001. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293: 2453-2455. [DOI] [PubMed] [Google Scholar]
- Lo W.S., Trievel, R.C., Rojas, J.R., Duggan, L.J., Hsu, J.Y., Allis, C.D., Marmorstein, R., and Berger, S.L. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5: 917-926. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 10: 953-961. [DOI] [PubMed] [Google Scholar]
- Martens J.A., Genereaux, J., Saleh, A., and Brandl, C.J. 1996. Transcriptional activation by yeast PDR1p is inhibited by its association with NGG1p/ADA3p. J. Biol. Chem. 271: 15884-15890. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Robert, F., Young, R.A., and Struhl, K. 2003a. Targeted recruitment of Set1 histone methylase by elongating PolII provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709-719. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Ciccone, D.N., Morshead, K.B., Oettinger, M.G., and Struhl, K. 2003b. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. 100: 1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Dole, S., and Struhl, K. 2003c. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278: 33625-33628. [DOI] [PubMed] [Google Scholar]
- Nickel B.E., Allis, C.D., and Davie, J.R. 1989. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry 28: 958-963. [DOI] [PubMed] [Google Scholar]
- Noma K., Allis, C.D., and Grewal, S.I. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293: 1150-1155. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. 2002. HDAC's at work: Everyone doing their part. Mol. Cell 9: 921-922. [DOI] [PubMed] [Google Scholar]
- Pollard K.J. and Peterson, C.L. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17: 6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant M.G., Schieltz, D., McMahon, S.J., Wood, J.M., Kennedy, E.L., Cook, R.G., Workman, J.L., Yates III, J.R., and Grant, P.A. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22: 8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218-229. [DOI] [PubMed] [Google Scholar]
- Reinke H. and Hörz, W. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11: 1599-1607. [DOI] [PubMed] [Google Scholar]
- Robyr D., Suka, Y., Xenarios, I., Kurdistani, S.K., Wang, A., Suka, N., and Grunstein, M. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437-446. [DOI] [PubMed] [Google Scholar]
- Robzyk K., Recht, J., and Osley, M.A. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501-504. [DOI] [PubMed] [Google Scholar]
- Saleh A., Collart, M., Martens, J.A., Genereaux, J., Allard, S., Cote, J., and Brandl, C.J. 1998. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J. Mol. Biol. 282: 933-946. [DOI] [PubMed] [Google Scholar]
- Salghetti S.E., Caudy, A.A., Chenoweth, J.G., and Tansey, W.P. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293: 1651-1653. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Jennings, J., Canutescu, A., Link, A.J., and Weil, P.A. 2002. Proteomics of the eukaryotic transcription machinery: Identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22: 4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider, R., Bannister, A.J., Sherriff, J., Berstein, B.E., Emre, N.C., Schreiver, S.L., Mellor, J., and Kousarides, T. 2002. Active genes are trimethylated at K4 of histone H3. Nature 419: 407-411. [DOI] [PubMed] [Google Scholar]
- Schaft D., Roguev, A., Kotovic, K.M., Shevchenko, A., Sarov, M., Shevchenko, A., Neugebauer, K.M., and Stewart, A.F. 2003. The histone H3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31: 2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller H.J. 2002. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Gen. 43: 139-160. [DOI] [PubMed] [Google Scholar]
- Sharon G., Raboy, B., Parag, H.A., Dimitrovsky, D., and Kulka, R.G. 1991. RAD6 gene product of Saccharomyces cerevisiae requires a putative ubiquitin protein ligase (E3) for the ubiquitination of certain proteins. J. Biol. Chem. 266: 15890-15894. [PubMed] [Google Scholar]
- Shi X., Finkelstein, A., Wolf, A.J., Wade, P.A., Burton, Z.F., and Jaehning, J.A. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16: 669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant, P.A., Roberts, S.M., Duggan, L.J., Belotserkovskaya, R., Pacella, L.A., Winston, F., Workman, J.L., and Berger, S.L. 1999. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19: 86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Belotserkovskaya, R., and Berger, S.L. 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. 99: 11622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature 403: 41-45. [DOI] [PubMed] [Google Scholar]
- Sun Z.W. and Allis, C.D. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104-108. [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash, S., and Prakash, L. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes & Dev. 2: 1476-1485. [DOI] [PubMed] [Google Scholar]
- Turner S.D., Ricci, A.R., Petropoulos, H., Genereaux, J., Skerjanc, I.S., and Brandl, C.J. 2002. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol. Cell. Biol. 22: 4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken, P.R., and Gottschling, D.E. 2002. Dot1p modulated silencing in yeast by methylation of the nucleosome core. Cell 109: 745-756. [DOI] [PubMed] [Google Scholar]
- Wang A., Kurdistani, S.K., and Grunstein, M. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298: 1412-1414. [DOI] [PubMed] [Google Scholar]
- Wood A., Krogan, N.J., Dover, J., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J.F., et al. 2003a. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11: 267-274. [DOI] [PubMed] [Google Scholar]
- Wood A., Schneider, J., Dover, J., Johnston, M., and Shilatifard, A. 2003b. The Paf1 complex is essential for histone monoubiquitination by the Rad6/Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278: 34739-34742. [DOI] [PubMed] [Google Scholar]
- Wu P.Y. and Winston, F. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22: 5367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Hall, H., Kizer, K.O., Shibata, Y., Hall, M.C., Borchers, C.H., and Strahl, B.D. 2003. Phosphorylation of RNA polymerase II CTD regulated H3 methylation in yeast. Genes & Dev. 17: 654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]