Abstract
The electroneutral cation-chloride cotransporter gene family, SLC12, contains nine members in vertebrates. These include seven sodium and/or potassium-coupled chloride transporters and two membrane proteins of unknown function. Although SLC12 family members have been identified in a number of lower species, the functional properties of these proteins are unknown. There are five SLC12 homologues in Drosophila melanogaster, including at least one member on each of the four main branches of the vertebrate phylogenetic tree. We have employed in situ hybridization to study the expression patterns of the Drosophila SLC12 proteins during embryonic development. Our studies indicate that all five members of this family are expressed during early embryogenesis (stages 1–6), but that spatial and temporal expression patterns become more refined as development proceeds. Expression during late embryogenesis was seen predominantly in the ventral nerve cord, salivary gland, gut, and anal pad. In parallel studies, we have carried out transport assays on each of the five Drosophila homologues, expressed as recombinant proteins in the cultured insect cell line High Five. Under our experimental conditions, we found that only one of these proteins, CG4357, transported the potassium congener 86Rb. Additional experiments established that rubidium transport via CG4357 was saturable (Km = 0.29 ± 0.05 mM), sodium-dependent (half-saturation constant = 53 ± 11 mM), chloride-dependent (half-saturation constant = 48 ± 5 mM), and potently inhibited by bumetanide (inhibitor constant = 1.17 ± 0.08 μM), a specific inhibitor of Na+-K+-2Cl− cotransporters. Taken together, our results provide strong evidence that CG4357 is an insect ortholog of the vertebrate Na+-K+-2Cl− cotransporters.
Keywords: Drosophila embryogenesis, in situ hybridization, Na+-K+-2Cl− cotransport, cation-chloride cotransport, chloride transport
slc12 is a small gene family of integral membrane proteins. To date, all of the SLC12 family members that have been successfully functionally characterized have proven to be electroneutral cation-chloride cotransporters (2, 4). The existence and physiological significance of a number of these transporters was established in functional studies that began to appear in the early 1980's (13), long before it was realized that they were genetically related proteins. The first of the SLC12 proteins to be identified at the molecular level was the Na+-K+-2Cl− cotransporter (NKCC) 1 of the shark rectal gland, which was cloned in 1994 (18). It is now known that there are nine SLC12 family members in vertebrates (2, 4); two NKCCs (NKCC1 and NKCC2), a Na+-Cl− cotransporter (NCC), four K+-Cl− cotransporters (KCC1, KCC2, KCC3 and KCC4), and two additional proteins, commonly referred to as CIP and CCC9, whose function remains uncertain. In addition, SLC12 sequences have been identified in a number of lower species, including crustaceans, insects, worms, plants, fungi, and some bacteria.
In vertebrates, SLC12 family members are known to play important roles in numerous physiological processes, including exocrine fluid secretion, renal salt and water absorption, hearing, olfaction, spermatogenesis, pain perception, visual processing, and other neuronal functions (2, 4, 5, 8). However, little is known about the properties or significance of the SLC12 proteins in lower species. There are five SLC12 homologues in the model organism Drosophila melanogaster, including at least one member on each of the four main branches of the vertebrate phylogenetic tree. Our purpose here is to begin to characterize these Drosophila SLC12 proteins to understand their significance in Drosophila, as well as to eventually better understand the properties of their vertebrate orthologs. We have used in situ hybridization to study the expression patterns of the Drosophila SLC12 family members during embryonic development. In addition, we have explored their functional properties when they are expressed in the cultured insect cell line High Five. Interestingly one of these proteins, CG4357, was found to have the functional properties of a NKCC, with kinetic properties very similar to vertebrate NKCCs.
MATERIALS AND METHODS
Molecular biology.
Molecular biological procedures employed standard methods. Ligations of PCR products were carried out using restriction sites incorporated into the PCR primers. The correctness of all clones used in the paper was confirmed by direct sequencing.
Preparation of Drosophila SLC12 RNA probes.
Full-length cDNA clones of the five Drosophila SLC12 proteins, CG4357 (clone GH27027), CG31547-PB (clone GH09711), CG12773 (clone LD15480), CG10413 (clone GH08340), and CG5594-PB (clone GH09271), were purchased from the Drosophila Genomics Resource Center. The complete coding sequences of CG4357, CG12773, and CG10413 were amplified by PCR and ligated into pBlueScript SK+ (Stratagene) between EcoRI and NotI, EcoRI and NotI, and XhoI and NotI, respectively. The nucleotide sequences corresponding to amino acids 93–1068 of CG31547-PB and amino acids 31–898 of CG5594-PB were amplified by PCR and ligated into pBlueScript SK+ between EcoRI and NotI, and XhoI and NotI, respectively; these regions of CG31547 and CG5594 were common to all alternatively spliced versions of their respective genes. Riboprobes were synthesized using the digoxigenin (DIG) RNA labeling kit (Roche). To generate sense probes, plasmids were linearized with NotI and transcribed with T7 RNA polymerase, and, to generate antisense probes, plasmids were linearized with EcoRI or XhoI, as appropriate, and transcribed with T3 RNA polymerase.
Whole mount in situ hybridization.
Embryos were collected at 6 and 18 h after egg lay on grape juice agar plates, then dechorionated, and fixed according to Tautz and Pfeifle (14). Briefly, embryos were dechorionated in 100% bleach for 1–3 min and then fixed in 2.5 ml 4% formaldehyde, 160 mM KCl, 40 mM NaCl, 4 mM EGTA, and 30 mM PIPES. To this mixture, 2.5 ml heptane were added, and the embryos were vortexed vigorously. The lower aqueous phase was removed, and methanol was added. Fixed embryos were stored in methanol at −20°C or used immediately for in situ hybridization. For in situ hybridization, embryos were treated with formaldehyde/PBT (PBS with 0.1% Tween 20), followed by 3 μg/ml proteinase K for 3–5 min, and then fixed again with 4% formaldehyde/PBT. Hybridization was performed at 58°C overnight using 0.1 μg/ml DIG-labeled antisense or sense probes, as described previously (16). Washing was performed using serial dilutions of the hybridization buffer in PBT. Embryos were then incubated with anti-DIG-AP antibody (Roche) (1:2,000 in PBT) overnight at 4°C. Wash and color development were performed using PBT and BM Purple AP substrate (Roche). Embryos were equilibrated in 70% glycerol/PBT and stored at 4°C. Embryo staging was performed according to Hartenstein (6).
Drosophila strains and crosses.
Crosses were performed using the following Drosophila melanogaster stocks obtained from the Bloomington Stock Center (Indiana University): no. 20377 (y1, w67c23; P{wHy}Ncc69DG04311), which contains a P-element transposon insertion in the 5′-UTR of the CG4357 gene; and no. 5492 [w*; Df(3L)eygC1/TM3, P{ftz/lacC}SC1, ryRK Sb1 Ser1], which deletes the 69A4–69D6 region of chromosome 3L. The wild-type strain used was Oregon R. All Drosophila crosses were performed at 25°C on MM media (KD Medical).
Quantitative PCR.
Parents and progeny from the Drosophila crosses were collected to quantitate CG4357 gene expression using quantitative PCR (QPCR). DNase-free RNA and cDNA were prepared from five adult females of each genotype using the FastRNA Pro Green kit (Q-BIOgene) with DNase removal treatment (Ambion) and reverse transcription (Bio-Rad), according to the manufacturers' instructions. Primers for QPCR were designed using Beacon Designer software (Bio-Rad). Primer sequences used to detect CG4357 gene expression are as follows: TACTCCCGCAGCCGCAAATACC (antisense), ACCAGCAGCAGGGCAACTCTC (sense). QPCR was performed for 40 cycles (95°C for 10 s and 62°C for 30 s) using SYBR-GREEN PCR Master Mix, 1 ng of each cDNA sample, and a My IQ real-time PCR thermocycler (Bio-Rad). Gene expression was normalized to 18S rRNA. Reactions were run in triplicate, and each experiment was repeated three times.
Insect expression vectors for Drosophila SLC12 proteins.
The complete coding sequences, including their native stop codons, of CG4357, CG31547-PB, CG12773, CG10413, and CG5594-PB were amplified by PCR and ligated into the insect expression vector pIB/V5-His (Invitrogen) between HindIII and NotI (CG4357, CG31547-PB, CG10413, and CG5594-PB) or SacI and NotI (CG12773). A CG4357 clone without its native stop codon and in frame with the V5 COOH-terminal tag of the pIB/V5-His vector was also constructed. All clones included an upstream Kozak consensus sequence. pIB/V5-His-CAT, a control vector expressing V5-tagged chloramphenicol acetyltransferase (CAT) was used as a control in some experiments.
Anti-CG4357 antibody production.
The nucleotide sequence corresponding to amino acids 899–1171 of CG4357 was amplified by PCR and ligated into the bacterial expression vector pQE30 (Qiagen) between BamHI and HindIII in frame with a 6xHis NH2-terminal tag. The resulting 6xHis fusion protein was expressed in E. coli and purified on Ni-NTA Agarose (Qiagen), following the manufacturer's instructions. Anti-CG4357 antibody was raised in rabbits against this recombinant protein by Sigma-Genosys. The ability of the antibody to detect the CG4357 protein expressed in the insect cell line High Five was confirmed by Western blotting (see below).
High five cell culture, transfection, and preparation of crude membranes.
The High Five insect cell line was grown in Express Five SFM medium supplemented with 25 μg/ml gentamycin sulfate and 2 mM glutamine at 27°C (the High Five cells and all media were from Invitrogen). Cells were grown and maintained in 6-cm culture dishes and vigorously pipetted into suspension for splitting. High Five cells growing in 6-cm culture dishes were transfected with the plasmids indicated using Cellfection (Invitrogen), according to the manufacturer's instructions. Crude membranes were prepared from High Five cells transiently transfected with the plasmids indicated as previously described (9).
Western blotting and confocal microscopy.
SDS-polyacrylamide gel electrophoresis and Western blotting were carried out as previously described (12). The primary antibodies used were anti-CG4357 (dilution 1:5,000) or an anti-V5 monoclonal (Invitrogen; dilution 1:1,000). Horseradish peroxidase-conjugated secondary antibodies (Pierce) were used at a dilution of 1:10,000, and detection was carried out using the ECL kit from Amersham Biosciences.
For the confocal studies, High Five cells transiently transfected with the plasmids indicated were replated onto glass coverslips. Immunostaining was carried out as previously described (10) using anti-CG4357 (dilution 1:600) or the anti-V5 monoclonal (dilution 1:100) as primary antibodies. The secondary antibodies (diluted 1:200) were Alexa Flour 594 donkey anti-rabbit IgG (H+L) and Alexa Flour 488 goat anti-mouse IgG (H+L), respectively (both from Invitrogen).
86Rb flux assay.
The following media were used in the flux experiments. “Preincubation medium” contained 50 mM NaCl, 4 mM CaCl2, 11 mM MgCl2, 15 mM Na-HEPES (pH 7.3 with NaOH), 5 mM glucose, and 170 mM sucrose. Unless indicated otherwise “uptake medium” was preincubation medium containing, in addition, 20 μM RbCl and ∼0.3 μCi/ml 86Rb (Perkin Elmer). “Termination medium” was preincubation medium containing, in addition, 5 mM RbCl and 250 μM bumetanide. All media are ∼340 mosmol/kgH2O.
The procedure for the 86Rb flux assays was similar to the one previously described from our laboratory for human embryonic kidney-293 cells (1) with some modifications. High Five cells growing in 6-cm culture dishes were transfected with the plasmids indicated. The next day, the plasmid was removed, and the cells were replated into 24-well dishes in fresh culture medium. The flux assay was then carried out the following day, as described below (variations in the flux protocol, if any, are noted in the figure legends). A row of four wells was used for each experimental point, three wells were used to measure 86Rb flux (i.e., fluxes were carried out in triplicate), and the fourth well was used to determine the protein/well using the bicinchoninic acid protein assay kit (Pierce). All of the wells were treated identically, with the exception that 86Rb was omitted from the uptake medium in the fourth well. For each row of wells, the culture medium was removed, the wells were washed twice in preincubation medium (in every case, additions to the wells were 0.5 ml), and then were incubated in preincubation medium for 40 min at room temperature (20°C, all steps hereafter were carried out at room temperature). 86Rb uptake was initiated by replacing the preincubation medium with uptake medium. After 7 min of incubation, the uptake medium was removed, and the well was washed three times with termination medium. Finally, a 0.5-ml aliquot of 1% Triton X-100 was added to each well, and samples were taken for liquid scintillation counting and protein determination. In control experiments (data not shown), we have verified that 86Rb uptake is linear with time for at least 7 min under the experimental conditions described here.
Data presentation.
All quantitative results are expressed as means ± SE for three or more independent experiments. Theoretical fits to the data were carried out by nonlinear least squares regression using the program SigmaPlot.
RESULTS
Phylogenetic analysis of the SLC12 gene family.
Blast searches of the Drosophila melanogaster genome reveal five members of the SLC12 gene family: CG4357 (also called Ncc69), CG31547 (occurring in two alternatively spliced versions, CG31547-PA and CG31547-PB), CG12773, CG10413, and CG5594 (occurring in four alternatively spliced versions, CG5594-PA, CG5594-PB, CG5594-PC, and CG5594-PD). Figure 1 shows a phylogenetic tree of SLC12 family members from human (shaded in gray), Drosophila melanogaster (in bold), C. elegans (C el 1–6), and several other representative species (see legend for further details). It can be seen that the vertebrate (human) family members lie on four main branches: one containing the sodium-dependent transporters NKCC1, NKCC2, and NCC; one containing the potassium-dependent transporters KCC1, KCC2, KCC3, and KCC4; and two other branches, one containing CIP and the other CCC9, both of whose function is presently unknown. There is at least one Drosophila ortholog and one C. elegans ortholog on each of these four branches.
Fig. 1.
Phylogenetic tree of representative members of the SLC12 family. Amino acid sequences were aligned using the program AlignX (a component of the Vector NTI Advance package from Invitrogen), which uses the Clustal W algorithm (15). The tree was displayed using the program Treeview (11). Human sequences are shaded in gray and indicate the 4 main branches of the vertebrate phylogenetic tree. Drosophila SLC12 family members are shown in bold. In addition to the Drosophila proteins, the SLC12 amino acid sequences included are Crab (Callinectes sapidus; AF190129), C el 1 (Caenorhabditis elegans; T22488), C el 2 (Caenorhabditis elegans; NP_501141), C el 3 (Caenorhabditis elegans; H16O14), C el 4 (Caenorhabditis elegans; NP_495469), C el 5 (Caenorhabditis elegans; P34261), C el 6 (Caenorhabditis elegans; NP_495555), C el 7 (Caenorhabditis elegans; NP_493773), cyanobacterium (Trichodesmium erythraeum; ZP_00074455), CCC9 (human; AAO49174), CIP (human; AAK21008), KCC1 (human; NP_005063), KCC2 (human; NP_065759), KCC3 (human; AAF24986), KCC4 (human; AAD39741), NCC (human; NP_000330), NKCC1 (human; NP_001037), NKCC2 (human; AAB07364), Archaea (Methanosarcina acetivorans; NP_619366), rice (Oryza sativa; BAB20646), and yeast (Saccharomyces cerevisiae; NP_009794). NKCC, Na+-K+-2Cl− cotransporter; NCC, Na+-Cl− cotransporter; KCC, K+-Cl− cotransporter.
Expression patterns of the SLC12 family during Drosophila embryogenesis.
To determine the temporal and spatial expression pattern of members of the SLC12 gene family during Drosophila development, we performed whole mount in situ hybridization using nonradioactive DIG-labeled RNA probes. In general, members of the SLC12 gene family exhibited unique expression patterns during Drosophila embryonic development, suggesting distinct biological roles for the membrane proteins they encode.
Expression of CG4357 was first detectable during early embryonic stages 1–3 (Fig. 2). Expression at these stages usually represents maternal RNA contributed to the oocyte by the mother. CG4357 expression was then detected at the cellular blastoderm stages (stages 4–6), when individual cells surround the central yolk sac and zygotic expression is first observed (Fig. 2). As development proceeds, three germ layers are generated during gastrulation (stages 6–8), followed by germ band elongation at stages 9–11 and germ band retraction at stages 12–13. CG4357 expression was seen in the developing gut and nervous system during germ band elongation and retraction stages (stages 9–13; Fig. 2). During stages 14–15, which represents the completion of a number of different morphogenic movements, CG4357 expression is detected predominantly in the foregut and anterior region of the developing embryo. The expression pattern of CG4357 persists through stages 16–17, during which organ system differentiation is completed just before the onset of larval development.
Fig. 2.
CG4357 gene expression during Drosophila embryogenesis by whole mount in situ hybridization. Expression patterns using antisense riboprobes and sense riboprobes are shown. Orientation of the embryo is anterior to the left and posterior to the right. Dorsal is up in lateral images. amg, Anterior midgut rudiment; fg, foregut; hg, hindgut; pmg, posterior midgut rudiment; vne, ventral neurogenic region; vnc, ventral nerve cord.
CG12773 showed patterns of expression similar to those of CG4357 during embryogenesis (Fig. 3). CG12773 was expressed broadly during stages 1–6, followed by expression in the developing gut and nervous system throughout most of the later stages of embryonic development (stages 9–17).
Fig. 3.
CG12773 gene expression during Drosophila embryogenesis by whole mount in situ hybridization. Expression patterns using antisense riboprobes and sense riboprobes are shown. Orientation of the embryo is anterior to the left and posterior to the right. Dorsal is up in lateral images.
CG5594 displayed maternal expression at stages 1–3, followed by expression during cellular blastoderm (stages 4–6) (Fig. 4). Expression in the anterior and posterior midgut, hindgut, anal pads, and the ventral nerve cord were seen during stages 12–13. Expression in the hindgut, anal pads, and nervous system continued throughout stages 14–17.
Fig. 4.
CG5594 gene expression during Drosophila embryogenesis by whole mount in situ hybridization. Expression patterns using antisense riboprobes and sense riboprobes are shown. Orientation of the embryo is anterior to the left and posterior to the right. Dorsal is up in lateral images. ap, Anal pads; mg, midgut.
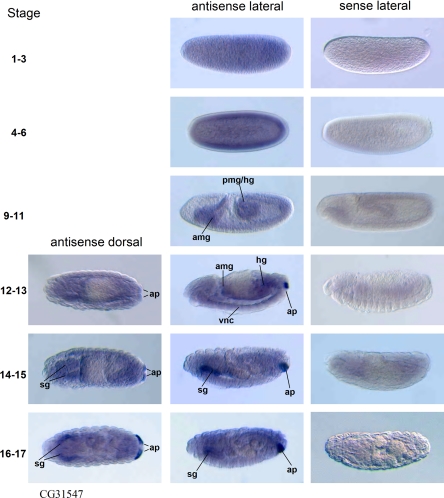
CG31547 was expressed during stages 1–6 (Fig. 5). At stages 9–11, expression was seen in the developing anterior and posterior midgut and the hindgut. At stages 12–13, expression of CG31547 was seen in the nervous system and gut, with very intense expression in the anal pads (Fig. 4). At later stages (stages 14–17), expression was most intense in the salivary glands and the anal pads.
Fig. 5.
CG31547 gene expression during Drosophila embryogenesis by whole mount in situ hybridization. Expression patterns using antisense riboprobes and sense riboprobes are shown. Orientation of the embryo is anterior to the left and posterior to the right. Dorsal is up in lateral images. sg, Salivary glands.
Finally, expression of CG10413 was distinct from that seen for other family members. Maternally contributed CG10413 transcripts were seen during stages 1–3, followed by expression during cellular blastoderm (stages 4–6) (Fig. 6). Thereafter, specific expression of CG10413 during later stages of embryogenesis was not detected.
Fig. 6.
CG10413 gene expression during Drosophila embryogenesis by whole mount in situ hybridization. Expression patterns using antisense riboprobes and sense riboprobes are shown. Orientation of the embryo is anterior to the left and posterior to the right. Dorsal is up in lateral images.
Functional activity.
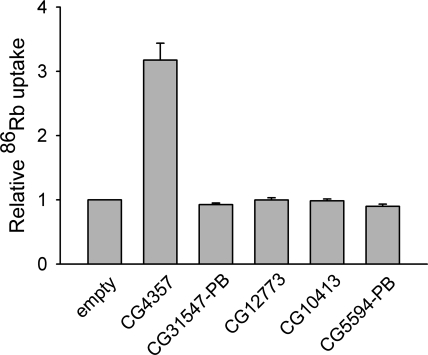
We tested for functional activity of the five Drosophila SLC12 family members by measuring the uptake of 86Rb into High Five cells transiently transfected with CG4357, CG31547-PB, CG12773, CG10413, and CG5594-PB, each cloned into the insect expression vector pIB/V5-His (see materials and methods). Rubidium is a potassium congener that is known to substitute for potassium on a number of biological membrane transporters, including the vertebrate KCCs and NKCCs. For this reason, we felt that 86Rb transport was an appropriate first functional test of the Drosophila SLC12 family. As shown in Fig. 7, under our experimental conditions, only the protein CG4357 was found to exhibit 86Rb transport levels significantly above those seen with the empty vector pIB/V5-His.
Fig. 7.
Uptake of 86Rb into High Five cells transiently transfected with the Drosophila SLC12 proteins indicated. The Drosophila SLC12 proteins were cloned into the insect expression vector pIB/V5-His and transfected into the High Five cells, as described in materials and methods. All clones included their native stop codons so that the COOH-terminal V5-His tag of the pIB/V5-His vector was not appended to the proteins. 86Rb uptake was measured as described in materials and methods, except that 20 μM unlabeled RbCl was omitted from the uptake medium so that only tracer 86Rb was present. The results of 3 independent experiments were averaged to produce the figure. In each experiment, fluxes were normalized to that observed in cells transfected with the empty vector pIB/V5-His (empty).
Characterization of CG4357 expression in High Five cells.
To further characterize the CG4357 protein, we first examined its expression and localization in the High Five cells. In Western blots, anti-CG4357, an antibody raised against the COOH-terminal 273 residues of CG4357 (see materials and methods), showed no reaction with membranes prepared from High Five cells transfected with the empty vector pIB/V5-His, but recognized a single protein of molecular mass ∼130 kDa in membranes prepared from High Five cells transfected with either CG4357 or CG4357 tagged with the V5 epitope (Fig. 8A; the predicted molecular mass of CG4357 is 129 kDa). A protein of the same molecular mass was recognized by an anti-V5 antibody in cells transfected with V5-tagged CG4357 (Fig. 8A), confirming the specificity of the anti-CG4357 antibody.
Fig. 8.
Expression, location, and function of CG4357 and V5-tagged CG4357 in the cultured insect cell line High Five. A: Western blots of membranes prepared from High Five cells transiently transfected with the empty vector pIB/V5-His (empty), CG4357, or V5-tagged CG4357, as indicated. Left: probed with the anti-CG4357 antibody; right: probed with anti-V5 antibody (see materials and methods for details). B: confocal images of High Five cells transiently transfected with the empty vector pIB/V5-His (a), pIB/V5-His-CAT (b), CG4357 (c), and V5-tagged CG4357 (d). All cells were prepared as described in materials and methods and simultaneously probed with both anti-V5 (green) and anti-CG4357 (red) antibodies. C: 86Rb fluxes measured in High Five cells transiently transfected with the same plasmids used in B. Fluxes, measured as described in materials and methods, have been normalized to that observed in cells transfected with the empty vector pIB/V5-His. The results of 3 independent experiments were averaged to produce the figure. CAT, chloramphenicol acetyltransferase.
Figure 8B shows confocal micrographs of High Five cells transfected with the empty vector pIB/V5-His (panel a), pIB/V5-His-CAT, a control vector expressing the cytosolic protein V5-tagged chloramphenicol acetyltransferase (panel b), CG4357 (panel c), and V5-tagged CG4357 (panel d). All cells were simultaneously probed with both anti-V5 (green) and anti-CG4357 (red) antibodies. It is clear from Fig. 8B, c and d, that both CG4357 and V5-tagged CG4357 are localized at or very near the plasma membrane of the High Five cells (the yellow staining in Fig. 8Bd is presumably due to the labeling of V5-tagged CG4357 by both anti-CG4357 and anti-V5 antibodies). In Fig. 8C, we examined the transport of 86Rb in High Five cells transiently transfected as in Fig. 8B. Interestingly, although both CG4357 and V5-tagged CG4357 appear to be trafficked to the plasma membrane of the High Five cells (Fig. 8B), only cells expressing the untagged protein exhibit CG4357-dependent 86Rb transport activity.
Characterization of transport via CG4357 in High Five cells.
We next examined the properties of 86Rb flux via CG4357. Because the High Five cells exhibit significant endogenous rubidium transport, we always measured 86Rb uptake in both cells transfected with empty vector (pIB/V5-His) and cells transfected with CG4357 on the same day and under the same experimental conditions. Specific fluxes via CG4357 could then be determined by subtracting the fluxes observed from cells transfected with empty vector. In Fig. 9A, we illustrate the rubidium concentration dependence of 86Rb uptake into High Five cells transfected with empty vector. This endogenous flux is clearly saturable, and analysis of these data via nonlinear least squares regression reveals an excellent fit to the Michaelis-Menten equation, indicating the presence of a single endogenous rubidium transport system in the High Five cells with Km = 2.56 ± 0.15 mM (see Fig. 9 legend). The flux induced by expression of CG4357 is likewise saturable and also conforms well to Michaelis-Menten kinetics with Km = 0.29 ± 0.05 mM (Fig. 9 legend).
Fig. 9.
Rubidium concentration ([Rb]) dependence of 86Rb fluxes via the endogenous High Five cell transport system and via CG4357. 86Rb fluxes were measured as a function of [Rb] in High Five cells transiently transfected with the empty vector pIB/V5-His (A) or with CG4357 (B), as indicated. Other experimental details are as described in materials and methods. Each data point represents the average ± SE for 3 or more independent determinations. Fluxes in B have been corrected for 86Rb transport via the endogenous High Five system (A) by subtracting uptakes measured in cells transfected with empty vector from those measured in cells transfected with CG4357 for each experimental condition (measurements were carried out on the same cell passage on the same day). Fluxes were fit to the Michaelis-Menton equation via nonlinear least squares regression. The fit to the data in A yielded Km = 2.56 ± 0.15 mM with r = 0.999, while that to the data in B yielded Km = 0.29 ± 0.05 mM with r = 0.990. The lines drawn through the data points correspond to these theoretical fits.
In Fig. 10, we show the sodium concentration dependence of 86Rb uptake into High Five cells transfected with empty vector (panel A) and CG4357 (panel B). No significant sodium dependence is seen for the endogenous High Five rubidium transport system (Fig. 10A), but the flux via CG4357 is clearly sodium dependent (Fig. 10B) and conforms well to the Michaelis-Menten equation with half-saturation constant (K1/2) = 53 ± 11 mM (Fig. 10 legend).
Fig. 10.
Sodium concentration ([Na]) dependence of 86Rb fluxes via the endogenous High Five cell transport system and via CG4357. 86Rb fluxes were measured as a function of [Na] in High Five cells transiently transfected with the empty vector pIB/V5-His (A) or with CG4357 (B), as indicated. In these experiments, Na-HEPES (pH 7.3) was replaced with N-methyl-d-glucamine-HEPES (pH 7.3) in all media. To obtain an uptake medium containing 100 mM NaCl, 100 mM sucrose were replaced with an additional 50 mM NaCl (see materials and methods for composition of uptake medium). The ionic strength of the uptake medium was then held constant by replacing sodium with N-methyl-d-glucamine to obtain the various [Na] indicated. Other experimental details are as described in materials and methods. The fluxes in B have been corrected for 86Rb transport via the endogenous High Five system (A), as described in Fig. 9 legend. The results of 3 independent experiments were averaged to produce the figures. In each experiment, fluxes were normalized to those observed at 100 mM NaCl. The dashed line in A indicates a relative flux of 1.0 (i.e., no dependence on [Na]). The data in B were fit to the Michaelis-Menton equation via nonlinear least squares regression, yielding half-saturation constant (K1/2) = 53 ± 11 mM with r = 0.998. The line drawn through the data in B corresponds to this theoretical fit.
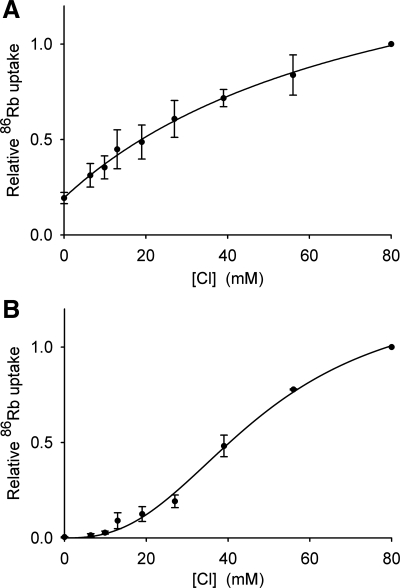
The effect of chloride concentration is illustrated in Fig. 11. The endogenous High Five transport system shows a saturable dependence on chloride (Fig. 11A), which can be modeled by the Michaelis-Menten equation with K1/2 = 80 ± 13 mM (see figure legend). In contrast, the dependence of rubidium transport via CG4357 on chloride concentration is clearly sigmoidal (Fig. 11B), indicating the involvement of more than one chloride ion in the rubidium transport event. These latter data conform well to the Hill equation, with K1/2 = 48 ± 5 mM and n = 2.6 ± 0.3 (see Fig. 11 legend), where n is a measure of the Cl/Rb stoichiometry (17).
Fig. 11.
Chloride concentration ([Cl]) dependence of 86Rb fluxes via the endogenous High Five cell transport system and via CG4357. 86Rb fluxes were measured as a function of [Cl] in High Five cells transiently transfected with the empty vector pIB/V5-His (A) or with CG4357 (B), as indicated. To obtain the [Cl] indicated while keeping the ionic strength of the uptake medium (normally 80 mM chloride; see materials and methods) constant, chloride was replaced with gluconate. Other experimental details are as described in materials and methods. The fluxes in B have been corrected for 86Rb transport via the endogenous High Five system (A), as described in Fig. 9 legend. In each data set, fluxes were normalized to those observed at 80 mM chloride. The results of 3 independent experiments were averaged to produce the figures. The data in A were fit to the Michaelis-Menton equation via nonlinear least squares regression, yielding K1/2 = 80 ± 13 mM with r = 0.998. The data in B were fit to the Hill equation, yielding K1/2 = 48 ± 5 mM and n = 2.6 ± 0.3 with r = 0.998. The lines drawn through the data points correspond to these theoretical fits.
Effects of SLC12 inhibitors.
In Fig. 12, we examine the effects of the SLC12 inhibitors bumetanide (open bars), a specific inhibitor of NKCCs, furosemide (dark shaded bars), an inhibitor of NKCCs and KCCs, and metolazone (light shaded bars), a specific inhibitor of NCCs; in each case, fluxes have been normalized to that observed in the absence of inhibitors (solid bars). All three of the compounds tested are employed clinically as diuretics in humans. These data indicate that metolazone has no significant inhibitory effect on either endogenous High Five 86Rb transport (Fig. 12A) or on 86Rb transport via GC4357 (Fig. 12B). On the other hand, furosemide and bumetanide inhibit both transporters, with bumetanide appearing to be somewhat more effective on the endogenous system and dramatically more effective on CG4357. The bumetanide dose response for inhibition of CG4357 is shown in Fig. 13. These data indicate that the inhibitor constant (KI) for bumetanide inhibition of CG4357 is 1.17 ± 0.08 μM (see figure legend), a value very similar to that found for vertebrate NKCC1s (1, 7).
Fig. 12.
Effects of SLC12 inhibitors on 86Rb fluxes via the endogenous High Five cell transport system and via CG4357. 86Rb fluxes were measured in the presence of bumetanide (open bars), furosemide (dark shaded bars) or metolazone (light shaded bars), at the concentrations indicated, in High Five cells transiently transfected with the empty vector pIB/V5-His (A) or with CG4357 (B). Fluxes have been normalized to that observed in the absence of inhibitors (solid bars). Cells were incubated with the inhibitors in preincubation medium for 10 min before the flux measurements; other experimental details are as described in materials and methods. The fluxes in B have been corrected for 86Rb transport via the endogenous High Five system (A), as described in Fig. 9 legend. Each data point represents the average ± SE for 3 independent determinations.
Fig. 13.
CG4357 bumetanide dose response. 86Rb fluxes were measured in High Five cells transiently transfected with CG4357 in the presence of bumetanide at the concentrations indicated. Experimental details are as given in Fig. 12 legend. Fluxes were corrected for 86Rb transport via the endogenous High Five system, as described in Fig. 9 legend, and then normalized to that observed in the absence of bumetanide. The data were fit to a model that assumes a single inhibitory site for bumetanide. This fit yields inhibitor constant (KI) = 1.17 ± 0.08 μM for bumetanide with r = 0.995. The line drawn through the data points corresponds to this theoretical fit.
Loss of CG4357 gene expression does not affect viability.
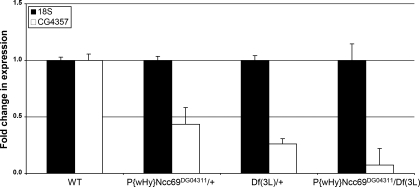
To further investigate the role of the SLC12 family member CG4357 during Drosophila development, we generated flies deficient in CG4357 expression by crossing a Drosophila strain that contains a P-element transposon insertion mutation in the 5′-UTR of CG4357 (P{wHy}Ncc69DG04311) and a strain that contains a chromosomal deletion that encompasses CG4357 [Df(3L)]. Progeny from this cross that were heterozygous for either the transposon (P{wHy}Ncc69DG04311/+) or the deletion [Df(3L)/+] were viable and had an ∼50% reduction in the level of CG4357 expression relative to wild type, as determined by QPCR (Fig. 14). This indicates that both the transposon and the deletion decrease CG4357 expression. Progeny from this cross that contained both the transposon insertion and the chromosomal deletion together [P{wHy}Ncc69DG04311/Df(3L)] had greatly reduced CG4357 expression relative to wild type (Fig. 14). However, no significant decrease in viability relative to their heterozygous siblings was observed (data not shown).
Fig. 14.
CG4357 gene expression in Drosophila lines containing mutations in CG4357. Quantitative PCR analysis of CG4357 gene expression was performed using the primer pairs described in materials and methods. CG4357 gene expression levels were determined for wild-type (WT) flies, flies heterozygous for the transposon insertion mutation (P{wHy}Ncc69DG04311/+), flies heterozygous for the chromosomal deletion encompassing CG4357 [Df(3L)/+], and flies that were homozygous mutants for CG4357, containing both the transposon insertion and the chromosomal deletion [P{wHy}Ncc69DG04311/Df(3L)]. RNA levels were normalized to 18S rRNA. Values shown are the average of one experiment performed in triplicate ± SD.
DISCUSSION
The long-term goal of our studies is to understand the developmental and functional roles of the cation-chloride-coupled cotransporter gene family SLC12 in the model organism Drosophila melanogaster. We anticipate that this information will not only lead to a better appreciation of the significance of these proteins in Drosophila, but also to increased understanding of their vertebrate orthologs. Here we employed in situ hybridization to study the expression patterns of the five Drosophila SLC12 family members, CG4357, CG12773, CG5594, CG31547, and CG10413, during embryonic development (Figs. 2–6). We found that all five members of this family are expressed during early embryogenesis (stages 1–6). Specific expression of CG10413 during later stages of embryogenesis was not detected. Expression of the remaining family members during late embryogenesis varied, but was seen predominantly in the ventral nerve cord, salivary gland, gut, and anal pad. In parallel studies, we carried out functional assays on each of the five Drosophila homologs by measuring the uptake of 86Rb into transiently transfected High Five cells. Rubidium is known to be able to substitute for potassium on a number of vertebrate SLC12 transporters, but, under our experimental conditions, we found that only one of the Drosophila SLC12 proteins, CG4357, exhibited significant 86Rb transport activity (Fig. 7).
In confocal micrographs, we found that CG4357 was expressed primarily in, or very close to, the plasma membrane of the High Five cells (Fig. 8B). In additional experiments, we found that 86Rb transport via CG4357 was saturable with Km = 0.29 ± 0.05 mM (Fig. 9B). 86Rb flux via CG4357 was also sodium and chloride dependent. A plot of 86Rb uptake vs. sodium concentration conformed well to the Michaelis-Menten equation, with K1/2 = 53 ± 11 mM (Fig. 10B), indicating a Na-Rb stoichiometry of 1:1 (17). In contrast, a plot of 86Rb flux via CG4357 vs. chloride concentration was sigmoidal, indicating the involvement of more than one chloride ion in the Rb transport event (Fig. 11B). A fit of these data to the Hill equation yielded K1/2 = 48 ± 5 mM and n = 2.6 ± 0.3, suggesting a Cl-Rb stoichiometry of ≥2:1. 86Rb flux via CG4357 was also inhibited by bumetanide (KI = 1.17 ± 0.08 μM; Figs. 12 and 13), a potent and specific inhibitor of vertebrate NKCC. In previous studies from our laboratory carried out on rat NKCC1 expressed in the human embryonic kidney cell line human embryonic kidney-293 (1), we found Km = 1.85 mM ± 0.26 mM for rubidium, K1/2 = 60.8 ± 4.0 mM for sodium, K1/2 = 48.1 ± 4.7 mM with n = 2.47 ± 0.45 for chloride, and K1/2 = 2.4 ± 0.7 μM for bumetanide. Thus the kinetic parameters for CG4357 and rat NKCC1 are quite similar, with the most notable difference being the higher affinity of CG4357 for rubidium. Taken together, these results provide strong evidence that CG4357 is an insect ortholog of the vertebrate NKCC.
The functional properties of the remaining four Drosophila SLC12 proteins have yet to be determined. We note, however, that the endogenous 86Rb transport system that we observe in the High Five cells is saturable (Fig. 9A), sodium independent (Fig. 10A), chloride dependent (Fig. 11A), and furosemide dependent (Fig. 12A), and thus appears to be a KCC. It is possible that corresponding Drosophila protein (possibly CG5594, which lies closest to the vertebrate KCCs phylogenetically, see Fig. 1) is inactive in the High Five cells under our experimental conditions or that an SLC12 homolog is found in the High Five cells (derived from the cabbage looper, Trichoplusia ni) that is not found in Drosophila. Further studies to explore these questions were beyond the scope of the present paper.
Expression of CG4357, the one SLC12 family member for which transporter activity was detected, was seen to change throughout Drosophila development, indicating dynamic temporal regulation of this gene (Fig. 2). Upon organ formation, CG4357 expression was found predominantly in the gut and nervous system, suggesting a role for this transporter in the ionic homeostasis of these developing organ systems. Indeed, the gut, anal pads, and malpighian tubules are intimately involved in maintaining the proper ionic balance in insects, suggesting that expression of transporters in these tissues may be crucial for both development and homeostasis. Support for this is seen when examining the expression patterns of other members of the SLC12 family. Three additional family members (CG31547, CG12773, and CG5594) are also expressed in the gut, and two of these (CG31547 and CG5594) have very intense expression in the anal pad, a structure present at the posterior end of the hindgut that is involved in regulating salt and water balance. Multiple family members are also expressed in the developing nervous system. Taken together, our data suggest that members of this transporter family are likely to be involved in the ion transport and osmoregulation that occurs in the digestive system of the insect, as well as in nervous system function. Finally, the expression of multiple family members in the gut and nervous system also suggests that there may be functional redundancy built into these organ systems. The potential for functional redundancy within this family is supported by our genetic experiments examining mutations in CG4357. Flies that are homozygous mutants for CG4357 were viable and fertile. While these data suggest that CG4357 is not required for viability, it remains possible that subtle defects in ionic concentrations, digestion, or nervous system function may still be present. Interestingly, Filippov et al. (3) also found that disruption of another family member, CG10413, did not produce an obvious phenotype or affect viability. Future studies will be required to examine the effects of mutations in multiple coexpressed family members to address the issue of redundancy.
GRANTS
This research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda MD.
DISCLOSURES
No conflicts of interest are declared by the author(s).
ACKNOWLEDGMENTS
We thank Drs. William D. Swaim, Marilyn Moore-Hoon, and Most. Nahid Parvin for advice and help with the confocal and flux studies.
REFERENCES
- 1.Dehaye JP, Nagy A, Premkumar A, Turner RJ. Identification of a functionally important conformation-sensitive region of the secretory Na+-K+-2Cl- cotransporter (NKCC1). J Biol Chem 278: 11811–11817, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Delpire E, Mount DB. Human and murine phenotypes associated with defects in cation-chloride cotransport. Annu Rev Physiol 64: 803–843, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Filippov V, Aimanova K, Gill SS. Expression of an Aedes aegypti cation-chloride cotransporter and its Drosophila homologues. Insect Mol Biol 12: 319–331, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Gamba G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev 85: 423–493, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Gavrikov KE, Nilson JE, Dmitriev AV, Zucker CL, Mangel SC. Dendritic compartmentalization of chloride cotransporters underlies directional responses of starburst amacrine cells in retina. Proc Natl Acad Sci USA 103: 18793–18798, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartenstein V. Atlas of Drosophila Development New York: Cold Spring Harbour Laboratory Press, 1993, p. 2–52 [Google Scholar]
- 7.Isenring P, Jacoby SC, Payne JA, Forbush B., III Comparison of Na-K-Cl cotransporters. NKCC1, NKCC2, and the HEK cell Na-L-Cl cotransporter. J Biol Chem 273: 11295–11301, 1998 [DOI] [PubMed] [Google Scholar]
- 8.Kaneko H, Putzier I, Frings S, Kaupp UB, Gensch T. Chloride accumulation in mammalian olfactory sensory neurons. J Neurosci 24: 7931–7938, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore-Hoon ML, Turner RJ. The structural unit of the secretory Na+-K+-2Cl- cotransporter (NKCC1) is a homodimer. Biochemistry 39: 3718–3724, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Nezu A, Parvin MN, Turner RJ. A conserved hydrophobic tetrad near the C terminus of the secretory Na+-K+-2Cl- cotransporter (NKCC1) is required for its correct intracellular processing. J Biol Chem 284: 6869–6876, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Parvin MN, Gerelsaikhan T, Turner RJ. Regions in the cytosolic C-terminus of the secretory Na(+)-K(+)-2Cl(-) cotransporter NKCC1 are required for its homodimerization. Biochemistry 46: 9630–9637, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Russell JM. Sodium-potassium-chloride cotransport. Physiol Rev 80: 211–276, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85, 1989 [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian E, Ten Hagen KG. Expression of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology 16: 83–95, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Turner RJ. Stoichiometry of coupled transport systems in vesicles. Methods Enzymol 191: 479–494, 1990 [DOI] [PubMed] [Google Scholar]
- 18.Xu JC, Lytle C, Zhu TT, Payne JA, Benz E, Jr, Forbush B., III Molecular cloning and functional expression of the bumetanide-sensitive Na-K-Cl cotransporter. Proc Natl Acad Sci USA 91: 2201–2205, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]