Abstract
Riboswitches are metabolite-responsive genetic control elements that reside in the untranslated regions (UTRs) of certain messenger RNAs. Herein, we report that the 5′-UTR of the lysC gene of Bacillus subtilis carries a conserved RNA element that serves as a lysine-responsive riboswitch. The ligand-binding domain of the riboswitch binds to L-lysine with an apparent dissociation constant (KD) of ∼1 µM, and exhibits a high level of molecular discrimination against closely related analogs, including D-lysine and ornithine. Furthermore, we provide evidence that this widespread class of riboswitches serves as a target for the antimetabolite S-(2-aminoethyl)-L-cysteine (AEC). These findings add support to the hypotheses that direct sensing of metabolites by messenger RNAs is a fundamental form of genetic control and that riboswitches represent a new class of antimicrobial drug targets.
Keywords: Allosteric RNA, molecular recognition, riboswitch, RNA aptamer, transcription termination
The precise control of gene expression in response to changes in the chemical and physical environment of cells requires selective interactions between biochemical sensor elements and the molecules that carry or interpret genetic information. Most known genetic factors that respond to such environmental changes are proteins (Ptashne and Gann 2002). However, a number of studies (e.g., see Mironov et al. 2002; Nahvi et al. 2002; Winkler et al. 2002a,b) have demonstrated that natural RNA molecules can also recognize small organic compounds and harness allosteric changes to control the expression of adjacent genes. These metabolite-binding RNA domains, termed riboswitches, typically are embedded within the 5′ untranslated regions (UTRs) of mRNAs, and control the expression of proteins involved in the biosynthesis or import of the target compound. Recently, it has been established that riboswitches also play an important role in controlling fundamental metabolic pathways in bacteria involved in sulfur metabolism (Epshtein et al. 2003; McDaniel et al. 2003; Winkler et al. 2003) and in the biosynthesis of various coenzymes and purines (Mandal et al. 2003). Furthermore, riboswitches are phylogenetically widespread among eubacterial organisms, and both sequence and biochemical data suggest that riboswitches are also present in the genes of eukaryotes (Sudarsan et al. 2003).
These observations indicate that riboswitches likely comprise a widely used mechanism of genetic control in living systems, and thus, we speculate that additional riboswitches might remain undiscovered. We were intrigued by reports that transcription of the lysC genes of Bacillus subtilis and Escherichia coli are repressed by high concentrations of lysine (Kochhar and Paulus 1996; Patte 1996; Patte et al. 1998; Mäder et al. 2002), but no protein factor in B. subtilis had been identified that served as the genetic regulator (Liao and Hseu 1998). The lysC gene encodes aspartokinase II, which catalyzes the first step in the metabolic pathway that converts L-aspartic acid into L-lysine (Belitsky 2002). Interestingly, several efforts have been successful in generating mutants that exhibit constitutive expression of the aspartokinase II enzyme, and all mutations map to the 5′-UTR of the lysC mRNA (Boy et al. 1979; Lu et al. 1991, 1992). Furthermore, a significant level of sequence similarity was identified between the B. subtilis and E. coli lysC 5′-UTRs (Patte et al. 1998). These characteristics strongly suggest that a lysine-responsive riboswitch might be serving as the genetic control element for this gene.
Results
The L box: a conserved mRNA element that is important for genetic control
Riboswitches are typically formed by close juxtaposition of a metabolite-binding domain (aptamer) with a second domain (expression platform) that encompasses mRNA elements necessary for gene expression. Although the RNA sequences and structural components that serve as the expression platform change significantly throughout evolution, the aptamer domain largely retains the sequence composition of its ligand-binding core along with the major secondary-structure features. This permits the use of phylogenetic analyses to identify related RNA domains and to establish a consensus sequence and structure for a given class of riboswitches.
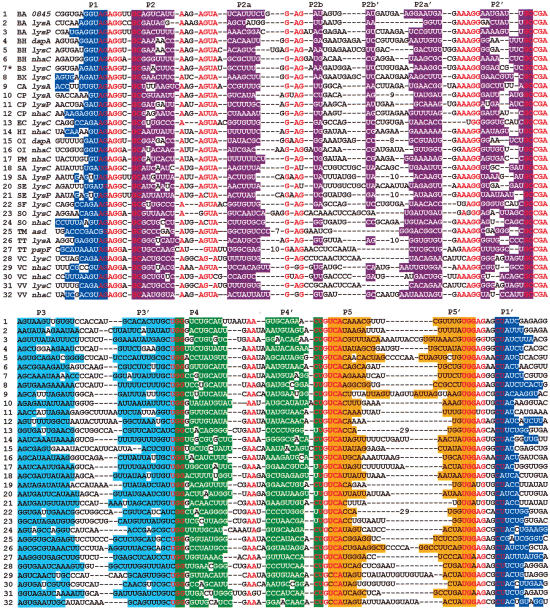
Beginning with the sequence homology reported to exist between the lysC 5′-UTRs of three bacterial species (Patte et al. 1998), the number of representatives was expanded by using an algorithm that searches for related sequences and secondary structures (e.g., see Mandal et al. 2003; Sudarsan et al. 2003). We identified 32 representatives of this RNA domain, termed the L box, in the 5′-UTRs of lysC homologs and other genes related to lysine biosynthesis from a number of Gram-positive and Gram-negative organisms (Fig. 1). The sequence alignment reveals that the RNA forms a five-stem junction wherein major base-paired domains are interspersed with 56 highly conserved nucleotides (Fig. 2A). Furthermore, the base-paired elements P2, P2a, P2b, P3, and P4 each appear to conform to specific length restrictions, suggesting that they are integral participants in the formation of a highly structured RNA. We also noticed that conserved sequences in the junction between stems P2 and P2a conform to a “loop E” motif, which is an RNA element that occurs frequently in other highly structured RNAs (e.g., see Leontis and Westhof 1998).
Figure 1.
The L box, a highly conserved sequence and structural domain, is present in the 5′-UTRs of Gram-positive and Gram-negative bacterial mRNAs that are related to lysine metabolism. Conserved portions of the L-box sequence and secondary structure were identified by alignment of 32 representative mRNAs as noted. Base-pairing potential representing P1-P5 are individually colored. Nucleotides in red are conserved in >80% of the examples. The asterisk identifies the representative (B. subtilis lysC 5′-UTR) that was examined in this study. Gene names are as annotated in GenBank or were derived by protein sequence similarity. Organism abbreviations are as follows: (BA) Bacillus anthracis; (BH) Bacillus halodurans; (BS) Bacillus subtilis; (BX) Bacillus sp.; (CA) Clostridium acetobutylicum; (CP) Clostridium perfringens; (EC) Escherichia coli; (HI) Haemophilus influenzae; (OI) Oceanobacillus iheyensis; (PM) Pasteurella multocida; (SA) Staphylococcus aureus; (SE) Staphylococcus epidermidis; (SF) Shigella flexneri; (SO) Shewanella oneidensis; (TM) Thermatoga maritima; (TT) Thermoanaerobacter tengcongensis; (VC) Vibrio cholerae; (VV) Vibrio vulnificus.
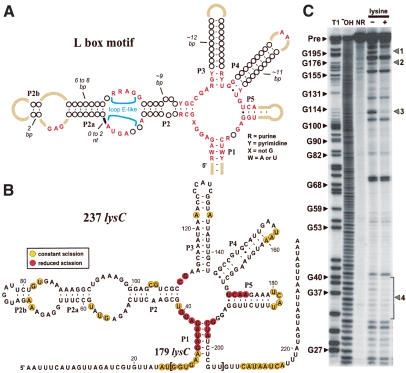
Figure 2.
The consensus L-box motif from the lysC 5′-UTR of B. subtilis undergoes allosteric rearrangement in the presence of L-lysine. (A) Consensus sequence and structure of the L-box domain as derived by using a phylogeny of 32 representative sequences from prokaryotic organisms (Fig. 1). Nucleotides depicted in red are present in at least 80% of the representatives, open circles identify nucleotide positions of variable identity, and tan lines denote variable nucleotide identity and chain length. (B) Sequence, secondary structure model, and lysine-induced structural modulation of the lysC 5′-UTR of B. subtilis. An additional 94 nucleotides (not depicted) reside between nucleotide 237 and the AUG start codon. Structural modulation sites (red-encircled nucleotides) were established by using 237 lysC RNA by monitoring spontaneous RNA cleavage, as depicted in C. (C) In-line probing of the 237 lysC RNA reveals lysine-induced modulation of RNA structure. Patterns of spontaneous cleavage, revealed by product separation using denaturing 10% PAGE, are altered at four major sites (denoted 1-4) in the presence (+) of 10 µM L-lysine (L) relative to that observed in the absence (-) of lysine. (T1) Partial digest with RNase T1; (-OH), partial digest with alkali; (NR) no reaction. Selected bands in the T1 lane (G-specific cleavage) are identified by nucleotide position. See Materials and Methods for experimental details.
The L-box domain of the B. subtilis lysC mRNA resides immediately upstream of a putative transcription terminator stem (Kochhar and Paulus 1996; Patte et al. 1998). In several other riboswitches with similar arrangements (e.g., Mironov et al. 2002; Winkler et al. 2002b; Mandal et al. 2003), the 5′-UTR can be trimmed to separate the minimal aptamer domain from the adjacent expression platform. An RNA fragment (237 lysC; Fig. 2B), encompassing nucleotides 1-237 of the lysC 5′-UTR, was generated and examined for allosteric function. This construct, which excludes the putative transcription terminator stem, was subjected to structural analysis by in-line probing (Soukup and Breaker 1999) to determine whether the presence of lysine alters RNA structure. The in-line probing assay takes advantage of the natural chemical instability of RNA to map the structural context of nucleotides and to assess allosteric changes. We observed that 237 lysC exhibits a pattern of spontaneous RNA cleavage (Fig. 2C) that is consistent with the secondary structure model of the L-box motif constructed from phylogenetic sequence data. Furthermore, we find that the addition of 10 µM L-lysine causes significant changes in the cleavage pattern at four locations along the RNA chain, indicating that allosteric modulation of the 5′-UTR fragment is occurring. In addition, we observe the same pattern of spontaneous cleavage and amino acid-dependent structural modulation when using the 179 lysC RNA construct, which encompasses only the most highly conserved portion of the L-box motif (nucleotides 27-205 of the lysC 5′-UTR).
A reduction of spontaneous cleavage is observed in each of the four sites of metabolite-induced structural modulation. In most instances, a reduction in spontaneous cleavage is due to the nucleotides becoming more ordered in the complex formed between RNA and its ligand (Soukup and Breaker 1999). Interestingly, these four groups of nucleotides are located at the center of the five-stem junction of the L-box secondary structure model (Fig. 2B), implying that these nucleotides are directly involved in recognizing the amino acid target. Similar patterns of ligand-induced structural modulation have been observed with the aptamer domains of other riboswitches (Nahvi et al. 2002; Winkler et al. 2002a,b; Mandal et al. 2003).
The lysine aptamer exhibits high specificity for L-lysine and discriminates against closely related analogs
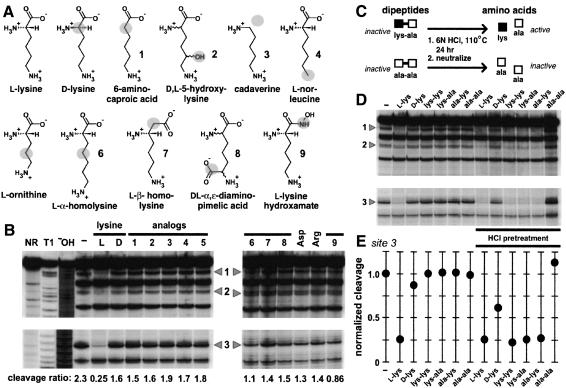
Riboswitches, like their counterpart genetic factors made of protein, must exhibit sufficient specificity and affinity for their target metabolite in order to achieve precision genetic control. To examine the molecular recognition characteristics of the lysC L-box domain, a series of in-line probing assays were performed using various analogs of lysine at 100 µM. The properties of a lysine analog collection were examined, wherein each compound carries minimal chemical changes relative to L-lysine (Fig. 3A). Nearly every chemical alteration to the amino acid renders the compound incapable of causing a structural modulation of the 179 lysC RNA (Fig. 3B). Perhaps most striking is that the RNA does not undergo structural modulation in the presence of D-lysine, which differs from L-lysine by the stereochemical configuration at a single carbon center.
Figure 3.
Molecular recognition characteristics of the lysine aptamer and the use of caged lysine. (A) Chemical structures of L-lysine, D-lysine, and nine closely related analogs. Shaded atoms identify chemical differences between L-lysine and the analog depicted. (B) In-line probing analysis of the 179 lysC RNA in the absence (-) of ligand, or in the presence of 10 µM L-lysine or 100 µM of various analogs as indicated for each lane. For each lane, the relative extent of spontaneous cleavage at site 3 is compared with that of the zone of constant cleavage immediately below this site, in which a cleavage ratio significantly below ∼1.5 reflects modulation. (C) Schematic representation of dipeptide digestion by hydrochloric acid. All dipetide forms are expected to be incapable of binding the lysine aptamer (inactive), whereas lysine-containing dipeptides should induce conformational changes in the aptamer (active) upon acid digestion. (D) In-line probing analysis of the 179 lysC RNA in the absence of lysine (-) or in the presence of various amino acids and dipeptides. Underlined lanes carry dipeptide preparations that were pretreated with HCl, as depicted in A. (E) The fraction of spontaneous cleavage at site 3 in D is plotted after normalization to the extent of processing in the absence of added ligand.
The absence of significant structural modulation in the presence of D-lysine and of other analogs indicates that at least three points of contact are being made between the RNA and its amino acid target. Specifically, the observation that analogs 1, 3, and 4 fail to induce structural modulation is consistent with contacts being made to the amino and carboxy groups of the chain atoms, and to the amino group of the side-chain, respectively. Moreover, the failures of compounds 2, 5, 6, 7, and 8 to induce conformational change in the RNA indicate that the aptamer forms a highly discriminating binding pocket that can measure the length and the integrity of the alkyl side-chain. This high level of molecular discrimination is of particular biological significance, as a genetic switch for lysine most likely must respond exclusively to L-lysine and not closely related natural compounds.
Similarly, the allosteric response of the 179 lysC RNA to various dipeptides and acid-hydrolyzed dipeptides was examined. This effort was undertaken to determine definitively whether lysine triggers allosteric modulation, and to test whether lysine in the context of a peptide could be recognized by the RNA. We speculated that dipeptides should not induce allosteric modulation of RNA structure, but that acid-mediated hydrolysis of dipeptides (Fig. 3C) carrying at least one lysyl residue should restore activity. As predicted, 179 lysC does not undergo allosteric modulation upon the addition of the dipeptides Lys-Lys, Lys-Ala, Ala-Lys, or Ala-Ala (Fig. 3D). However, the three dipeptides that carry at least one lysyl residue induce structural modulation of RNA upon pretreatment of the dipeptides with 6 N HCl for 23 h at 115°C, followed by evaporation and neutralization. The extent of structural modulation (Fig. 3E) indicates that the samples containing the hydrolyzed lysine-containing dipeptides fully saturate the lysC aptamer, which is in accordance with the acid-mediated release of saturating amounts (>1 µM; see below) of L-lysine.
We also observed that an intermediate level of structural modulation occurs when D-lysine is pretreated with HCl. Interestingly, the published rate of racemization between D- and L-lysine (Engel and Hare 1982) is sufficient to account for the ∼1 µM of L-lysine that is needed to produce half-maximal structural modulation (Fig. 3E). These results are consistent with our hypothesis that lysine is the molecular ligand for the lysC aptamer, and that RNA conformational changes are not due to unknown contaminants of the commercial L-lysine preparation, or are otherwise reflective of a protein binding site.
Binding affinity and stoichiometry of the B. subtilis L-lysine aptamer
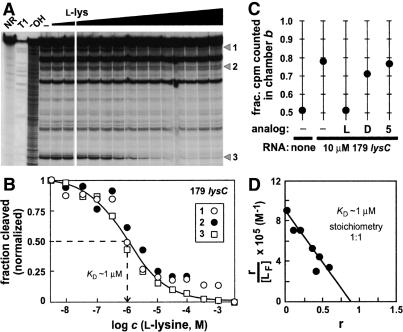
An approximation of the dissociation constant (KD) was made by conducting in-line probing assays with 179 lysC by using various concentrations of L-lysine (Fig. 4A). The sites of structural modulation exhibit progressively lower levels of spontaneous cleavage in response to increasing concentrations of ligand. A plot of the extent of RNA cleavage versus concentration of L-lysine (Fig. 4B) indicates that half-maximal structural modulation occurs when ∼1 µM amino acid is present in the mixture, thus reflecting the apparent KD of the 179 lysC for its target ligand.
Figure 4.
Determination of the dissociation constant and stoichiometry for L-lysine binding to the 179 lysC RNA. (A) In-line probing with increasing concentrations of L-lysine ranging from 3 nM to 3 mM. Details are as defined for Figure 2C. (B) Plot depicting the normalized fraction of RNA undergoing spontaneous cleavage versus the concentration of amino acid for sites 1-3. The broken line identifies the concentration of L-lysine required to bring about half-maximal structural modulation, which indicates the apparent KD for ligand binding. (C) The 179 lysC RNA (10 µM) shifts the equilibrium of tritiated L-lysine (50 nM) in an equilibrium dialysis chamber. To investigate competitive binding, unlabeled L-lysine (L), unlabeled D-lysine (D), or L-ornithine (5) was added to a final concentration of 50 µM each to one chamber of a pre-equilibrated assay as indicated. (D) Scatchard analysis of L-lysine binding by the 179 lysC RNA. The variable r represents the ratio of bound ligand concentration versus the total RNA concentration, and the variable [LF] represents the concentration of free ligand.
The apparent KD value for a longer construct that encompasses structural elements predicted to be involved in transcription termination exhibits a significantly poorer affinity for L-lysine (data not shown). Specifically, an RNA construct encompassing nucleotides 1-315 of the lysC 5′-UTR was found by in-line probing to exhibit an apparent KD of ∼500 µM. Similar differences in ligand affinities for other riboswitches have been observed, wherein the minimized aptamer binds more tightly its cognate ligand compared with the same aptamer in the context of the complete riboswitch (aptamer plus the adjoining expression platform). This is most likely due to the presence of competing secondary or tertiary structures that reduce the fraction of nascent mRNAs that are correctly folded in the ligand-receptive form at any given point in time. Some of these alternate folds might be important for the function of the riboswitch as a genetic control element, but they would reduce ligand binding affinity by reducing preorganization of the aptamer domain.
Equilibrium dialysis also was used to examine the affinity and specificity of the 179 lysC aptamer for its target (Fig. 4C). In the absence of RNA, tritiated L-lysine is expected to distribute equally between the two chambers (a and b) of an equilibrium dialysis apparatus. However, the addition of excess aptamer to one chamber of the system should shift the distribution of tritium toward this chamber as a result of complex formation. This asymmetric distribution of tritium is expected to be restored to unity by the addition of a large excess of unlabeled competitor ligand, which displaces the bulk of the tritiated lysine from the RNA. As expected, the fraction of tritiated L-lysine in chamber b of the equilibrium dialysis apparatus is ∼0.5 in the absence of RNA (Fig. 4C) after a 10-h incubation. This fraction is altered to ∼0.8 after incubation when a 200-fold excess of 179 lysC (10 µM) is added to chamber b, whereas this symmetric distribution of tritium is restored upon incubation for an additional 10 h after the introduction of excess (50 µM) unlabeled L-lysine. Furthermore, D-lysine and L-ornithine do not restore equal distribution of tritium, which is consistent with their failure to modulate RNA structure as determined by in-line probing.
A Scatchard plot also was created by using data from a series of equilibrium dialysis experiments conducted under various concentrations of tritiated L-lysine (Fig. 4D). The slope of the resulting line indicates that the 179 lysC RNA binds to L-lysine with an apparent KD of ∼1 µM, which is consistent with that observed by using in-line probing. Furthermore, the x-intercept of the line occurs near an r value of one, which demonstrates that the RNA forms a 1:1 complex with its ligand.
The lysine aptamer and adjacent sequences function as an amino acid-dependent riboswitch
With a number of riboswitches examined to date, there is a discernable set of structures residing immediately downstream of the aptamer domain that serve to control gene expression in response to ligand binding. Typically, the structure of this “expression platform” is modulated by metabolite binding to the aptamer domain. The alternative structure subsequently leads to modulation of transcription or translation processes. For example, the TPP riboswitch on the thiM mRNA of E. coli carries an expression platform that appears to preclude ribosome binding to the Shine-Dalgarno sequence of the adjacent coding region (Winkler et al. 2002a). Similarly, the expression platforms of various riboswitches from B. subtilis undergo ligand-induced formation of a stem-loop structure that induces transcription termination (e.g., Mironov et al. 2002; Winkler et al. 2002b; Mandal et al. 2003; McDaniel et al. 2003).
It has been reported that the lysC mRNA undergoes transcription termination in cultured B. subtilis cells grown in the presence of excess L-lysine (Kochhar and Paulus 1996). We observed that a sequence domain that participates in forming the P1 stem of the lysC aptamer is complementary to a portion of the putative terminator hairpin that resides ∼30 nucleotides downstream (Fig. 5A). This architecture is similar to that of several other riboswitches, some of which exhibit termination of transcription in vitro upon addition of the corresponding ligand as cited above. Therefore, the lysC leader sequence appears to serve as a L-lysine-specific riboswitch that induces transcription termination by modulating the formation of a terminator stem.
Figure 5.
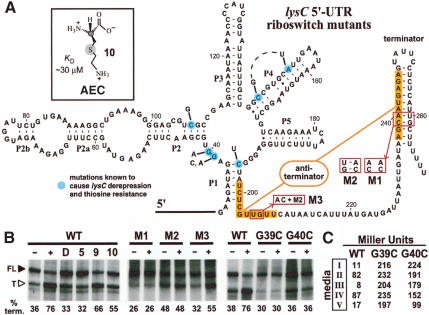
The B. subtilis lysC riboswitch and its mechanism for metabolite-induced transcription termination. (A) Sequence and repressed-state model for the lysC riboswitch secondary structure. The nucleotides highlighted in orange identify the putative antiterminator interaction that could form in the absence of L-lysine. Boxed nucleotides identify sites of disruption (M1) and compensatory mutations for the terminator stem (M2) and for the terminator and antiterminator stems (M3). Nucleotides shaded in light blue identify some of the positions in which mutations exhibit lysC derepression that were reported previously (Vold et al. 1975; Lu et al. 1992). These mutations confer resistance to AEC (inset). (B) Single-round in vitro transcription termination assays conducted in the absence (-) or presence (+) of 10 mM L-lysine or other analogs as indicated. (FL) Full-length transcripts; (T) terminated transcripts. The percent of the terminated RNAs relative to the total terminated and full-length transcripts are provided for each lane (% term.). (C) In vivo expression of a β-galactosidase reporter gene fused to wild-type (WT), G39A, and G40A mutant lysC 5′-UTR fragments. Media conditions are as follows: (I) normal medium (0.27 mM lysine); (II) minimal medium (0.012 mM); (III) lysine-supplemented minimal medium (1 mM); (IV) lysine hydroxamate-supplemented (medium II plus 1 mM lysine hydroxamate) minimal media; (V) AEC-supplemented (medium II plus 1 mM AEC) minimal medium.
To examine this hypothesis, in vitro transcription assays were conducted in the absence and presence of L-lysine and several analogs (Fig. 5B, left). In the absence of added ligand, single-round transcription in vitro using E. coli RNA polymerase produces terminated product corresponding to ∼36% of the total transcription yield. In contrast, the amount of terminated product increases to ∼76% when 10 mM L-lysine is present during in vitro transcription. We explored several concentrations of ligand and observed similar transcription termination extents with concentrations of L-lysine >10 mM, indicating that this concentration induces maximal modulation under our transcription conditions in vitro. Neither D-lysine nor L-ornithine induces termination at this concentration, which is consistent with the fact that these compounds are not recognized by the lysine aptamer domain and thus are not expected to trigger transcription termination.
It is important to note that our in vitro transcription assays were conducted by using a concentration of ligand that is substantially higher than that required to induce structural modulation of the 179 lysC aptamer. Concentrations of L-lysine in the low micromolar range are not sufficient to modulate transcription termination in vitro. There are several possible explanations for this observation. For example, we have established herein that the larger 315 lysC construct (most likely experiencing greater structural heterogeneity) exhibits a much poorer apparent KD. Therefore, it is possible that an RNA construct of ∼265 nucleotides representing the nascent RNA emerging from the RNA polymerase active site as it approaches the transcription termination site would have characteristics that are more reflective of the biologically relevant amino acid receptor. This construct, when placed in the complex with the RNA polymerase and its DNA template, might have a KD value that more closely matches the concentration of L-lysine needed to induce transcription termination. Alternatively, the process of genetic control might be a kinetically driven system in which the aptamer does not reach equilibrium between bound and unbound states, but in which the timing of RNA folding, ligand association, and speed of transcription are more directly involved in the decision whether to express the gene.
The configuration of the expression platform for the lysC gene in B. subtilis strongly implicates a transcription termination mechanism, wherein the binding of L-lysine is expected to stabilize the P1 stem, thus permitting formation of the terminator hairpin (Fig. 5A). This proposed mechanism was examined by placing mutations within the critical pairing elements and by assessing lysine-induced transcription termination (Fig. 5B, center). Specifically, variant M1 carries two mutations that disrupt the formation of the terminator stem. This variant loses lysine-dependent modulation of transcription termination, and produces greater transcriptional read-through relative to the wild-type construct. M2 carries a total of four mutations that compensate for the disruption of the terminator stem but cause disruption of the antiterminator stem. This construct also loses lysine-dependent modulation, whereas the amount of the terminated product expectedly becomes greater. Finally, the six-nucleotide variant M3 that carries the same mutations as M2 plus two additional mutations to restore the antiterminator base-pairing potential results in near wild-type performance with regards to lysine-mediated modulation of transcription termination. These findings are consistent with a riboswitch mechanism wherein lysine binding precludes formation of an antiterminator stem, thus increasing transcription termination by formation of an intrinsic terminator structure.
Evidence that riboswitches serve as drug targets
Unlike other lysine analogs, both L-lysine hydroxymate and the antimetabolite compound (AEC; Fig. 5A, inset) cause an increase in transcription termination (Fig. 5B, left). These two compounds exhibit the best apparent KD values of any of the analogs tested with 179 lysC, with values for L-lysine hydroxymate and AEC of ∼100 and ∼30 µM, respectively (data not shown). In previous studies, a series of mutants were identified in B. subtilis (Vold et al. 1975; Lu et al. 1992) and E. coli (Patte et al. 1998) that cause resistance to AEC and cause derepression of lysC expression. These mutations all map to the lysine aptamer domain (see Fig. 5A for select B. subilis mutants), and all appear to cause disruptions in the conserved elements or the base-pairing integrity of the structure.
The functional integrity of two AEC-resistant mutants (G39A and G40A) was examined by equilibrium dialysis and by in-line probing, and both mutants fail to exhibit lysine-binding activity (data not shown). Furthermore, RNA constructs that carry mutations in the otherwise conserved P1-P2 junction fail to undergo lysine-dependent transcription termination in vitro (Fig. 5B, right). It has been established that AEC is incorporated into the proteins of E. coli (e.g., see Hirshfield and Zamecnik 1972; Di Girolamo et al. 1990), most likely with toxic effects to the cell. However, our findings suggest that the toxic action of AEC might at least partially be due to direct binding of the analog to the lysine riboswitch, thus causing repression of aspartokinase expression to a level that is deleterious to cell growth.
To explore this possibility, the function of the wild-type 5′-UTR of lysC and of the two AEC-resistant mutants were examined in vivo by fusion to a lacZ reporter gene. Previously, lysine was shown to cause repression of B. subtilis lysC expression in both lysine prototroph and auxotroph strains (Lu et al. 1991). In the current study, we chose to use a lysine auxotroph strain (1A40) to ensure that lysine concentrations in vivo could be manipulated over a wide range. The wild-type riboswitch domain exhibits ligand-dependent modulation upon addition of L-lysine, whereas the G39A and G40A mutants fail to regulate β-galactosidase expression (Fig. 5C, medium II vs. III). In contrast, lysine hydroxymate fails to repress expression of the reporter gene in vivo (medium IV), indicating that this compound might not attain a sufficiently high concentration inside cells to trigger transcription termination.
As with lysine, AEC also substantially represses β-galactosidase expression for the wild-type construct, but causes very little reduction with the two derepression mutants (medium V). Because AEC shows little repression activity when the riboswitch is defective, it is not likely that AEC incorporation into the β-galactosidase enzyme is the cause for the substantial repression in the wild-type construct. Therefore, our observations are consistent with the hypothesis that the toxic effects of AEC could be due in part to its function as an effector for the lysine riboswitch. In other words, a compound that only targeted the lysine riboswitch, and that was not incorporated into proteins, might still be deleterious to cell growth under limiting conditions because lysine would not be synthesized in sufficient quantities.
Discussion
The first mutants that caused deregulation of lysine biosynthesis in B. subtilis were identified nearly three decades ago (Vold et al. 1975); however, the mechanism of genetic regulation has remained unresolved. In this study, we demonstrate that the 5′-UTR of the lysC mRNA from B. subtilis serves as a riboswitch that responds to the amino acid lysine. The derepressed mutants isolated in the original study cause disruption of the aptamer domain of the riboswitch, such that the ligand is no longer bound by the RNA. Furthermore, in vivo expression studies using mutant lysC fragment-reporter gene fusions indicate that these riboswitch mutations most likely cause unregulated overexpression of aspartokinase, which catalyzes the first step in the biosynthetic pathway to lysine and several other amino acids.
Bacteria use various mechanisms to respond genetically to amino acid concentrations. Two of the more prominent mechanisms, translation-mediated transcription attenuation and T-box-dependent mechanisms (Henkin and Yanofsky 2002), both sense the presence of nonaminoacylated tRNAs. Indeed, 18 of the 20 common amino acids in B. subtilis appear to be detected indirectly through the use of T-box elements. Interestingly, there is no known tRNAlys-dependent T box in any organism, and presumably the lysine riboswitch described herein serves as the genetic sensor for this amino acid in the absence of a corresponding T box. The genetic distribution of lysine riboswitches affiliated with the nhaC gene from several organisms (Fig. 1) indicates that this RNA genetic element might be a key regulator of cellular pH. In certain organisms, the activity of lysine decarboxylase, which consumes a proton upon conversion of L-lysine to cadaverine, is used to control pH (Meng and Bennett 1992). The nhaC gene encodes a proton antiporter (Na+/H+) that would not be necessary if lysine decarboxylase is used to regulate pH instead. Therefore, deactivation of nhaC might occur when an adequately high level of L-lysine is present.
Because the lysC mRNA functions as receptor for L-lysine, it seems reasonable to speculate that riboswitches might serve as targets for drug compounds (Hesselberth and Ellington 2002; Sudarsan et al. 2003). The lysine riboswitch, and perhaps other classes of riboswitches as well, could be targeted by analogs that selectively bind to the riboswitch and induce genetic modulation. In B. subtilis, an analog of lysine that triggers the riboswitch would be expected to function as an antimicrobial agent, because the reduction of aspartokinase expression should induce starvation for lysine and other critical metabolites. Our finding that AEC binds to the lysine aptamer in vitro to cause down-regulation of a reporter construct fused to the wild-type riboswitch provides support for this hypothesis. In addition, we have evidence that the antimicrobial compound pyrithiamine function by binding the thiamine pyrophosphate riboswitch (data not shown). Therefore, riboswitches could serve as a new class of targets for drug discovery efforts.
Recent discoveries have been elucidating the roles of small RNAs in guiding gene expression in a wide range of organisms (for review, see Gottesman 2002). It is apparent that small RNAs, including riboswitch domains embedded within mRNAs, can control gene expression by a wide range of mechanisms. Unlike other RNA genetic control elements, riboswitches directly bind to metabolites and control the expression of genes that are involved in the import and biosynthesis of a number of fundamental metabolites (Mandal et al. 2003). Riboswitches examined previously respond to compounds that are chemically related to nucleotides. However, the existence of a class of riboswitches that responds to a small amino acid with high selectivity serves as proof that natural RNA switches can detect and respond to a greater range of metabolite classes. This should provide encouragement to those who seek additional natural riboswitch classes, or for those who intend to create designer riboswitches by using molecular engineering strategies.
Materials and methods
Chemicals and oligonucleotides
L-lysine, all analogs with the exception of L-α-homolysine (compound 6, Fig. 3A), tritiated lysine (L-Lysine-[4,5-3H(N)]), and the four dipeptides were purchased from Sigma. A protocol adapted from that reported previously (Dong 1992) was used to synthesize L-α-homolysine. Purity and integrity of synthetic L-α-homolysine was confirmed by thin-layer chromatography (TLC) and nuclear magnetic resonance (NMR).
DNA oligonucleotides were synthesized by the Howard Hughes Medical Institute Keck Foundation Biotechnology Resource Center at Yale University, purified by denaturing polyacrylamide gel electrophoresis (PAGE), and eluted from the gel by crush-soaking in 10 mM Tris-HCl (pH 7.5 at 23°C), 200 mM NaCl, and 1 mM EDTA. Oligonucleotides were recovered from solution by precipitation with ethanol.
Phylogenetic analyses
L-box domains were identified by sequence similarity to the B. subtilis lysC 5′-UTR by using the program SequenceSniffer (unpublished algorithm) to search completed genomic sequences of prokaryotes. Ultimately, the program was used to search for degenerate matches to the pattern (WAGAGGNGC [10] A [3] RKTA [50] RRGR [10] CCGARR [40] GG [13] VAA [13] YTGTCA [36] TGRWG [2] CTWY); however, less complete versions of this pattern were used with iterative refinements to identify the consensus sequence and structure of the L-box motif. Bracketed numbers are variable gaps with constrained maximum lengths denoted. Nucleotide notations are as follows: Y indicates pyrimidine; R indicates purine; W indicates A or T; K indicates G or T; and V indicates A, G, or C. Up to six violations of this pattern were permitted when forming the phylogeny depicted in Figure 1. One representative (BX; Fig. 1) was obtained from a previous publication (Schendel and Flickinger 1992).
In-line probing ofRNA constructs
The B. subtilis 315 lysC, 237 lysC, and 179 lysC RNAs were prepared by in vitro transcription using T7 RNA polymerase and the appropriate PCR DNA templates. RNA transcripts were dephosphorylated and subsequently 5′ 32P-labeled by using a protocol similar to that described previously (Seetharaman et al. 2001). Labeled precursor RNAs (∼2 nM) were subjected to inline probing using conditions similar to those described previously (Soukup and Breaker 1999; Winkler et al. 2002b). Reactions (10 µL) were incubated at 25°C for 40 h in a buffer containing 50 mM Tris (pH 8.5 at 25°C), 20 mM MgCl2, and 100 mM KCl in the presence or absence of L-lysine or various analogs as indicated for each experiment. Denaturing 10% PAGE was used to separate spontaneous cleavage products, which were detected and quantitated by using a Molecular Dynamics PhosphorImager and ImageQuaNT software.
Equilibrium dialysis and scatchard analyses
Equilibrium dialysis assays were conducted by using a Dispo-Equilibrium Dialyzer (ED-1, Harvard Bioscience), wherein two chambers a and b were separated by a 5000 MWCO membrane. The final composition of buffer included 50 mM Tris-HCl (pH 8.5 at 25°C), 20 mM MgCl2, and 100 mM KCl (30 µL delivered to each chamber). Assays were initiated by the addition of 3H-lysine (50 nM initial concentration prior to equilibration; 40 Ci/mmole; 15,000 cpm) to chamber a. When present, RNA (179 lysC) was introduced into chamber b to yield a concentration of 10 µM. After 10 h of equilibration at 25°C, a 3 µL aliquot from each chamber was removed for quantitation by liquid scintillation counter. Competition assays were established by delivering an additional 3 µL of buffer to a and an equivalent volume of buffer containing 50 µM unlabeled L-lysine, D-lysine, L-ornithine, or L-lysine hydroxamate as indicated to b. After 10 h of additional incubation at 25°C, 3 µL aliquots were again drawn for quantitation of tritium distribution.
Scatchard data points were generated as described above with the following exceptions. RNA was added to chamber b to yield a concentration of 1 µM RNA and equilibration of the dialysis mixtures proceeded for 20 h. In addition, 3H-lysine concentrations were varied from 50 nM to 2.5 µM. Calculation of points on the Scatchard plot from the equilibrium dialysis data was carried out as described elsewhere (Winkler et al. 2003).
In vitro transcription termination assays
Transcription termination assays were conducted by using a method of single-round transcription adapted from that described previously (Landick et al. 1996). The template for lysC 5′-UTR transcription was altered (C6G of the RNA) such that the first C residue of the nascent RNA is not encountered until position 17. Polymerization was initiated by the addition of a mixture of ApA dinucleotide (1.35 µM), GTP and UTP (2.5 µM each) plus unlabeled ATP (1 µM), and [α-32P]-ATP (4 µCi), which was incubated for 10 min. Halted complexes were restarted by the addition of 150 µM each of the four NTPs, and heparin (0.1 mg/mL) was simultaneously added to prevent polymerases from initiating transcription on new templates. Transcription mixtures also contained 20 mM Tris-HCl (pH 8.0at 23°C), 20 mM NaCl, 14 mM MgCl2, 0.1 mM EDTA, 0.01 mg/mL BSA, 1% v/v glycerol, 4 pmole DNA template, 0.045 U/µL E. coli RNA polymerase (Epicenter), and 10 mM of L-lysine or the lysine analog as indicated for each experiment. Reactions were incubated at 37°C for an additional 20 min, and the products were examined by denaturing 6% PAGE followed by analysis using a Phosphor-Imager. At least three replicates were conducted for each assay with essentially identical results, and the data shown are representative of these experiments.
In vivo analysis of lysC genetic variants
Fusions of the lysC 5′-UTR with a lacZ reporter gene were used to assess the function of the lysine riboswitch in vivo using methods similar to those described previously (Winkler et al. 2003). Briefly, the lysC 5′-UTR, comprising the promoter and the first 315 nucleotides of the transcription template, was prepared as an EcoRI-BamHI fragment by PCR. Sequence variants M1-M3, G39A, and G40A were generated by PCR amplification of the wild-type construct by using primers that carried the desired mutations. The PCR products were cloned into pDG1661 immediately upstream of the lacZ reporter gene, and the integrity of the resulting clones were confirmed by sequencing. Transformations of pDG1661 variants into B. subtilis strain 1A40 (obtained from the Bacillus Genetic Stock Center) were performed, and the correct transformants (integration into the amyE locus) were identified by selecting for chloramphenicol resistance and screening for spectinomycin sensitivity.
Cells were grown with shaking at 37°C either in rich medium (2XYT broth or tryptose blood agar base) or defined medium (0.5% w/v glucose, 2 g/L [NH4]2SO4, 18.3 g/L K2HPO4 · 3H2O, 6 g/L KH2PO4, 1 g/L sodium citrate, 0.2 g/L MgSO4 · 7H2O, 5 µM MnCl2, and 5 µM CaCl2). Methionine, lysine, and tryptophan were added to 50 µg/mL for routine growth. Growth under lysine-limiting conditions was established by incubation under routine growth conditions in defined medium to an A595 of 0.1, at which time the cells were pelleted by centrifugation, resuspended in minimal medium, split into five aliquots, and supplemented with five different media types, as defined in the legend to Figure 5C. Cultures were incubated for an additional 3 h before performing β-galactosidase assays.
Acknowledgments
We thank Wade C. Winkler for pointing out the similarity between the loop E motif and a portion of the L-box domain. We also thank other members of the Breaker laboratory for comments on the manuscript. This work was supported by grants from the NIH (GM 559343) and the NSF (EIA-0129939; EIA-0323510; EIA-0324045). R.R.B. is also supported by a fellowship from the David and Lucile Packard Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1140003.
References
- Belitsky B.R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines. In Bacillus subtilis and its closest relatives: From genes to cells. (eds. A.L. Sonenshein, et al.), pp. 203-231. ASM Press, Washington, D.C.
- Boy E., Borne, F., and Patte, J.C. 1979. Isolation and identification of mutants constitutive for aspartokinase III synthesis in Escherichia coli K12. Biochimie 61: 1151-1160. [DOI] [PubMed] [Google Scholar]
- Di Girolamo M., Busiello, V., Coccia, R., and Foppoli, C. 1990. Aspartokinase III repression and lysine analogs utilization for protein synthesis. Physiol. Chem. Phys. Med. NMR 22: 241-245. [PubMed] [Google Scholar]
- Dong Z. 1992. An efficient asymmetric synthesis of L-α-ω-diaminoalkanoic acids. Tetrahedron Lett. 33: 7725-7726. [Google Scholar]
- Engel M.H. and Hare, P.E. 1982. Racemization rates of the basic amino acids. Year Book Carnegie Inst. Washington 81: 422-425. [Google Scholar]
- Epshtein V., Mironov, A.S., and Nudler, E. 2003. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. 100: 5052-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. 2002. Stealth regulation: Biological circuits with small RNA switches. Genes & Dev. 16: 2829-2842. [DOI] [PubMed] [Google Scholar]
- Henkin T.M., and Yanofsky, C. 2002. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. BioEssays 24: 700-707. [DOI] [PubMed] [Google Scholar]
- Hesselberth J.R. and Ellington, A.D. 2002. A (ribo) switch in the paradigms of genetic regulation. Nature Struct. Biol. 9: 891-893. [DOI] [PubMed] [Google Scholar]
- Hirshfield I.N. and Zamecnik, P.C. 1972. Thiosine-resistant mutants of Escherichia coli K-12 with growth-medium-dependent lysyl-tRNA synthetases activity, I: Isolation and physiological characterization. Biochim. Biophys. Acta 259: 330-343. [PubMed] [Google Scholar]
- Kochhar S. and Paulus, H. 1996. Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiology 142: 1635-1639. [DOI] [PubMed] [Google Scholar]
- Landick R., Wang, D., and Chan, C.L. 1996. Quantitative analysis of transcriptional pausing by E. coli RNA polymerase: his leader pause sites as a paradigm. Methods Enzymol. 274: 334-353. [DOI] [PubMed] [Google Scholar]
- Leontis N.B. and Westhof, E. 1998. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 283: 571-583. [DOI] [PubMed] [Google Scholar]
- Liao H.-H. and Hseu, T.-H. 1998. Analysis of the regulatory region of the lysC gene of Escherichia coli. FEMS Microbiol. Lett. 168: 31-36. [DOI] [PubMed] [Google Scholar]
- Lu Y., Chen, N.Y., and Paulus, H. 1991. Identification of aecA mutations in Bacillus subtilis as nucleotide substitutions in the untranslated leader region of the aspartokinase II operon. J. Gen. Microbiol. 137: 1135-1141. [DOI] [PubMed] [Google Scholar]
- Lu Y., Shevtchenko, T.N., and Paulus, H. 1992. Fine structure mapping of cis-acting control sites in the lysC operon of Bacillus subtilis. FEMS Microbiol. Lett. 92: 23-27. [DOI] [PubMed] [Google Scholar]
- Mäder U., Homuth, G., Scharf, C., Büttner, K., Bode, R., and Hecker. M. 2002. Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability. J. Bacteriol. 184: 4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Boese, B., Barrick, J.E., Winkler, W.C., and Breaker, R.R. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577-586. [DOI] [PubMed] [Google Scholar]
- McDaniel B.A.M., Grundy, F.J., Artsimovitch, I., and Henkin, T.M. 2003. Transcription termination control of the S-box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. 100: 3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S.Y. and Bennett, G.N. 1992. Nucleotide sequence of the Escherichia coli cad operon: A system for neutralization of low extracellular pH. J. Bacteriol. 174: 2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., and Nudler, E. 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747-756. [DOI] [PubMed] [Google Scholar]
- Nahvi A., Sudarsan, N., Ebert, M.S., Zou, X., Brown, K.L., and Breaker, R.R. 2002. Genetic control by a metabolite binding RNA. Chem. Biol. 9: 1043-1049. [DOI] [PubMed] [Google Scholar]
- Patte J.C. 1996. Biosynthesis of lysine and threonine. In Escherichia coli and Salmonella: Cellular and molecular biology, vol. 1 (eds. F.C. Neidhardt, et al.), pp. 528-541. ASM Press, Washington, DC. [Google Scholar]
- Patte J.-C., Akrim, M., and Méjean, V. 1998. The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol. Lett. 169: 165-170. [DOI] [PubMed] [Google Scholar]
- Ptashne M. and Gann, A. 2002. Genes and signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schendel F.J. and Flickinger, M.C. 1992. Cloning and nucleotide sequence of the gene coding for aspartokinase II from a thermophilic methylotrophic Bacillus sp. Appl. Environ. Microbiol. 58: 2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman S. Zivarts, M., Sudarsan, N., and Breaker, R.R. 2001. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nature Biotechnol. 19: 336-341. [DOI] [PubMed] [Google Scholar]
- Soukup G.A. and Breaker, R.R. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N., Barrick, J.E., and Breaker, R.R. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B., Szulmajster, J., and Carbone, A. 1975. Regulation of dihydrodipicolinate synthase and aspartate kinase in Bacillus subtilis. J. Bacteriol. 121: 970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W., Nahvi, A., and Breaker, R.R. 2002a. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952-956. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Cohen-Chalamish, S., and Breaker, R.R. 2002b. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 99: 15908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., and Breaker, R.R. 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10: 701-707. [DOI] [PubMed] [Google Scholar]