Summary
Bcl2 can enhance susceptibility to carcinogenesis but the mechanism(s) remains fragmentary. Here we discovered that Bcl2 suppresses DNA double-strand break (DSB) repair and V(D)J recombination by down-regulating Ku DNA binding activity, which is associated with increased genetic instability. Exposure of cells to ionizing radiation enhances Bcl2 expression in the nucleus which interacts with both Ku70 and Ku86 via its BH1 and BH4 domains. Removal of the BH1 or BH4 domain abrogates the inhibitory effect of Bcl2 on Ku DNA binding, DNA-PK and DNA end-joining activities, which results in failure of Bcl2 to block DSB repair as well as V(D)J recombination. Intriguingly, Bcl2 directly disrupts the Ku/DNA-PKcs complex in vivo and in vitro. Thus, Bcl2 suppression of the general DSB repair and V(D)J recombination may occur in a mechanism by inhibiting the non-homologous end-joining pathway, which may lead to an accumulation of DNA damage and genetic instability.
Introduction
DNA double-strand breaks (DSBs) are particularly detrimental because both strands are damaged (Hefferin et al., 2005), which can result from exogenous agents (i.e. ionizing radiation (IR) and certain chemotherapeutic drugs) or endogenously generated reactive oxygen species (Khanna et al., 2001; Van Gent et al., 2001). Estimates put the number of endogenous DSBs anywhere between 10 and 100 per nucleus per day (Burma et al., 2006). Inability to repair DSBs can lead to the accumulation of genomic rearrangements that potentially promote tumorigenesis (Khanna et al., 2001; Van Gent et al., 2001). DSBs can be repaired by two pathways: homologous recombination (HR) and non-homologous end joining (NHEJ) (Hefferin et al., 2005; Pasttwa et al., 2003). HR appears to be the predominant mechanism of DSB repair in yeast, which is operative only in the S/G2 phases of the cell cycle when a sister chromatid is available. By contrast, NHEJ, which simply pieces together the broken DNA ends, can function in all phases of the cell cycle. Therefore, NHEJ is the major pathway in mammalian cells for repairing DSBs (Burma et al., 2006; Hefferin et al., 2005). There are six major factors which unequivocally function in this pathway: two subunits of the DNA end binding heterodimer Ku70/Ku86, one catalytic subunit of DNA-PK (DNA-PKcs), Artemis and the XRCC4/DNA ligase IV complex (Burma et al., 2006; Hefferin et al., 2005). The Ku70/86 heterodimer initially recognizes and binds to broken ends of double-strand DNA and acts as an alignment factor to promote DNA end joining (Feldmann et al., 2000). DNA-PKcs is recruited to DNA whose end is bound to Ku which forms the active DNA-PK complex (Lees-Miller et al., 2003). After processing of the DNA ends, the XRCC4/DNA ligase IV complex is recruited to the DNA-PK complex and the DSBs are repaired, although that often occurs with a loss of nucleotides that may result in an inaccurate repair. Programmed DSBs are generated during V(D)J recombination which is necessary for generating antibody and T cell receptor diversity (Burma et al., 2006; Jones et al., 2004). Completion of the V(D)J recombination reaction, including joining of coding or recombination signal (RS) ends, is mediated by the NHEJ pathway (Bassing et al., 2002), which requires multiple members of the DSB repair machinery including Ku 70/86, Artemis and DNA-PKcs. The Ku- or DNA-PKcs-deficient cells have defects in their ability to support the V(D)J recombination reaction (Gu et al., 1997; Rooney et al., 2002; Zhu et al., 1996). Recombination-activating gene proteins 1 and 2 (RAG1 and RAG2) generate DSBs precisely between conserved recombination signal sequences (RSS) and flanking V, D or J coding sequences of two gene segments. This reaction results in two distinct intermediates: RS ends with blunt 5′ phosphorylated DSBs and coding ends in the form of covalently sealed hairpin. Two coding ends or two RS ends are then joined to form coding and RS joins, respectively, in a reaction that employs the NHEJ pathway (Rooney et al., 2002).
Bcl2 is a cellular proto-oncogene that functions as a potent anti-apoptotic molecule and tumor promoter (Linette et al., 1995). Overexpression of Bcl2, which results from the translocation of t(14;18), leads to the dysregulation of apoptotic cell death during the transformation process in follicular B-cell lymphoma and lymphomagenesis in transgenic mice (McDonnell et al., 1991; Tsujimoto et al., 1985). Several studies have suggested that Bcl2-mediated oncogenesis is associated with the enhancement of genetic instability and mutagenesis that are correlated with increased DNA damage and impaired DNA repair (Chebonnel-Lasserrre et al., 1996; Kuo et al., 1999; Liu et al., 1997; Youn et al., 2005). We have recently discovered that Bcl2 potently attenuates the repair of nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone -induced a basic sites of DNA lesions by enhancing c-Myc transcriptional activity (Jin et al., 2006). However, it is currently unclear whether and how Bcl2 affects DSB repair in promoting genomic instability and carcinogenesis. Here we provide strong evidence that Bcl2 can inhibit DSB repair as well as formation of signal and coding joins in the V(D)J recombination in a mechanism involving a Ku/DNA-PKcs-mediated NHEJ pathway.
Results
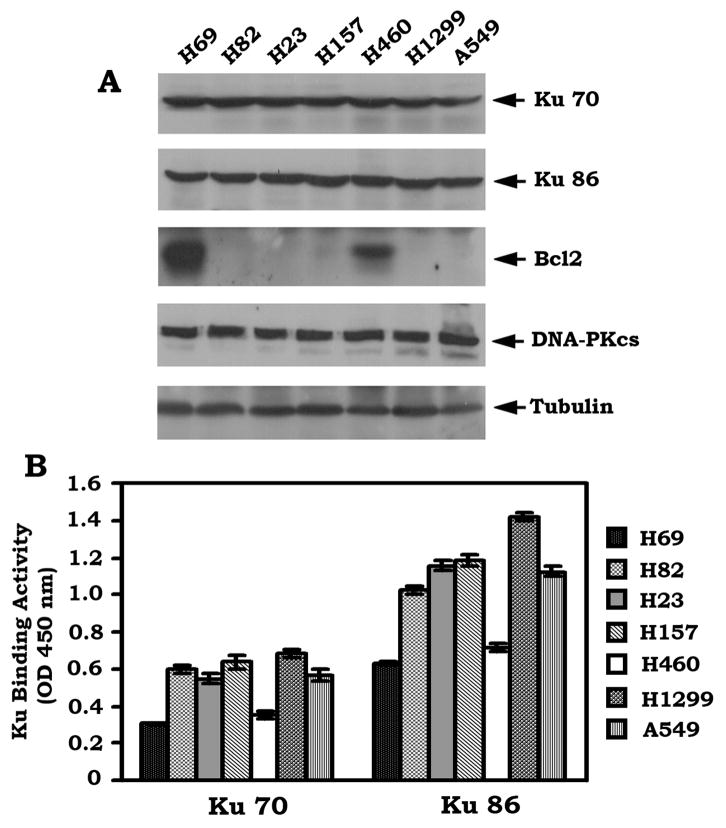
Expression of endogenous Bcl2 is associated with decreased levels of Ku activity in various human lung cancer cells
Bcl2 is involved in regulating DNA damage and repair, which may be associated with genomic instability and mutagenesis that promote tumor development (Jin et al., 2006; Kuo et al., 1999; Youn et al., 2005). Ku70, Ku86 and DNA-PKcs are the major components of the NHEJ machinery required for DSB repair and are widely expressed in both small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) cells (Figure 1A). Interestingly, Bcl2 is co-expressed with the NHEJ proteins in H69 and H460 but not in other lung cancer cells tested (Figure 1A). To test whether Bcl2 can affect the function of Ku proteins, the Ku DNA binding activity was assessed using a Ku70/86 DNA Repair Kit. A decreased level of Ku activity was observed in both H460 and H69 cells that express high levels of endogenous Bcl2 as compared to the other cell lines that express undetectable levels of Bcl2 (Figure 1B). Thus, Bcl2 expression appears to be correlated with a reduced Ku activity in human lung cancer cells, suggesting that Bcl2 may be involved in regulating the NHEJ pathway in the development of lung cancer.
Figure 1. Expression of endogenous Bcl2 is associated with decreased Ku DNA binding activity.
(A) Expression levels of endogenous Bcl2, Ku 70, Ku 86 and DNA-PKcs in human lung cancer cells were analyzed by Western blot. (B) DNA binding activity of Ku 70 or Ku 86 was measured using a Ku70/86 DNA Repair Kit. Error bars represent ± S.D.
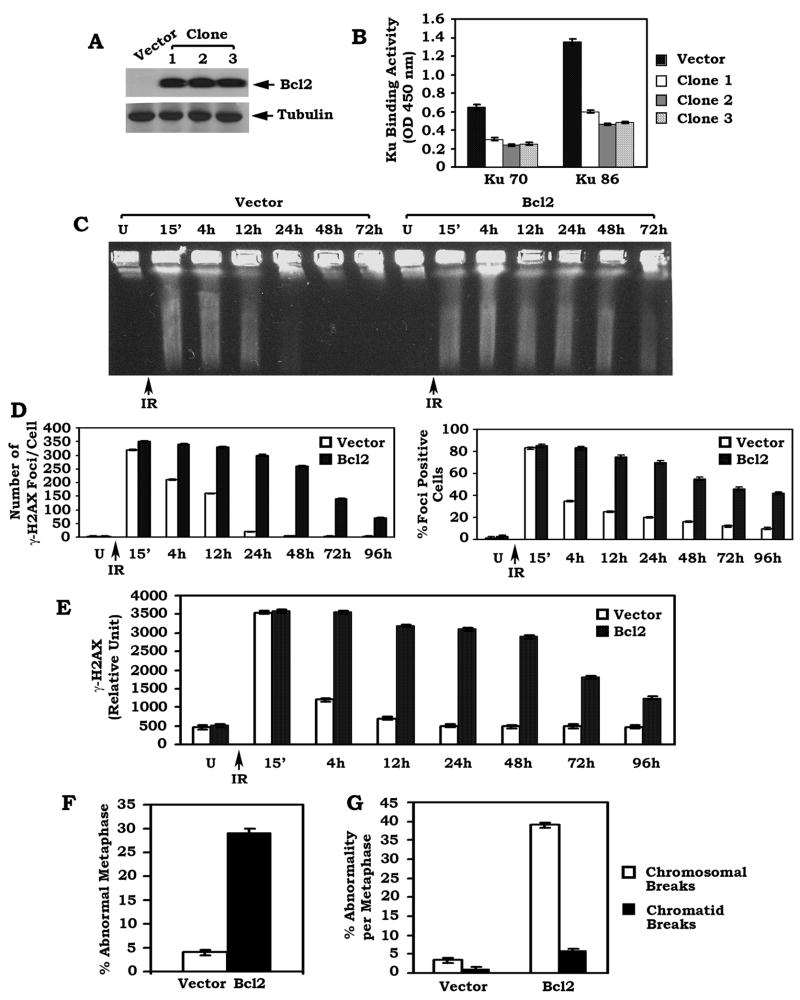
Overexpression of Bcl2 down-regulates Ku DNA binding activity in association with suppression of DSB repair and increased genomic instability
To directly test whether Bcl2 may negatively regulate Ku function and DSB repair, Bcl2 was stably expressed in H1299 cells that do not express detectable levels of endogenous Bcl2. Results indicate that overexpression of Bcl2 significantly inhibits the DNA binding activity of both Ku70 and Ku86 more than 50% as compared to vector-only control (Figure 2A–B). Similar results were obtained from three independent clones expressing similar levels of exogenous Bcl2, indicating that these findings are reliable. Since the NHEJ mechanism is considered the major repair pathway of DSBs in mammalian cells (Critchlow et al., 1998; Pastink et al., 2001; Pierce et al., 2001), Bcl2-mediated inhibition of Ku activity may functionally regulate DSB repair. To test this possibility, a pulsed-field gel electrophoresis (PFGE) under neutral conditions (a direct method for DSB measurement) was performed as described (Ager et al., 1990). About 15 min after ionizing radiation (IR), a significant amount of genomic DNA from all cells ran out of the wells and migrated into the gel, which indicates DNA fragmentation (i.e. DSBs). By 24h after IR, the majority of genomic DNA from vector-only control cells was retained in the well (Figure 2C), indicating that majority of DSBs had been repaired. In contrast, DNA fragmentation can still be observed after 72h in cells overexpressing Bcl2 (Figure 2C).
Figure 2. Overexpression of exogenous Bcl2 down-regulates Ku activity in association with decreased DSB repair and increased genetic instability.
(A) WT Bcl2/pCIneo DNA construct was stably transfected into H1299 human lung cancer cells. Expression of Bcl2 was determined by Western blot. (B) DNA binding activity of Ku 70 or Ku 86 was measured using a Ku70/86 DNA Repair Kit. Error bars represent ± S.D. (C–E) H1299 cells overexpressing WT Bcl2 or vector control were exposed to 1 Gy of ionizing radiation (IR). Cells were harvested at various time points. DSBs were determined by PFGE or analysis of γ-H2AX by immunostaining and Western blot. The number of γ-H2AX foci per cell was determined on a cell to cell basis by the quantitative analysis of at least 30 randomly chosen cells as described (Balajee et al., 2004). The percentage of γ-H2AX foci positive cells was determined by analyzing 100 randomly chosen cells as described (Chowdhury et al., 2005). Levels of γ-H2AX protein from Western blot were quantified by densitometry. Error bars represent ± S.D. (F) The percentage of abnormal metaphases from Bcl2 overexpressing cells or vector-only control cells was quantified by T-FISH analysis. At least 30 metaphases per culture were analyzed. Error bars represent ± S.D. (G) Frequency of cytogenetic abnormality per metaphase in Bcl2 overexpressing cells or vector-only control cells. Each experiment was repeated three times and error bars represent ± S.D.
Formation of a chromatin-associated γ-H2AX focus is considered to be a sensitive and selective signal for the existence of DSBs (Modesti et al., 2001; Rogakou et al., 1999). IR potently induces formation of γ-H2AX foci in both vector-only control and Bcl2 overexpressing cells after exposure of cells to IR (Figure S1A). The intensity and number of γ-H2AX foci per cell as well as the percentage of foci-positive cells are significantly reduced by 12 ~24h in vector-only control cells. Intriguingly, an increased number of γ-H2AX foci with stronger intensity per cell and an higher percentage of foci-positive cells are observed in cells overexpressing WT Bcl2 as compared to vector-only control after 12~24h (Figure S1A and Figure 2D–E). Western blot analysis using γ-H2AX provides further evidence that expression of Bcl2 significantly prolongs the persistence of γ-H2AX in cells after IR (Figure S1B and Figure 2E). Since similar results were also obtained in another cell type (i.e. MCF7, a breast cancer cell line) (Figure S2), this suggests that the inhibitory effect of Bcl2 on DSB repair is a general reaction and not a cell type specific phenomenon.
To test the effect of Bcl2 on chromosomal aberrations, we employed a fluorescence in situ hybridization (FISH) assay that combines DAPI staining with a telomere-specific peptide nucleic acid (PNA) probe (T-FISH) to assess metaphase chromosomes for instability as described (Zha et al., 2007). Increased levels of cytogenetic abnormalities, including chromosomal and chromatid breaks, were observed in Bcl2 overexpressing cells as compared to vector-only control cells (Figure S1C and Figure 2F–G). These findings reveal that Bcl2-mediated inhibition of DSB repair may promote genetic instability.
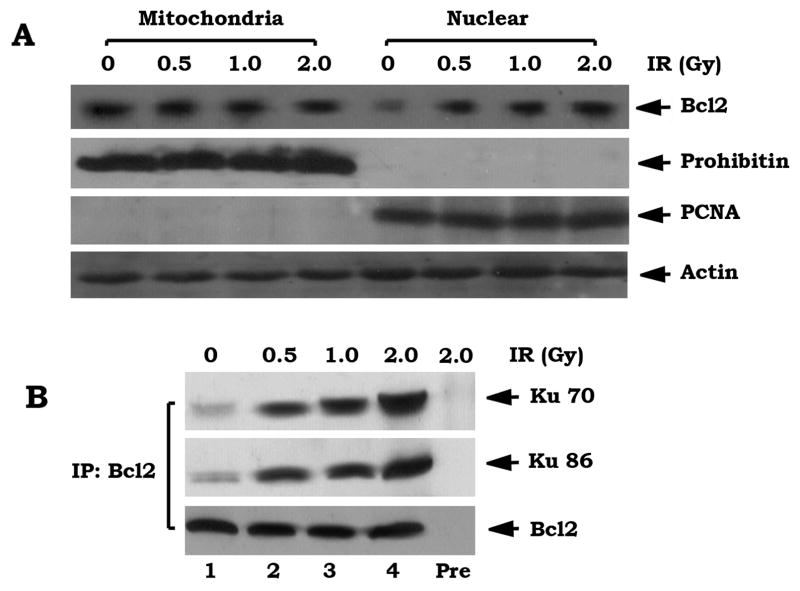
IR-induced DSB signal stimulates accumulation of Bcl2 in nucleus, which interacts with Ku proteins
Bcl2 is primarily localized in the outer mitochondrial membranes with minor expression in the nucleus (Hockenbery et al., 1990). Recent reports indicate that Bcl2 also resides in the nucleoplasm and may even function within the nucleus (Jin et al., 2006; Schardle et al., 1999). In contrast, the Ku protein mainly localizes and functions in nucleus (Bertinato et al., 2000). To test how Bcl2 regulates DSB repair and whether a DSB signal may affect subcellular expression of Bcl2, H460 cells expressing high levels of endogenous Bcl2 were exposed to increasing doses of IR. Subcellular distribution of Bcl2 was then examined by subcellular fractionation. As shown in Figure 3A, IR-induced DSBs enhance Bcl2 expression in nucleus in a dose-dependent manner.
Figure 3. IR-induced DSB signal promotes accumulation of Bcl2 in nucleus and interaction with Ku 70 and Ku 86.
(A) H460 cells were treated with increasing doses of IR. Subcellular fractionation was performed in H460 cells to isolate nuclear (Nuc) and mitochondrial (Mito) fractions. Western blot analysis of subcellular fractions was performed to detect Bcl2. Prohibitin or PCNA was used as a mitochondrial or nuclear marker, respectively. (B) A co-IP experiment was performed using isolated nuclear fraction and an agarose-conjugated Bcl2 antibody. Bcl2 and the Bcl2-associated Ku 70 or Ku 86 were analyzed by Western blot. Rabbit preimmune serum (Pre) was used as a control.
To test for a direct interaction between Bcl2 and Ku, a co-immunoprecipitaion (co-IP) using isolated nuclear extracts and an agarose-conjugated Bcl2 antibody was performed. Results reveal that the IR-induced DSB signal promotes Bcl2 to directly associate with Ku 70 or Ku 86 in nucleus (Figure 3B). Importantly, no detectable levels of Bcl2/Ku complexes were observed in the cytosolic fraction (data not shown), indicating that IR-stimulated Bcl2/Ku interaction may only occur in the nucleus. Thus, finding a direct interaction between Bcl2 and Ku proteins in the nucleus, strongly suggests that Bcl2 may act directly on the NHEJ factor Ku 70/86.
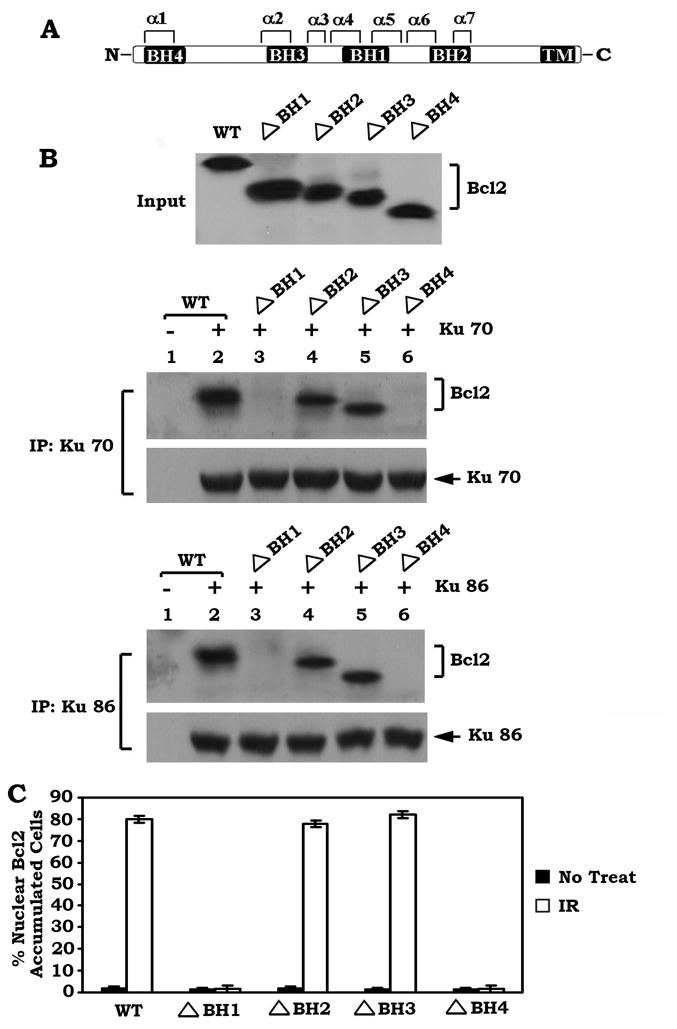
Bcl2 directly interacts with Ku via its BH1 and BH4 domains which are required for Ku mediated nuclear sequestration of Bcl2
Bcl2 family members share homology in the Bcl2 homology (BH) domains including BH1, BH2, BH3 and BH4 (Kelekar et al., 1998). To directly assess whether Bcl2 can bind to Ku at these BH domains, purified Ku70 or Ku86 protein was incubated with purified recombinant WT or each of the BH deletion Bcl2 mutants in 1% CHAPS lysis buffer at 4 °C for 1–2h. Intriguingly, Ku70 or Ku86 is able to associate with WT, ΔBH2 and ΔBH3 but not with the ΔBH1 and ΔBH4 Bcl2 mutants (Figure 4B), indicating that the BH1 and BH4 domains are the Ku binding sites on Bcl2. To test whether the Ku binding capacity is essential for the nuclear sequestration of Bcl2, a series of BH domain deletion mutants were created and stably expressed in H1299 cells. Results indicate that approximately 80% of the nuclear Bcl2 accumulated cells were observed in cells expressing WT, ΔBH2 and ΔBH3 Bcl2 mutants but not in those cells expressing the Ku binding deficient Bcl2 mutants (ΔBH1 and ΔBH4) after IR (Figure S3 and Figure 4C). Thus, the Ku binding sites (i.e. the BH1 and BH4 domains) may be required for the nuclear sequestration of Bcl2.
Figure 4. Bcl2 directly interacts with Ku via the BH1 and BH4 domains.
(A) Schematic representation of the Bcl2 homology domains (BH) in Bcl2 protein. (B) Purified Ku 70 or Ku 86 (0.1μg) was incubated with purified WT or each of the BH deletion Bcl2 mutants (0.1μg each) in 1% CHAPS lysis buffer at 4°C for 1–2h. The Ku-associated Bcl2 was co-immunoprecipitated with a Ku 70 or Ku 86 antibody and analyzed by Western blot. (C) H1299 cells expressing WT or each of various Bcl2 BH deletion mutants were exposed to IR (1Gy). Subcellular distribution of Bcl2 was analyzed by immunofluorescent staining. The percentage of nuclear Bcl2 accumulated cells was determined by analyzing 300 randomly chosen cells. Error bars represent ± S.D.
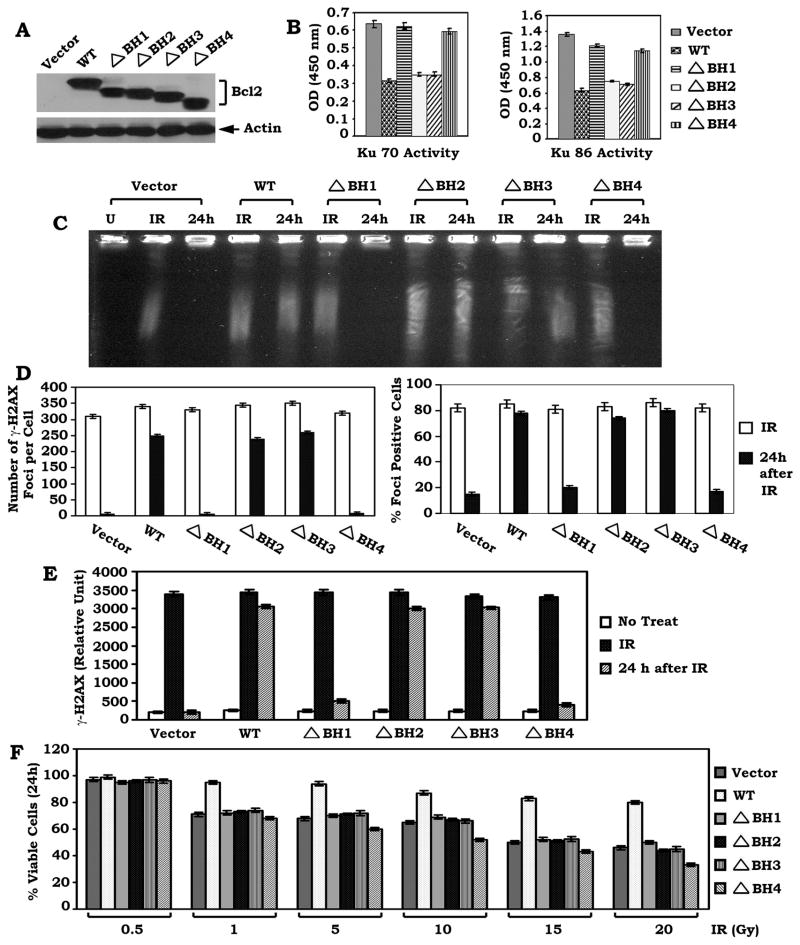
BH1 and BH4 domains are required for Bcl2 suppression of Ku DNA binding activity and DSB repair
To test the functional role of the Bcl2/Ku binding in Ku activity and DSB repair, the Ku activity and DSB repair were compared in H1299 cells expressing similar levels of WT or each of the BH deletion Bcl2 mutants. Results reveal that expression of WT, ΔBH2 and ΔBH3 but not the ΔBH1 or ΔBH4 Bcl2 mutant significantly suppresses Ku70 or Ku86 activity (Figure 5A–B). Functionally, deletion of either the BH1 or BH4 domain from Bcl2 results in failure of Bcl2 to inhibit the repair of IR-induced DSBs as determined by PFGE or analysis of γ-H2AX by immunostaining and Western blot (Figure 5C–E and Figure S4). In contrast, removal of the BH2 or BH3 domain does not affect Bcl2’s ability to suppress DSB repair. This supports the notion that the binding of Bcl2 to Ku via the BH1 or BH4 domain may be required for the inhibitory effect of Bcl2 on either Ku70/86 activity or DSB repair. To uncover a functional relationship between the antiapoptotic activity of Bcl2 and its inhibitory role in DSB repair, the effect of WT or each of the BH deletion Bcl2 mutants on survival was measured by a clonogenic survival assay following IR treatment. Interestingly, all H1299 cells expressing the ΔBH1, ΔBH2, ΔBH3 or ΔBH4 Bcl2 mutant display less viability as compared to WT Bcl2 expressing cells (Figure 5F). These results indicate that all of the BH domains in Bcl2 are required for Bcl2’s full and potent antiapoptotic function, which is consistent with previous reports (Castelli et al., 2004; Cheng et al., 1997; Reed et al., 1996). However, only the BH1 and BH4 domains are essential for the effect of Bcl2 on DSB repair (Figure 5), suggesting that the antiapoptotic function of Bcl2 may be not required for Bcl2 mediated suppression of DSB repair.
Figure 5. BH1 and BH4 domains are essential for Bcl2 to suppress Ku DNA binding activity and DSB repair.
(A) WT or each of the BH deletion Bcl2 mutants was stably transfected into H1299 cells. Expression of Bcl2 was analyzed by Western Blot. (B) DNA binding activity of Ku 70 or Ku 86 was measured using a Ku70/86 DNA Repair Kit. Error bars represent ± S.D. (C–E) H1299 cells expressing WT or each of the BH deletion Bcl2 mutants were exposed to 1 Gy of IR. After 24h, DSBs were measured by PFGE or analysis of γ-H2AX by immunostaining or Western blot using a γ-H2AX antibody with quantitative evaluation as described in the legend of Figure 2C–E. (F) H1299 cells expressing WT or each of the BH deletion Bcl2 mutants were treated with increasing doses of IR (0.5 to 20 Gy). After 24h, cell viability was determined by clonogenic survival assay. Error bars represent ± S.D.
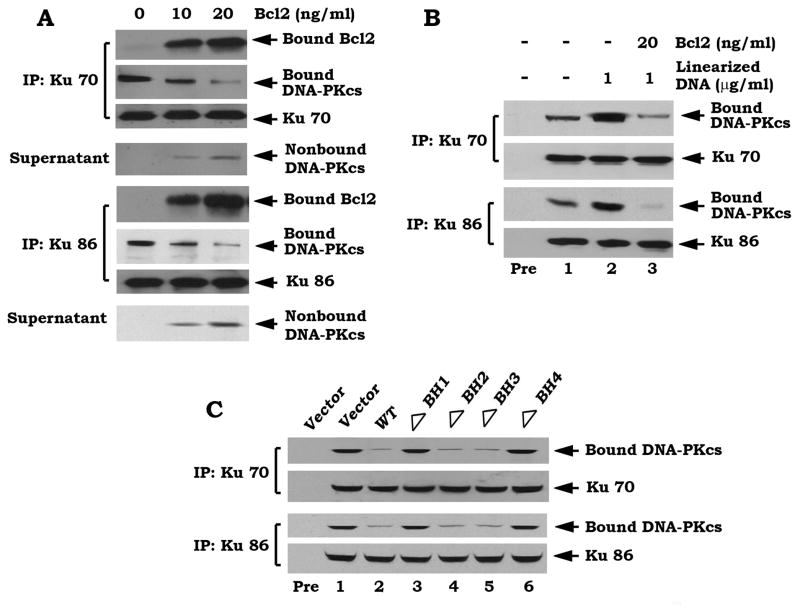
Bcl2 directly disrupts the Ku/DNA-PKcs complex
It is known that DNA-PK is composed of the Ku70/86 heterodimer and DNA-PK catalytic subunit (DNA-PKcs) which exists as a large complex (Yaneva et al., 1997). Since Bcl2 can directly interact with Ku70 or Ku86 via its BH1 and BH4 domains (Figures 3–4), Bcl2 may affect the functional Ku/DNA-PKcs complex by binding to Ku. To directly test this, the Ku70/DNA-PKcs or Ku86/DNA-PKcs complex was co-immunoprecipitated from H1299 parental cells. The immune complex was incubated with purified, recombinant Bcl2 at 4°C for 1–2 h and proteins released from the complex were identified in the supernatant following centrifugation. Interestingly, Bcl2 can directly disrupt the Ku70/DNA-PKcs or Ku86/DNA-PKcs complex in vitro since addition of purified Bcl2 results in decreased levels of bound DNA-PKcs and increased levels of nonbound DNA-PKcs (Figure 6A). Because the protein kinase activity of DNA-PK can be activated when Ku binds to altered DNA structures including DSB, nicks or hairpin loops (Zhu et al., 1996), the linearized double-strand DNA may facilitate the Ku/DNA-PKcs interaction. To test the effect of linearized double-strand DNA on the physiological interactions of these proteins in vitro, we generated a linearized double-strand DNA using the pGL3 plasmid and the restriction endonuclease NarI. As shown in Figure 6B, addition of linearized double-strand DNA significantly enhances the Ku70/DNA-PKcs or Ku86/DNA-PKcs association, which is inhibited by addition of purified Bcl2 protein. Intriguingly, overexpression of Bcl2 does not affect expression levels of either Ku or DNA-PKcs but significantly inhibits Ku/DNA-PKcs association (Figure S5). Since overexpression of WT, the ΔBH2 or ΔBH3 but not the BH1 or BH4 Bcl2 mutant potently suppresses the Ku/DNA-PKcs binding (Figure 6C), this indicates that deletion of the BH1 or BH4 domain from Bcl2 results in failure of Bcl2 to disrupt the Ku/DNA-PKcs binding. Thus, the inhibitory effect of Bcl2 on Ku/DNA-PKcs binding requires the BH1 and BH4 domains (i.e. Ku binding sites).
Figure 6. Bcl2 directly disrupts the Ku/DNA-PKcs complex.
(A) The Ku 70/DNA-PKcs or Ku 86/DNA-PKcs complex was co-immunoprecipitated from H1299 parental cells and incubated with purified Bcl2 (10–20ng/ml) in 1% CHAPS lysis buffer at 4 °C for 1h. After centrifugation, the resulting supernatant and immuno-complex beads were subjected to SDS-PAGE. Ku, Bcl2 and DNA-PKcs that bound to Ku or unbound DNA-PKcs present in the supernatant were then analyzed by Western blot. (B) The Ku 70/DNA-PKcs or Ku 86/DNA-PKcs complex was incubated with linearized DNA in the absence or presence of purified WT Bcl2 (20ng/ml) in 1% CHAPS lysis buffer at 4 °C for 1h. The Ku/DNA-PKcs complex was analyzed as described above. (C) H1299 cells expressing WT or each of the BH deletion Bcl2 mutants were disrupted in 1% CHAPS lysis buffer. Co-IP was performed using an agarose-conjugated Ku 70 or Ku 86 antibody, respectively. The Ku-associated DNA-PKcs was then analyzed by Western blot. Rabbit preimmune serum (Pre) was used as a control.
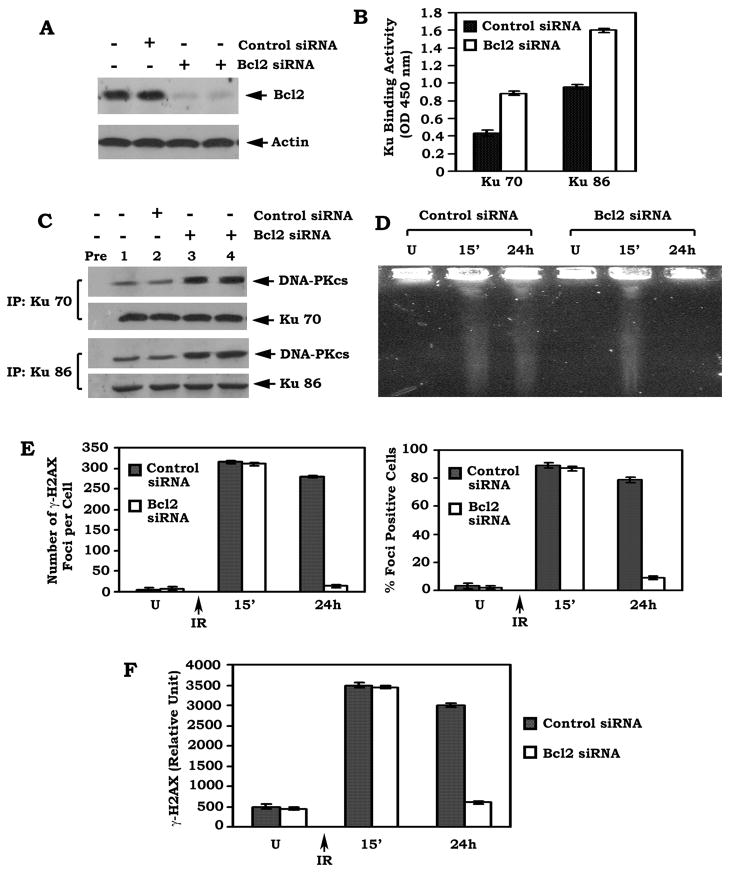
Depletion of Bcl2 by RNAi enhances Ku DNA binding activity and formation of the Ku/DNA-PKcs complex in association with accelerated DSB repair
To physiologically test a role of Bcl2 in DSB repair, a gene silencing approach was employed. Transfection of Bcl2 siRNA significantly reduces the expression level of endogenous Bcl2 by more than 95% in H460 cells (Figure 7A). This effect of siRNA on Bcl2 expression is highly specific since the control siRNA has no effect. Functionally, depletion of Bcl2 by RNAi not only up-regulates Ku DNA binding activity but also enhances formation of the Ku/DNA-PKcs complexes in association with accelerated repair of IR-induced DSBs as determined by either PFGE or analysis of γ-H2AX by immunostaining and Western blot (Figure 7B–F and Figure S6). These findings provide strong evidence that physiologically expressed Bcl2 is able to suppress DSB repair by a mechanism involving the NHEJ pathway.
Figure 7. Depletion of endogenous Bcl2 by RNAi enhances Ku DNA binding activity and formation of the Ku/DNA-PKcs complex in association with accelerated DSB repair.
(A) H460 cells expressing high levels of endogenous Bcl2 were transfected with Bcl2 siRNA or control siRNA. Expression of Bcl2 was analyzed by Western blot. (B) Ku 70 or Ku 86 DNA binding activity in H460 cells expressing Bcl2 siRNA or control siRNA was analyzed. Error bars represent ± S.D. (C) H460 cells expressing Bcl2 siRNA or control siRNA were disrupted in 1% CHAPS lysis buffer. A co-IP was performed using an agarose-conjugated Ku 70 or Ku 86 antibody, respectively. The Ku-associated DNA-PKcs was then analyzed by Western blot. Rabbit preimmune serum (Pre) was used as a control. (D–F) H460 cells expressing Bcl2 siRNA or control siRNA were exposed to 1 Gy of IR. After 24h, DSBs were measured by PFGE or analysis of γ-H2AX by immunostaining or Western blot using a γ-H2AX antibody with quantitative evaluation as described in the legend of Fig. 2C–E. Error bars represent ± S.D.
Expression of Bcl2 inhibits DNA-PK activity in association with suppression of DNA end-joining and DSB repair
DNA-PK, including the Ku70/Ku86 heterodimer and DNA-PKcs, plays an important role in repairing DSBs (Smith et al., 1999; Yaneva et al., 1997). Since Bcl2 can potently impede formation of the Ku/DNA-PKcs complex via binding to Ku (Figure 6), DNA-PK activity was further assessed using a Signa TECT DNA-PK Assay Kit. Results indicate that stable expression of WT or either the ΔBH2 or ΔBH3 Bcl2 mutant results in a significant decrease of DNA-PK activity while expression of the ΔBH1 or ΔBH4 Bcl2 mutant has no effect (Figure S7A), indicating that Bcl2 mediated suppression of DNA-PK activity may occur through a mechanism involving binding to Ku via the BH1 and BH4 domains.
To test the effect of NU7026 (a DNA-PK specific inhibitor; Willmore et al., 2004) on Bcl2 mediated suppression of DSB repair, H1299 cells overexpressing WT or each of the BH deletion Bcl2 mutants were exposed to IR (1 Gy) in the absence or presence of NU7026 (10μM). In the absence of NU7026, expression of WT or the ΔBH2 or ΔBH3 but not the ΔBH1 or ΔBH4 Bcl2 mutants potently suppresses DSB repair (Figure S7B–D). In contrast, DSB repair is completely defective in the presence of NU7026 in all cell lines that express or do not express Bcl2 (Figure S7B–D). Thus, Bcl2 may have no additional inhibitory role on DSB repair after inhibition of DNA-PK activity. These findings support the notion that DNA-PK may function as a required downstream target of Bcl2 for its effect on DSB repair.
To further test whether Bcl2 affects DNA end-joining capacity, DNA nonhomologous end-joining activity was measured as described (Shin et al., 2004). Intriguingly, expression of WT Bcl2 significantly inhibits DNA end-joining activity and deletion of either BH1 or BH4 but not the BH2 or BH3 domain dramatically abolishes Bcl2’s ability to suppress DNA end-joining (Figure S7E), indicating that the binding of Bcl2 to Ku via the BH1 and BH4 domains may also be critical for its effect on DNA end-joining.
Expression of Bcl2 inhibits formation of RS and coding joins in V(D)J recombination
Recent reports have demonstrated that Ku and DNA-PKcs are required for formation of both RS and coding joins during V(D)J recombination (Gu et al., 1997; Rooney et al., 2002). To test the effects of Bcl2 on the levels and quality of V(D)J recombination, we assessed the ability of RAG-transfected H1299 cells expressing WT or each of the BH deletion Bcl2 mutants to undergo V(D)J recombination in the context of transiently transfected substrates designed to test the formation of either RS join (pJH200) or coding join (pJH290) as described previously (Gu et al., 1997). At least four independent transfection experiments were carried out with each cell line. Results reveal that expression of WT or the ΔBH2 or ΔBH3 Bcl2 mutant results in significant impairment (i.e. more than 50%) in the ability to form both RS and coding joins as compared to vector-only control (Table S1). The Ku binding capacity of Bcl2 may be essential for this effect since overexpression of the Ku binding deficient Bcl2 mutants (i.e. ΔBH1 or ΔBH4) have no effect on either RS or coding joining (Table S1). It is known that a precise RS join generates an ApaLI site and the loss of nucleotides from the RS ends will prevent the formation of the ApaLI site. Digestion of PCR products generated from the recombination substrate pJH 200 with this ApaLI restriction enzyme was carried out for the analysis of fidelity of the RS joins as described (Gu et al., 1997). Intriguingly, less than half of the RS joins recovered from H1299 cells expressing WT or the ΔBH2 or ΔBH3 Bcl2 mutant, were cut by ApaLI (i.e. 38–50%), indicating that expression of Bcl2 also reduces the fidelity of RS joins, which is dependent on Bcl2’s Ku binding capacity because expression of the Ku binding deficient Bcl2 mutants (i.e. ΔBH1 or ΔBH4) does not affect the fidelity of RS joins (Table S1). Thus, in addition to the general DSB repair process, Bcl2 may also negatively regulate the repair of DSBs generated by RAG1/2 during the V(D)J recombination reaction.
Discussion
DSBs are toxic genomic DNA lesions that, if unrepaired or repaired incorrectly, can lead to mutations, chromosomal breaks or translocations with malignant transformation (Pasttwa et al., 2003; Stucki et al., 2005). Cancer is very often associated with the accumulation of genomic aberrations including large deletions, inversions or translocations, which may be a consequence of DSBs (Lengauer et al., 1998). Bcl2 was originally discovered as a gene product at the chromosomal breakpoint of t (14;18) (Tsujimoto et al., 1985), which may play a role in genetic instability and tumor development by regulating DSB repair. Here we found that expression of Bcl2 results in decreased Ku DNA binding activity with suppression of DSB repair (Figure. 2), suggesting that the inhibitory effect of Bcl2 on DSB repair may occur through a mechanism that prevents the NHEJ factor (i.e. Ku70/86) from binding the broken DNA ends. Because IR-induced DSB signal potently stimulates Bcl2 accumulation in nucleus which efficiently interacts with Ku (Figures 3, 4 and S3), the NHEJ factor Ku 70/86 may function as a nuclear “receptor” associating with Bcl2 to retain Bcl2 in nucleus. DSB-enhanced expression of nuclear Bcl2 is not likely due to a translocation of Bcl2 from mitochondria because levels of mitochondrial Bcl2 have no significant change (Figure 3A). This may occur through a transcriptional or other unknown mechanism(s).
There are three possible fates of cells from IR-induced DSBs: (1) DSBs are rapidly repaired via the NHEJ pathway leading to cell survival; (2) Apoptosis; (3) Cell survival with unrepaired or partially repaired DSBs and/or subsequent genetic instability. Because the NHEJ factors that mediate DSB repair appear to promote cell survival, Bcl2, as a survival protein, is supposed to promote DSB repair. Unexpectedly, our data reveal that Bcl2 potently suppresses the repair of IR-induced DSBs in various cell types (Figures 2 and S2). Obviously, the survival function of Bcl2 is not a result from its inhibitory effect on DSB repair, which occurs in a mitochondria-dependent mechanism (Deng et al., 2004; Hockenbery et al, 1990). Thus, the inhibitory effect of Bcl2 on DSB repair, plus its role in supporting survival, may result in cell survival with unrepaired or partially repaired DSBs (i.e. the third fate). We propose that Bcl2 may have two separate functions (i.e. survival and inhibition of DSB repair) via separate mechanisms. The antiapoptotic function of Bcl2 mainly results from the mitochondrial Bcl2, which maintains mitochondrial integrity (Green et al., 1998). The inhibitory effect of Bcl2 on DSB repair may result mainly from the nuclear portion of Bcl2. The Ku/Bcl2 complex can be observed only in isolated nuclear extracts but not in cytosolic fraction (Figure 3 and data not shown), suggesting that Bcl2-mediated inhibition of Ku activity and DSB repair occurs in the nucleus. Since expression of nuclear-targeted Bcl2 has been reported to enhances the sensitivity of cells to apoptosis (Jin et al, 2006), the nuclear Bcl2-mediated inhibition of DSB repair may contribute to cell death. Thus, the antiapoptotic function of mitochondrial Bcl2 may be a good selection against the apoptotic effect from nuclear Bcl2-mediated inhibition of DSB repair, which results in accumulation of DSBs in living cells, leading to increased genetic instability (Figure 2F–G). NHEJ factors function as genomic caretakers to maintain genetic stability by repairing DSBs. Bcl2-enhanced cytogenetic abnormalities (i.e. chromosomal and chromatid breaks) may result from Bcl2’s negative effect on DSB repair. Because genomic instability is a characteristic, predisposing factor to tumorigenesis, our findings help solve the puzzle why Bcl2, an antiapoptotic protein, also functions as a potent oncogenic molecule (Linette, et al, 1995; McDonnell et al, 1991).
Structure-function studies reveal that Ku 70 or Ku 86 directly interacts with Bcl2 at its BH1 and BH4 domains (Figure 4B). Importantly, these two Ku binding sites on Bcl2 are essential for its nuclear localization after IR treatment (Figure 4C and Figure S3), which further supports the notion that Ku may act as a nuclear “receptor” associating with Bcl2 to sequester Bcl2 in the nucleus. Since the BH1 or BH4 domain deficient Bcl2 mutant not only fails to suppress Ku DNA binding activity but also loses Bcl2’s ability to suppress DSB repair (Figure 5), this suggests that the BH1 and BH4 domains are also required for Bcl2 to function in the attenuation of DSB repair.
Our findings and others’ have provided strong evidence that all BH domains are necessary for the full and potent antiapoptotic function of Bcl2 (Figure 5F; Castelli et al., 2004; Cheng et al., 1997). The BH4 domain is a known survival domain in Bcl2 because caspase-mediated cleavage or mutagenic removal of the BH4 domain abolishes the antiapoptotic activity of Bcl2 (Cheng et al., 1997; Reed et al., 1996). In addition, the BH1, BH2 and BH3 domains form the surface binding pocket of Bcl2 (Castelli et al., 2004). Removal of the BH1, BH2 or BH3 domain from Bcl2 not only impairs the integrity of the surface binding pocket but also results in decreased antiapoptotic activity of Bcl2 (Figure 5F). Since deletion of the BH2 or BH3 domain does not affect Bcl2’s capacity to suppress DSB repair, but significantly dampens its antiapoptotic activity (Figure 5), we propose that the antiapoptotic function of Bcl2 is not required for its effect on DSB repair and that Bcl2 mediated suppression of DSB repair is dependent on its ability to interact with Ku, which requires the BH1 and BH4 domains.
Recruitment of DNA-PKcs by the Ku complex to the broken DNA ends and formation of the functional Ku/DNA-PKcs complexes are essential for repairing DSBs (Hefferin et al., 2005). Our findings indicate that Bcl2 can directly interact with Ku via its BH1 and BH4 domains (Figures 3–4). This helps explain why overexpression of WT, ΔBH2 or ΔBH3 but not ΔBH1 or ΔBH4 Bcl2 mutants in cells significantly suppresses Ku/DNA-PKcs binding (Figure 6C). Furthermore, depletion of endogenous Bcl2 by RNAi from H460 cells not only enhances Ku DNA binding activity and formation of the Ku/DNA-PKcs complex but also accelerates DSB repair (Figure 7). Intriguingly, Bcl2 has no significant, additional inhibitory effect on DSB repair after inhibition of DNA-PK activity by the DNA-PK specific inhibitor, NU7026 (Figure S7B–D), suggesting that Bcl2 mediated suppression of DSB repair is dependent on DNA-PK activity.
The V(D)J recombination has been considered a physiological DSB repair process in which the NHEJ proteins (i.e. Ku, DNA-PKcs, etc.) are recruited to repair specifically broken ends such as the signal and hairpin coding ends generated by RAG1/2 (Bassing et al., 2002; Gu et al., 1997). Here we discovered that overexpression of Bcl2 results in decreased formation of coding and RS joins with low fidelity, as assayed by cotransfection of RAG1 and RAG2 expression vectors along with plasmid V(D)J recombination substrates (Table S1). This consequence may result from the inhibitory effect of Bcl2 on Ku and DNA-PK activities because expression of the Ku binding deficient Bcl2 mutant (i.e. ΔBH1 or ΔBH1) has no such effect. Since both general DSB repair and V(D)J recombination require Ku and DNA-PKcs, this explains why Bcl2 plays a role in general DSB repair similar to that seen in V(D)J recombination.
In summary, our findings have demonstrated that suppression of DSB repair and the V(D)J recombination by Bcl2 occurs through a mechanism involving the NHEJ pathway. DSBs enhance Bcl2 expression and association with Ku in nucleus. Direct interaction of Bcl2 with Ku via its BH1 and BH4 domains may be required for Bcl2 to inhibit Ku DNA binding, formation of Ku/DNA-PKcs complexes, DNA-PK activity and DNA end-joining, which lead to suppression of DSB repair as well as V(D)J recombination. The inhibitory effect of Bcl2 on DSB repair enhances genetic instability, which may contribute to the development of various malignancies, including lung cancer. Thus, Bcl2, in addition to its antiapoptotic property, may also function as an oncogenic molecule to inhibit the repair of DSBs through the NHEJ pathway.
Experimental Procedures
Materials
Bcl2, Ku70, Ku86, DNA-PKcs and tubulin antibodies as well as Bcl2 siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The γH2AX antibody was purchased from Upstate Biotech (Charlotteville, VA). Ku70/Ku86 DNA Repair Kit was obtained from Active Motif (Carlsbad, CA). Purified, recombinant WT and BH deletion Bcl2 mutant proteins were obtained from ProteinX Lab (San Diego, CA). The Signa TECT DNA-PK assay system was purchased from Promega (Madison, WI). The DNA-PK specific inhibitor NU7026 was obtained from SIGMA. The pJH290, pJH200, RAG1 and RAG2 constructs as well as Escherichia coli MC1061 were obtained from Dr. Frederick W. Alt (Harvard Medical School). All of the reagents used were obtained from commercial sources unless otherwise stated.
Generation of various Bcl2 deletion mutants
The BH1, BH2, BH3 and BH4 Bcl2 deletion mutants were created by using a mutagenesis kit (BD Biosciences) and confirmed by sequencing of the cDNA as described (Hou et al., 2007). The WT and various Bcl2 deletion mutants were then cloned into the pCIneo (Promega) mammalian expression vector.
Cell Lines, plasmids and transfections
H1299 and H460 cells were maintained in RPMI 1640 with 10% fetalbovine serum. WT or each of various Bcl2 deletion mutants were stably expressed in H1299 cells. The expression levels of exogenous Bcl2 were analyzed by Western blot analysis and three separate clones expressing similar amounts of exogenous Bcl2 were selected for further analysis.
Preparation of cell lysates
Cells were washed with 1 × PBS and resuspended in ice-cold 1% CHAPS lysis buffer (1% CHAPS, 50mM Tris, pH 7.6, 120mM NaCl, 1mM EDTA, 1mM Na3VO4, 50mM NaF and 1mM β-mercaptoethanol) with a cocktail of protease inhibitors (EMD Biosciences). Cells were lysed by sonication and centrifuged at 14,000 × g for 10 min at 4°C. The resulting supernatant was collected as the total cell lysate.
Pulsed field gel electrophoresis (PFGE)
PFGE was performed as described (Ager et al., 1990). Briefly, cells were harvested and resuspended in ice-cold buffer L (0.1M Na2-EDTA, 0.01M Tris, 0.02 M NaCl, pH8.0) at a concentration of 5× 106 cells/ml, and mixed with an equal volume of 1% low melting point agarose (Beckman) at 42 °C. The mixture was pipetted into a small length of Tygon tubing, clamped tight at both ends, and chilled to 0°C. The solidified agarose “snake” was extruded from the tubing and added to 10× volume of buffer L containing 1mg/ml proteinase K and 1% sarkosyl, and incubated 16 hours at 50°C. Following lysis, the agarose snake was washed four times with TE buffer and then cut into 0.5 cm plugs. The plugs were inserted into the wells of a pre-cooled 1% low melting point agarose gel (4 °C). PFGE (200-s pulse time, 150V, 15h at 14°C) was performed using clamped, homogenous electric fields (CHEF)-Mapper (Bio-Rad). After electrophoresis, the gel was stained with ethidium bromide for photography.
Telomere–FISH (T-FISH) analysis
T-FISH was performed using Telomere PNA FISH Kit/Cy3 (Dako Cytomation, Denmark) as described (Zha et al., 2007). Briefly, cells were incubated with colcemid (KaryoMAX, GIBCO) at 100ng/ml for 1 hour and then harvested by trypsinization. Cells were swollen in prewarmed 30mM sodium citrate for 30 min at 37 °C, fixed in methanol/acetic acid (3/1), and air dried on slides overnight. After pepsin digestion, slides were denatured at 80 °C for 5 min, hybridized with Cy3-labeled PNA telomeric probe (Cy3-[TTAGGG]3) in 70% formamide at RT for 3–4 hours, washed, dehydrated, and mounted in Vectashield with DAPI ( Vector Laboratories, CA). Metaphase images were captured using a fluorescent microscope (Zeiss) and 63× objective lens. At least 30 metaphases of each cell line were scored for chromosomal aberrations.
Immunofluorescence
The cell were washed with 1× PBS, fixed with methanol and acetone (1:1) for 5 minutes and then blocked with 10% normal mouse or rabbit serum for 20 min at room temperature. Cells were incubated with a mouse or rabbit primary antibody for 90 min. After washing, samples were incubated with Alex 488 (green) conjugated anti-mouse or Alexa 594 (red)-conjugated anti-rabbit secondary antibodies or DAPI for 60 min. Cells were washed with 1× PBS and observed under a fluorescent microscope (Zeiss). Pictures were taken and colored with the same exposure setting for each experiment. Individual green- and red-stained images derived from the same field were merged using Open lab 3.1.5 software from Improvision, Inc. (Lexington, MA).
Measurement of Ku70/86 activity
Ku70/Ku86 DNA binding activity was analyzed using a Ku70/86 DNA Repair Kit (Active Motif). Briefly, cells were washed and resuspended in hypotonic buffer. The nuclear extract was isolated for Ku activity analysis. 5μg of nuclear protein was loaded into the oligonucleotide coated 96-well plate and incubated for 60 min at room temperature. Ku70 or Ku86 antibody was added and incubated for another 60 min. After washing, HRP-conjugated secondary antibody was added and incubated for 60 min. After washing, developing solution and stop solution were added. Optical density was read on a spectrophotometer at 450 nm. Each experiment was repeated three times and data represent the mean ± S.D. of three separate determinations.
Bcl2 silence
H460 cells expressing high levels of endogenous Bcl2 were transfected with Bcl2 siRNA using Lipofect AMINE™ 2000 (Invitrogen). A control siRNA(nonhomologous to any known gene sequence) was used as a negative control. The levels of Bcl2 expression were analyzed by Western blotting. Specific silencing of the targeted Bcl2gene was confirmed by at least three independent experiments.
Subcellular fractionation
Cells (2 × 107) were washed once with cold 1× PBS and resuspended in isotonic mitochondrial buffer (210 mM mannitol, 70 mM sucrose, 1 mM EGTA, 10 mM Hepes, pH 7.5). The resuspended cells were homogenized with a polytron homogenizer operating for four bursts of 10 s each at a setting of 5. The mitochondrial and nuclear fractions were isolated as previously described (Jin et al., 2006). Protein from each fraction was analyzed by Western blot.
Transient V(D)J Recombination Assay
Recombination substrates pJH200 and pJH290 were used in the transient V(D)J recombination assay to test for recombination signal (RS) and coding joins as described (Gu et al., 1997). Briefly, cells were seeded onto 10-cm tissue culture plate and cultured for 24h before the transfection. pJH200 or pJH290 was cotransfected with the full-length RAG1 and RAG2 expression constructs under the control of the Rous sarcoma virus long terminal repeat promoter using Lipofectamine 2000™. Extra chromosomal DNA was recovered after 48h by an alkaline lysis method and electroporated into Escherichia coli MC1061. V(D)J recombination activity was measured by comparing the ratio of ampicillin-resistant (Ampr) chloramphenicol-resistant (Camr) colonies versus Ampr colonies among H1299 cells expressing WT or each of the BH deletion Bcl2 mutants. Because a precise signal join generates an ApaLI site, ApaLI digestion of PCR products from the recombinant substrate pJH 200 was used to measure the fidelity of the RS joins as described (Gu et al., 1997).
Clonogenic survival assay
A clonogenic survival assay was performed as described (Shih et al., 2005). Briefly, the log-phased cells were plated overnight. Following irradiation, cells were trypsinized, counted, and plated for colony formation. Following 7 to 10 days of incubation, colonies were fixed with methanol/acetic acid (3:1), stained with crystal violet and counted. Each experiment was repeated three times.
Supplementary Material
Acknowledgments
This work was supported by NCI, National Institutes of Health Grant R01 CA112183, by a Flight Attendant Medical Research Institute Clinical Innovator Award (to X.D.).
We are grateful to Dr. Frederick W. Alt (Harvard Medical School) for kindly providing the pJH200, pJH290, RAG1 and RAG2 constructs.
References
- Ager D, Dewey W, Gardiner K, Harver W, Johnson R, Waldren C. Measurement of radiation-induced DNA double-strand breaks by Pulsed-field gel electrophoresis. Radiation research. 1990;122:181–187. [PubMed] [Google Scholar]
- Balajee A, Geard C. Replication protein A and γ-H2AX foci assembly is triggered by cellular response to DNA double-strand breaks. Experimental Cell Research. 2004;300:320–334. doi: 10.1016/j.yexcr.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Bassing C, Swat W, Alt FW. The Mechanism and Regulation of Chromosomal V(D)J Recombination. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Bertinato J, Poulter C, Hache R. Nuclear localization of Ku antigen is promoted independently by basic motifs in the Ku 70 and Ku 80 subunits. Journal of Cell Science. 2000;114:89–99. doi: 10.1242/jcs.114.1.89. [DOI] [PubMed] [Google Scholar]
- Burma S, Chen B, Chen D. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair. 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Castelli M, Reiners J, Kessel D. A mechanism for the proapoptotic activity of ursodeoxycholic acid: effects on Bcl2 conformatiom. Cell Death Differ. 2004;11:906–914. doi: 10.1038/sj.cdd.4401433. [DOI] [PubMed] [Google Scholar]
- Chebonnel-Lasserrre C, Gauny S, Kronenberg A. Suppression of apoptosis of Bcl2 and Bcl-XL promotes susceptibility to mutagenesis. Oncogene. 1996;13:1489–1497. [PubMed] [Google Scholar]
- Cheng EH, Kirsch DG, Clem RJ, Ravi R, Kastan MB, Bedi A, Ueno K, Hardwick JM. Conversion of Bcl-2 to a Bax-like Death Effector by Caspases. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh M, Ishii H, Peterson C, Buratowski S, Lieberman J. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Critchlow S, Jackson S. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- Deng X, Gao F, Flagg T, May WS. Mono- and multi-site phosphorylation enhances Bcl2’s anti-apoptotic function and inhibition of cell cycle entry functions. Proc Natl Acad Sci USA. 2004;101:153–158. doi: 10.1073/pnas.2533920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strandn break repair in cell-free extracts from Ku80-deficiency cells: implication for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2595. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Gu Y, Jin S, Gao Y, Weaver D, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci U S A. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefferin M, Tomkinson A. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Hockenbery D, Nunez G, Milliman C, Schreiber R, Korsmeyer S. Bcl2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 1990;348:334–336. doi: 10.1038/348334a0. [DOI] [PubMed] [Google Scholar]
- Hou Y, Gao F, Wang Q, Zhao J, Flagg T, Zhang Y, Deng X. Bcl2 impedes DNA mismatch repair by directly regulating the hMSH2-hMSH6 heterodimeric complex. J Biol Chem. 2007;282:9279–9287. doi: 10.1074/jbc.M608523200. [DOI] [PubMed] [Google Scholar]
- Jin Z, May WS, Gao F, Flagg T, Deng X. Bcl2 suppresses DNA repair by enhancing c-Myc transcriptional Activity. J Biol Chem. 2006;281:14446–14456. doi: 10.1074/jbc.M511914200. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M. The taming of a transposon; V(D)J recombination and the immune system. Immunological Reviews. 2004;200:233–248. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Kelekar A, Thompson C. Bcl2-family proteins: the role of the BH3 domain in apoptosis. Trends in Cell Biology. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- Khanna K, Jackson S. DNA double-strand breaks: signal, repair and the cancer connection. Nature Genetics. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Kuo M, Shiah S, Wang C, Chuang S. Suppression of apoptosis by Bcl-2 to enhance benzene metabolites-induced oxidative DNA damage and mutagenesis: A possible mechanism of carcinogenesis. Mol Pharmacol. 1999;55:894–901. [PubMed] [Google Scholar]
- Lees-Miller S, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler K, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Linette G, Hess J, Sentman C, Korsmeyer S. Peripheral T-cell lymphoma in lckr-Bcl2 transgenic mice. Blood. 1995;86:1255–1260. [PubMed] [Google Scholar]
- Liu Y, Naumovski L, Hanawalt P. Nucleotide excision repair capacity is attenuated in human promyelocytic HL60 cells that overexpress Bcl2. Cancer Research. 1997;57:1650–1653. [PubMed] [Google Scholar]
- McDonnell T, Korsmeyer S. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18) Nature. 1991;349:254–256. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- Modesti M, Kanaar R. DNA repair: spot(light)s on chromatin. Curr Biol. 2001;11:R229–R232. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- Pastink A, JEeken J, Lohman P. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res. 2001;480:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- Pasttwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochimica Polonica. 2003;50:891–908. [PubMed] [Google Scholar]
- Pierce AJM, Stark J, Araujo F, Moynahan M, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:52–59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- Reed J, Zha H, Aime-Sempe C, Takayama S, Wang H. Structure-function analysis of Bcl2 family proteins. Regulators of programmed cell death. Adv Exp Med Biol. 1996;406:99–112. [PubMed] [Google Scholar]
- Rogakou E, Boon C, Redon C, Bonner W. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid Phenotype Associated with Defective V(D)J Coding End Processing in Artemis-Deficient Mice. Mol Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- Schardle CA, Li S, Re GG, Fan W, Willingham MC. Mitotic chromosomal Bcl2: II. Localization to interphase nuclei. J Histochem Cytochem. 1999;47:151–158. doi: 10.1177/002215549904700204. [DOI] [PubMed] [Google Scholar]
- Shih S, Erbele T, Chen A. Ku modulates DNA topoisomerase I-mediated radiosensitization, but not cytotoxicity, in mammalian cells. Cancer Res. 2005;65:9194–9199. doi: 10.1158/0008-5472.CAN-05-2387. [DOI] [PubMed] [Google Scholar]
- Shin K, Kang M, Dicterow E, Kameta A, Baluda M, Park N. Introduction of human telomerase reverse transcriptase to normal human fibroblasts enhances DNA repair capacity. Clinical cancer research. 2004;10:2551–2560. doi: 10.1158/1078-0432.ccr-0669-3. [DOI] [PubMed] [Google Scholar]
- Smith G, Jackson S. The DNA dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton J, Mohammad D, Yaffe M, Smerdon S, Jacson S. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science. 1985;229:1390–1393. doi: 10.1126/science.3874430. [DOI] [PubMed] [Google Scholar]
- Van Gent D, Hoeijmakers J, Kanaar R. Chromosomal stability and DNA double-strand break connection. Nature Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Willmore E, de Caux S, Sunter N, Tilby M, Jackson G, Austin C, Durkacz B. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103:4659–4665. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- Yaneva M, Kowalewski T, Lieber M. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5089–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn C, Cho H, Kim S, Kim H, Kim M, Chang I, Lee J, Chung M, Hahm K, You H. Bcl-2 expression suppresses mismatch repair activity through inhibition of E2F transcriptional activity. Nature Cell Biology. 2005;7:137–147. doi: 10.1038/ncb1215. [DOI] [PubMed] [Google Scholar]
- Zha S, Alt FW, Cheng H, Brush J, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci USA. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-Deficient Mice Exhibit Severe Combined Immunodeficiency and Defective Processing of V(D)J Recombination Intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.