Abstract
Calcineurin is a Ca2+/calmodulin-regulated protein phosphatase required for Saccharomyces cerevisiae to respond to a variety of environmental stresses. Calcineurin promotes cell survival during stress by dephosphorylating and activating the Zn-finger transcription factor Crz1p/Tcn1p. Using a high-throughput assay, we screened 119 yeast kinases for their ability to phosphorylate Crz1p in vitro and identified the casein kinase I homolog Hrr25p. Here we show that Hrr25p negatively regulates Crz1p activity and nuclear localization in vivo. Hrr25p binds to and phosphorylates Crz1p in vitro and in vivo. Overexpression of Hrr25p decreases Crz1p-dependent transcription and antagonizes its Ca2+-induced nuclear accumulation. In the absence of Hrr25p, activation of Crz1p by Ca2+/calcineurin is potentiated. These findings represent the first identification of a negative regulator for Crz1p, and establish a novel physiological role for Hrr25p in antagonizing calcineurin signaling.
Keywords: Calcineurin, calcium, casein kinase I, transcription, nuclear localization, signal transduction
Calcineurin is a conserved Ca2+/calmodulin-dependent serine/threonine phosphatase that plays a critical role in Ca2+ signaling (for review, see Aramburu et al. 2000). One of the primary functions of calcineurin in mammals is to regulate gene transcription through dephosphorylation and activation of the NFAT family of transcription factors (Crabtree and Olson 2002). Through NFAT, calcineurin regulates a variety of cellular processes including T-cell activation (for review, see Crabtree and Olson 2002), development of cardiac hypertrophy (Molkentin et al. 1998), angiogenesis (Graef et al. 2001), and embryonic axon outgrowth (Graef et al. 2003).
In the yeast Saccharomyces cerevisiae, calcineurin signaling is necessary for cells to adapt to a variety of environmental stresses. Exposure to conditions such as high salt, alkaline pH, high temperature, or mating pheromone results in elevated intracellular Ca2+, which activates calcineurin (Cyert 2003). Under these conditions, calcineurin induces the expression of genes whose products promote cell survival; these include proteins that function in small molecule transport, ion homeostasis, cell wall synthesis and maintenance, lipid metabolism, and vesicle transport (Rudolph et al. 1989; Haro et al. 1991; Cunningham and Fink 1994; Mendoza et al. 1994; Mazur et al. 1995; Yoshimoto et al. 2002). Although calcineurin is not essential under standard laboratory conditions, deletion of the catalytic subunits (CNA1/CNA2; Cyert et al. 1991; Liu et al. 1991) or the regulatory subunit (CNB1; Kuno et al. 1991; Cyert and Thorner 1992) results in sensitivity to pheromone, sodium, manganese, lithium, and high pH (Mendoza et al. 1994; Farcasanu et al. 1995; Moser et al. 1996; Pozos et al. 1996; Withee et al. 1997).
Calcineurin affects gene transcription through its regulation of the Zn-finger transcription factor Crz1p/Tcn1p/Hal8p (Matheos et al. 1997; Stathopoulos and Cyert 1997; Mendizabal et al. 1998). Calcineurin dephosphorylates Crz1p, causing it to translocate from the cytosol to the nucleus (Stathopoulos-Gerontides et al. 1999), and the accumulation of Crz1p in the nucleus is due to an increase in its rate of nuclear import and a concomitant decrease in its rate of nuclear export (Polizotto and Cyert 2001; Boustany and Cyert 2002). Once in the nucleus, Crz1p binds a DNAelement known as the CDRE (calcineurin-dependent response element) through which it activates gene transcription (Stathopoulos and Cyert 1997; Stathopoulos-Gerontides et al. 1999). All known calcineurin-dependent transcriptional changes are believed to be mediated through its activation of Crz1p (Yoshimoto et al. 2002).
Crz1p is a phosphoprotein, and because genetic analyses have failed to identify any Crz1p kinases, we screened for such a kinase on a proteomic scale, using a recently developed assay employing protein kinase microarrays. We simultaneously tested 119 yeast kinases for their ability to phosphorylate Crz1p in vitro and identified the casein kinase I homolog Hrr25p. Casein kinase I is a conserved family of kinases that regulate processes as diverse as membrane trafficking and circadian rhythm (Vielhaber and Virshup 2001). Previous studies showed that Hrr25p is a dual-specificity protein kinase that plays a role in the response to DNAdamage (Hoekstra et al. 1991, 1994; DeMaggio et al. 1992; Ho et al. 1997), meiosis (Hoekstra et al. 1991), and vesicle trafficking (Murakami et al. 1999).
In this study we establish a new role for Hrr25p as a regulator of Crz1p. We demonstrate that Hrr25p phosphorylates Crz1p and regulates both its activity and localization. Thus, Hrr25p acts in opposition to calcineurin and promotes termination of the Crz1p transcriptional response.
Results
Identification of Hrr25p as a Crz1p kinase
To identify potential Crz1p protein kinases, 119 of the 122 known and predicted yeast kinases were tested simultaneously for their ability to phosphorylate recombinant GST-Crz1p using a method developed by Zhu et al. (2000; see Materials and Methods). Briefly, GST-Crz1p was first covalently bound to glass slides. To each slide, a different GST-kinase purified from yeast was added and assayed for its ability to phosphorylate GST-Crz1p. In this assay, Hrr25p catalyzed 5.5-fold more phosphate incorporation into GST-Crz1p than into GST.
Crz1p and Hrr25p interact in vitro and in vivo
In the assay described above, GST-Crz1 was phosphorylated by GST-Hrr25p purified from yeast. Therefore, to establish that this activity was due to Hrr25p rather than Hrr25p-associated proteins, we tested recombinant Hrr25p for its ability to phosphorylate Crz1p. Wild-type Hrr25p and catalytically inactive Hrr25p-K38A(Murakami et al. 1999) were purified from bacteria and incubated with Crz1p in the presence of γ32P-ATP. Incubation with wild-type Hrr25p resulted in Crz1p phosphorylation (Fig. 1A). In contrast, incubation without kinase or with Hrr25p-K38A did not lead to phosphorylation. This result establishes that Hrr25p directly phosphorylates Crz1p in vitro.
Figure 1.
Hrr25p phosphorylates Crz1p in vitro and in vivo. (A) Hrr25p phosphorylates Crz1p in vitro. Recombinant ZZ-Crz1p was incubated with recombinant wild-type or catalytically inactive Hrr25p and γ32P-ATP. Phosphorylation of ZZ-Crz1 was analyzed by SDS-PAGE and autoradiography. The bottom panel shows ZZ-Crz1p stained by Coomassie. (B) Crz1p phosphorylated by Hrr25p is a target of calcineurin. ZZ-Crz1p was phosphorylated by Hrr25p as in A and incubated with buffer alone or calcineurin, calmodulin, and Ca2+ or EGTA. Samples were analyzed by SDS-PAGE and autoradiography. The bottom panel shows ZZ-Crz1p stained by Coomassie. (C) Hrr25p phosphorylates and binds to Crz1p in vivo. IgG sepharose was used to isolate ZZ-CRZ1p from extracts of cells (KKY271) expressing vector (pRS316), overexpressing wild-type (pAM4-1), or catalytically inactive HA-HRR25-K38A(pAM8-2). Samples were analyzed by SDS-PAGE and Western blotting. ZZ-Crz1p was visualized using an α-Crz1 antibody; HA-Hrr25p was visualized using an α-HAantibody.
If the phosphorylation of Crz1p by Hrr25p is relevant to the calcineurin pathway, then, once phosphorylated by Hrr25p, Crz1p should become a target for dephosphorylation by calcineurin. To test this hypothesis, Crz1p was phosphorylated by Hrr25p and incubated with calcineurin in the presence of calmodulin and Ca2+. We observed specific dephosphorylation of Crz1p that was inhibited by the addition of the Ca2+ chelator EGTA(Fig. 1B). These results suggest that Hrr25p plays a physiological role in the regulation of Crz1p.
Next we tested for physical association between Hrr25p and Crz1p in vivo. Crz1p was C-terminally tagged with the IgG binding domain from Protein A(ZZ) and then integrated into the genome (KKY271). Crz1p-ZZ was then isolated from extracts of cells overexpressing HA-HRR25, HA-HRR25-K38A, or carrying an empty vector. We examined the ability of Crz1p-ZZ to bind HA-Hrr25p by Western blot analysis and found that Crz1p-ZZ associated with both active and catalytically inactive HA-Hrr25p (Fig. 1C). Therefore, Crz1p interacts with Hrr25p independently of its kinase activity. We also observed that Crz1p-ZZ associated with active HAHrr25p displayed a shift in electrophoretic mobility characteristic of its hyperphosphorylated form (Fig. 1C; Stathopoulos-Gerontides et al. 1999). In contrast, Crz1p-ZZ is unphosphorylated in cells expressing an empty vector or when bound to HA-Hrr25p-K38A (Fig. 1C). These data show that Hrr25p associates with and phosphorylates Crz1p in vivo.
Hrr25p is localized diffusely throughout the cell and at the bud neck
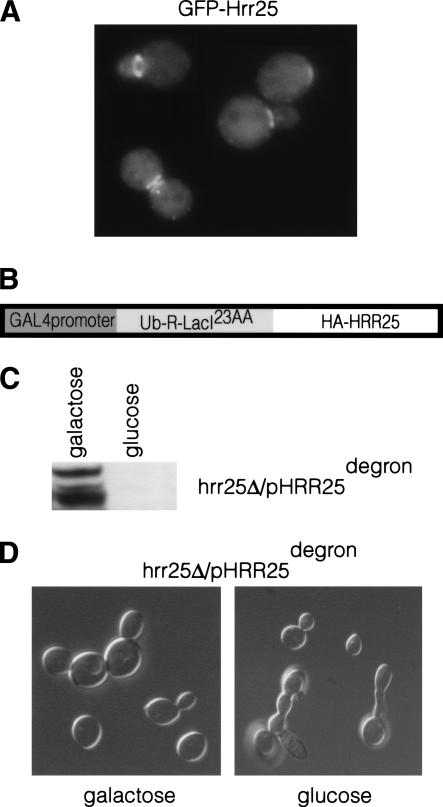
Crz1p translocates from the cytosol to the nucleus upon Ca2+ treatment; therefore, we investigated whether Hrr25p localization was similarly regulated. We fused GFP to the N terminus of HRR25 and expressed the fusion from the MET25 promoter (see Materials and Methods). This fusion complemented an hrr25Δ strain (KKY387), and Western blot analysis indicated that the fusion was stable (data not shown). GFP-Hrr25p localized diffusely throughout the entire cell (Fig. 2A). In addition, we noted that GFP-Hrr25p concentrated at the emerging bud tip and the bud neck (Fig. 2A). These results are consistent with subcellular fractionation experiments, which indicated the presence of Hrr25p in both soluble and particulate fractions (data not shown). GFP-Hrr25p localization was unchanged upon treatment with Ca2+. Aprevious study demonstrated that incubating cells with the DNAsynthesis inhibitor hydroxyurea activates Hrr25p (Ho et al. 1997). GFP-Hrr25p localization was also unaltered by hydroxyurea treatment (data not shown).
Figure 2.
Hrr25p localization and generation of a conditional HRR25. (A) GFP-Hrr25p is diffusely localized throughout the cell and concentrates at the bud tip and at the bud neck. YPH499 cells expressing GFP-Hrr25p (pKK191) were grown to log phase and examined (see Materials and Methods). (B) Schematic of the HRR25degron allele expressed under a galactoseinducible promoter (pKK204). The N-terminal ubiquitin is cleaved cotranslationally, revealing the destabilizing arginine (the N-degron). The 23 amino acids of LacI provide an internal lysine residue to which the polyubiquitin chain is attached. Hrr25pdegron is then degraded by the proteosome. (C) α-HA Western blot showing the disappearance of Hrr25pdegron 5 h after addition of glucose (KKY387). (D) Abnormal cell morphology characteristic of hrr25 mutant cells seen 5 h after the addition of glucose to KKY387. Cells displaying this abnormal morphology represent 5% of all cells.
Depending on the strain background, HRR25 mutants are either inviable or exhibit a severe growth defect (Hoekstra et al. 1991; Giaever et al. 2002). Therefore, to facilitate analysis of the role of Hrr25p in Crz1p regulation, we constructed a conditional allele of HRR25. Using an established method, we fused a protein module (degron) to the N terminus of HA-Hrr25p, the presence of which results in rapid degradation of the kinase (Bachmair et al. 1986). This unstable form of HA-Hrr25p (HAHrr25pdegron; pKK204) was expressed under control of a galactose-regulated promoter and introduced into cells lacking the genomic copy of HRR25 to create strain KKY387 (Fig. 2B). When grown in galactose, Hrr25pdegron is expressed and the cells are viable. When glucose is added, expression of Hrr25pdegron is terminated, and the protein is rapidly degraded; within 5 h of glucose addition, Hrr25pdegron is no longer detectable by Western blot (Fig. 2C), and a fraction of cells begin to display the characteristic morphology of hrr25Δ cells, including elongated buds and multibudded cells (Fig. 2D). We then used this conditional allele of HRR25 to investigate the role of Hrr25p in the regulation of Crz1p (see below).
Hrr25p regulates Crz1p transcriptional activity
We examined whether Hrr25p has a physiological role in Crz1p signaling by testing the effect of the kinase on Crz1p-dependent gene expression. We monitored Crz1p transcriptional activity using a reporter gene that contains four tandem copies of the Crz1p binding site placed upstream of β-galactosidase (4xCDRE::LacZ; ASY832; Stathopoulos and Cyert 1997). Addition of Ca2+ caused an increase in β-galactosidase activity indicative of Crz1p activation (Fig. 3A). In cells overexpressing HRR25, this Ca2+-induced gene expression was reduced 2.4-fold. We observed equivalent reductions in response to both low (10 mM) and high (200 mM) Ca2+ concentrations (Fig. 3A; data not shown). Overexpression of catalytically inactive HRR25-K38A had no effect on β-galactosidase levels (data not shown), indicating that the kinase activity of Hrr25p is necessary for negative regulation of Crz1p. HRR25 overexpression similarly decreased the expression of several Crz1p target genes, as determined by Northern analysis (data not shown).
Figure 3.
Hrr25p affects Crz1p transcriptional activity. (A) Overexpression of HRR25 decreases Crz1p-dependent transcription. Cells carrying an integrated 4xCDRE::LacZ reporter (ASY832) and either pCu423CUP1 or pKK194 (2µHRR25) were induced with 500 µM CuSO4 for 3 h. Where indicated, 200 mM CaCl2 was added for 3 h. β-galactosidase activity was measured and is reported as the maximum rate/microgram protein. (B) Depletion of Hrr25p results in an increase in Crz1p-dependent transcription. KKY401 cells were grown to log phase, and 2% glucose was added for 5 h where indicated. CaCl2 was added to the indicated concentrations, and cells were grown for an additional 30 min. Light bars represent galactose-grown cells (+Hrr25p); dark bars represent glucose-grown cells (-Hrr25p).
Using the Hrr25pdegron strain containing the 4xCDRE::LacZ reporter (KKY401), we next investigated the effect of Hrr25p depletion on Crz1p activity. In the absence of Ca2+, cells lacking Hrr25p displayed an increased basal level of CDRE::LacZ expression. Furthermore, in response to Ca2+ treatment, cells depleted of Hrr25p displayed elevated β-galactosidase activity compared to cells expressing the kinase (Fig. 3B). Interestingly, the lack of Hrr25p had a greater effect on Crz1p activity at low Ca2+ concentrations (10 mM vs. 50 mM). This increase in Crz1p-dependent transcription was caused by loss of Hrr25p rather than the change in carbon source used to deplete Hrr25p; wild-type cells (ASY832) grown in galactose exhibit levels of Ca2+-induced CDRE::LacZ activity similar to those of such cells grown in glucose (data not shown). These results suggest that loss of Hrr25p lowers the threshold of Crz1p activation, and that the effect of Hrr25p depletion is more pronounced under conditions where Crz1p is not fully activated.
Hrr25p affects Crz1p subcellular localization
Because localization of Crz1p is affected by its phosphorylation state, we investigated the effect of Hrr25p on Crz1p distribution. Cells overexpressing HRR25 or carrying an empty vector were treated with 200 mM CaCl2, and GFP-Crz1p localization was examined 5 and 25 min after treatment (Fig. 4A). Five minutes after Ca2+ addition, 72% of control cells exhibited exclusively nuclear localization of GFP-Crz1p. In contrast, when HRR25 was overexpressed, significantly fewer cells (25%) displayed nuclear localization at this time. Twenty-five minutes after Ca2+ addition, the percentage of control cells exhibiting exclusively nuclear localization decreased to 42%, reflecting the redistribution of GFP-Crz1p to the cytosol (27% cytosolic). HRR25 overexpression stimulated the return of GFP-Crz1p to the cytosol; GFP-Crz1p was predominantly cytosolic in 85% of these cells, whereas only 2% of cells showed strong nuclear accumulation. These results suggest that the effect of HRR25 overexpression on Crz1p transcriptional activity is due to decreased nuclear localization of Crz1p in the presence of Ca2+.
Figure 4.
Hrr25p promotes Crz1p cytosolic localization. (A) YPH499 cells expressing 3xGFP-Crz1p (pLMB127) and either pCu423CUP1 or pKK194 (2µHRR25) were induced for 3 h with 500 µM CuSO4. Then 200 mM CaCl2 was added, and cells were examined at the indicated times after CaCl2 addition. For each time point, 150-200 cells were counted, and the graph represents the average of two independent experiments. Gray bars represent cells with exclusively nuclear (N) GFP-Crz1p; white bars represent cells with intermediate nuclear/cytosolic localization (N/C); black bars represent cells with cytosolic localization (C). (B) Hrr25p depletion promotes Crz1p localization in low concentrations of Ca2+. KKY387 cells expressing 3xGFP-Crz1p were grown for 3 h with or without glucose. Then 50 mM CaCl2 was added where indicated, and cells were examined within 5 min of CaCl2 addition.
To explore this further, we also examined GFP-Crz1p localization using Hrr25pdegron to deplete the kinase. GFP-Crz1p was cytosolic in untreated cells, and this localization was unaffected by Hrr25p depletion (Fig. 4B). Addition of 50 mM CaCl2 caused strong nuclear localization of GFP-Crz1 in cells lacking Hrr25p. In contrast, this low concentration of Ca2+ failed to induce GFP-Crz1 nuclear accumulation in cells containing Hrr25p. This change in the response of GFP-Crz1p to 50 mM Ca2+ was a result of Hrr25p depletion, and not due to the altered carbon source, as GFP-Crz1p localization in wild-type cells (YPH499; data not shown) was identical in glucose or galactose. Therefore, Hrr25p depletion lowered the amount of Ca2+ required to trigger GFP-Crz1p nuclear accumulation. This is consistent with the β-galactosidase results, in which loss of Hrr25p led to increased Crz1p activation in response to low concentrations of Ca2+. Hrr25p therefore has a significant effect in regulating the subcellular localization of Crz1p.
Hrr25p is not the sole Crz1p kinase
In the experiments described above, depletion of Hrr25p did not result in constitutive activation of Crz1p, suggesting that additional proteins, most likely protein kinases, inhibit Crz1p activity in vivo. To investigate this issue further, we first compared the electrophoretic mobility of Crz1p in vivo and in vitro. When phosphorylated in vitro by Hrr25p, GST-Crz1p showed a decrease in its mobility relative to its unphosphorylated form (Fig. 5A). GST-Crz1p isolated from extracts of wild-type cells (YPH499) treated with the calcineurin inhibitor FK506 also showed a decrease in electrophoretic mobility. However, GST-Crz1p isolated from FK506-treated cells migrated slower than GST-Crz1p phosphorylated by Hrr25p in vitro. This suggests that, in vivo, additional kinases contribute to Crz1p phosphorylation. In addition, GST-Crz1p isolated from extracts of untreated yeast cells migrates slower than GST-Crz1p purified from bacteria, suggesting a basal level of Crz1p phosphorylation under these conditions.
Figure 5.
Hrr25p is not the only Crz1p kinase. (A) Crz1p phosphorylation state differs in vivo and in vitro. For the in vitro assay, 0.1 µg recombinant GST-Crz1p was incubated for 30 min ± 0.03 µg GST-Hrr25 and 200 µM ATP. For the in vivo experiment, wild-type yeast (YPH499) expressing GST-Crz1p (pLMB119) were treated for 30 min with 2 µg/mL FK506 or solvent, and extracts were prepared. Proteins were analyzed by SDS-PAGE followed by Western blotting with an α-GST antibody. (B) GST-Crz1p is phosphorylated to a lesser extent by Hrr25p-depleted extracts. Five micrograms recombinant GST-Crz1p immobilized on glutathione sepharose was incubated with 250 µg extract prepared from wild-type (YPH499) or KKY387 cells grown in galactose (expressing Hrr25p) or in glucose (Hrr25p-depleted), γ32P-ATP, and an ATP regenerating system, and incubated for 40 min. Samples were analyzed by SDS-PAGE and autoradiography. The lower panel shows the depletion of Hrr25p from the extracts by Western blot using an α-HA antibody.
We next examined the effects of Hrr25p depletion on Crz1p phosphorylation by investigating the phosphorylation of bacterially expressed GST-Crz1p by extracts lacking Hrr25p. GST-Crz1p was immobilized on glutathione sepharose and incubated with extract prepared from cells expressing or depleted of Hrr25p (KKY387). An ATP regenerating system and γ32P-ATP was added, and the incorporation of 32P into GST-Crz1p was determined. Although GST-Crz1p was phosphorylated under both conditions, depletion of Hrr25p resulted in a 4.5 ± 1.7-fold decrease in 32P incorporation (Fig. 5B). GST-Crz1p was phosphorylated to the same extent by wild-type and Hrr25p-containing extracts. In addition, GST-Crz1 incubated with Hrr25p-deficient extract migrated slightly faster than GST-Crz1p incubated with wild-type extract. Together these observations suggest that, although Hrr25p provides a significant amount of Crz1p phosphorylation, additional kinases act on Crz1p in vivo.
Discussion
Yeast cells adapt to a wide range of growth conditions through activation of signaling pathways that sense and respond to changes in the environment. Calcineurin, a Ca2+/calmodulin-dependent phosphatase, is activated in response to cell wall damage, increased salinity, and alkaline pH, and promotes cell survival under these conditions, in part by dephosphorylating the transcription factor Crz1p (Cyert 2003). Upon dephosphorylation, Crz1p translocates from the cytosol to the nucleus, where it activates gene expression. However, activation of Crz1p is readily reversible, and when cells are treated with the calcineurin inhibitor FK506, Crz1p is rapidly rephosphorylated and redistributed to the cytosol. Despite extensive genetic analyses, the identity of the kinase(s) responsible for termination of calcineurin/Crz1p signaling has remained elusive.
The sequencing of the yeast genome has identified the entire protein coding potential of this organism; new technologies are currently being developed to harness this information to determine the biological and biochemical functions of all yeast proteins. We took advantage of one such technique to search for Crz1p kinase(s) on a proteomic scale, and identified Hrr25p (Zhu et al. 2000). The data presented here establish that Hrr25p is a bona fide regulator of Crz1p in vivo. Furthermore, this study marks the first successful application of this proteome-based assay to identify a biologically relevant protein kinase for a specific substrate.
Hrr25p is a casein kinase I homolog with multiple cellular functions
Hrr25p was initially identified in a screen for mutants sensitive to continuous expression of HO endonuclease (Hoekstra et al. 1991). This finding, combined with the observation that these mutants are sensitive to X-ray irradiation, MMS, and hydroxyurea (Hoekstra et al. 1991; Ho et al. 1997) suggests that Hrr25p plays a role in DNA repair. In fact, the only other known in vivo substrate for Hrr25p, the transcription factor Swi6p, up-regulates expression of repair genes in response to DNAdamage (Ho et al. 1997). Hrr25p has also been implicated in secretory functions (Murakami et al. 1999) and meiosis (Hoekstra et al. 1991). The varied and severe defects exhibited by hrr25 mutants suggest that the kinase plays a role in many important cell functions. We have shown that Hrr25p works in opposition to calcineurin in regulating Crz1p; however, inhibition of calcineurin fails to suppress the growth defects of hrr25Δ cells (K. Kafadar and M. Cyert, unpubl.). Therefore, additional phosphatases must dephosphorylate Hrr25p substrates in vivo.
Hrr25p is one of four casein kinase I homologs in yeast. Yck1p and Yck2p are an essential gene pair and are involved in cell morphogenesis, cytokinesis, and endocytosis (Robinson et al. 1992, 1993). Yck3p forms an essential gene pair with Hrr25p (Wang et al. 1996), although we found no evidence of Yck3p involvement in Crz1p regulation (K. Kafadar and M. Cyert, unpubl.). Yck1p, Yck2p, and Yck3p all contain prenylation motifs and are tightly associated with the plasma membrane. Hrr25p lacks this modification, and we show here, using a GFP-Hrr25p fusion protein, that this kinase localizes throughout the cell, and concentrates at the emerging bud tip and bud neck. Taken together with the fact that hrr25 mutant cells have cytokinesis defects, this observation suggests that Hrr25p may play an important role in cell separation. Consistent with our localization data, GFP-Hrr25p is found in both soluble and membrane-associated pools following subcellular fractionation. These results differ somewhat from those of a previous study which found Hrr25p in plasma membrane and nuclear fractions, although the epitope-tagged version of Hrr25p used by those authors was not tested for functionality in vivo (Vancura et al. 1994).
Hrr25p negatively regulates Crz1p
Our results demonstrate that, in addition to its other functions, Hrr25p negatively regulates Crz1p. Overexpression of HRR25 prevents Crz1p from efficiently accumulating in the nucleus, and stimulates its rapid export from the nucleus, thus decreasing its ability to activate gene expression. In addition, overexpression of the kinase results in hyperphosphorylation of Crz1p. Consistent with these data, loss of Hrr25p results in increased basal and Ca2+-induced Crz1p-dependent CDRE activation, as well as increased nuclear accumulation in response to low concentrations of Ca2+. With the identification of a Crz1p kinase, we have added an additional component to the calcineurin pathway, which provides another mechanism by which Crz1p may be regulated in vivo. Regulation of Hrr25p activity could modulate Crz1p activity in response to Ca2+, thereby increasing or decreasing the efficacy of a given environmental stress in activating Crz1p-dependent gene expression. Although hydroxyurea treatment has been shown to activate Hrr25p in vivo (Ho et al. 1997), this treatment had no effect on GFP-Crz1p localization (K. Kafadar and M. Cyert, unpubl.). Crz1p phosphorylation alters its rate of nuclear import and export (Polizotto and Cyert 2001; Boustany and Cyert 2002), and Hrr25p may affect either, or both, of these rates.
Currently nothing is known of how Hrr25p activity is regulated in vivo. Hrr25p is most closely related to mammalian CKIδ and CKIε, both of which are inhibited by autophosphorylation of a C-terminal extension. Interestingly, calcineurin has been shown to alleviate this inhibition in vitro and in vivo by dephosphorylating CKIε (Cegielska et al. 1998; Lowrey et al. 2000; Liu et al. 2002). Although Hrr25p also contains an extended C terminus and is known to autophosphorylate (DeMaggio et al. 1992), we were unable to detect any dephosphorylation of Hrr25p by calcineurin in vitro (K. Kafadar and M. Cyert, unpubl.). Further insight into the in vivo regulation of Hrr25p will be required to fully appreciate its physiological role in modulating Crz1p activity.
Physical association of regulatory and effector proteins results in finely modulated and rapid responses to stimuli. In addition, the same effector protein bound to different regulators may respond differently to the same stimulus. We have shown that Crz1p forms a stable interaction with Hrr25p. The observation that Hrr25p binds Crz1p independently of its catalytic activity suggests that these two proteins form a long-lived complex, rather than interacting only during the phosphorylation reaction. This interaction was also seen in a high-throughput study to identify protein complexes in yeast (Ho et al. 2002). Crz1p has also been shown to bind calcineurin (Boustany and Cyert 2002). The interaction of calcineurin and Crz1p ensures that, upon activation, calcineurin rapidly desphosphorylates and activates the protein. Once calcineurin is inactivated due to falling Ca2+ levels, rephosphorylation by Hrr25p would result in the swift return of Crz1p to the cytosol, and termination of the response. Previous studies have demonstrated that Crz1p shuttles continuously between the cytosol and the nucleus (Stathopolous-Gerontides et al. 1999). Because Hrr25p forms a stable association with Crz1p, rephosphorylation of the transcription factor could occur in either cellular compartment. This method of rapid protein modification through physical association with regulators is observed in many signaling pathways. For example, NFAT is known to interact with calcineurin as well as several kinases. Dephosphorylation by calcineurin promotes NFAT translocation to the nucleus, whereas phosphorylation by several kinases results in the rapid return of NFAT to the cytosol (Crabtree and Olson 2002).
Additional kinases
The consensus sites for CKI have been extensively studied. The preferred phosphorylation site, S(P)-X(1-3)-S*/T* (Kennelly and Krebs 1991), suggests that CKI typically phosphorylates targets that have been previously phosphorylated, although the phosphoserine can be replaced with a negatively charged residue (Kennelly and Krebs 1991). We have shown, using purified proteins, that Hrr25p efficiently phosphorylates Crz1p in vitro. Preliminary analysis suggests as many as eight residues are phosphorylated (K. Kafadar and M. Cyert, unpubl.). Analysis of the Crz1p sequence reveals several clusters of CKI consensus sites, and future studies will examine which of these are bona fide targets of Hrr25p. Because Hrr25p efficiently phosphorylates Crz1p in vitro, the kinase may not require previous phosphorylation; it may begin by phosphorylating a site with an acidic amino acid, and thus initiate a cascade of downstream phosphorylation events. However, it is possible that a priming kinase or an additional regulatory step is required for efficient phosphorylation of Crz1p by Hrr25p in vivo but not in vitro.
Although significant, the effects of Hrr25p on Crz1p activity, localization, and phosphorylation are partial, suggesting that additional kinase(s) are involved in Crz1p regulation. This hypothesis is supported by the observation that Crz1p phosphorylated by Hrr25p in vitro is not phosphorylated to the same extent as Crz1p phosphorylated in vivo, as determined by electrophoretic mobility. Also, in the absence of Hrr25p, Crz1p is phosphorylated, albeit to a lesser degree than when Hrr25p is present. As mentioned above, this additional phosphorylation may be due to a priming kinase for Hrr25p, although other kinases may also phosphorylate distinct regions of Crz1p. Further observations indicate that Crz1p activity is not regulated solely through its localization. When tethered permanently in the nucleus, Crz1p is still activated by Ca2+ in a calcineurin-dependent manner, although the activation is less than that of wild-type Crz1p (K. Kafadar and M. Cyert, unpubl.). This activation is unaffected by Hrr25p, providing further evidence for the existence of additional Crz1p kinases.
Several other kinases emerged as potential Crz1p kinases from the high-throughput screen described here, and we are currently investigating whether they affect Crz1p in vivo. However, this assay may fail to identify additional Crz1p kinases, due to several limitations. For example, specific physiological conditions may be required to isolate active forms of some kinases. Alternatively, a kinase may require prior phosphorylation of a consensus site, or biochemical conditions not provided by the assay for proper activity.
The identification of Hrr25p as a Crz1 kinase represents a significant advance in our understanding of the calcineurin pathway. Extensive genetic analyses failed to identify any such kinase(s). Failure to recover HRR25 from genetic screens is most likely due to the severe growth defect of hrr25 mutant cells and the partial effect of its overexpression on Crz1p activity. Use of the high-throughput proteome-based kinase assay described here has allowed the first identification of a biologically relevant negative regulator for Crz1p.
In summary, we have identified a novel physiological role for the casein kinase I homolog Hrr25p in regulating the calcineurin signaling pathway. By phosphorylating Crz1p, Hrr25p inhibits Crz1p accumulation in the nucleus, thereby preventing the activation of Crz1p-dependent transcription. The addition of Hrr25 to the calcineurin/Crz1p pathway suggests new possible mechanisms for Crz1p regulation in response to a changing environment, and may provide insight into the coordination of the calcineurin response with other stress-induced pathways.
Materials and methods
Yeast media and general methods
Standard media and culture conditions were used (Sherman 1991) except that twice the level of amino acids and nucleotides were added to synthetic media. Selective media used for calcium induction were made with 3.5% NH4Cl instead of NH4SO4. FK506 was generously provided by Fujisawa Corp. Yeast cells were transformed using the lithium acetate method, and bacterial cells were transformed by electroporation (Ausubel et al. 1987). All recombinant DNA manipulations were performed according to standard protocols (Ausubel et al. 1987). Plasmids were prepared using Qiagen spin mini-prep kits (Qiagen).
Yeast strains
The yeast strains used in this study are described in Table 1. Strain KKY271 was created from BJ5459 by homologous recombination using a C-terminal CRZ1::ZZ-tev-S::kanMX knock-in cassette generated by PCR (gift from Karsten Weiss, Department of Cell and Developmental Biology, University of California at Berkeley); “ZZ” represents the protein Abinding domain of IgG, “tev” is a cleavage site for TEV site-specific protease, and “S” is a 15-amino acid S-tag derived from RNase Awhich binds the S-protein. KKY387 was derived from YPH499; degrontagged HRR25 (pKK204) was introduced into cells under a galactose-inducible promoter, and the genomic HRR25 was then deleted using an HRR25::loxP-kanMX-loxP disruption cassette generated by PCR (Guldener et al. 1996).
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | Silorski and Hieter 1989 |
| ASY832 | Same as YPH499 but TRP1-4xCDRE-LacZ::ura3-52 | A. Stathopolous (unpubl.) |
| BJ5459 | MATaura3-52 trp1 lys2-801 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1-Δ-Δ1.6R can1 GAL | Jones 1991 |
| KKY271 | Same as BJ5459 except CRZ1::ZZ-tev-S::kanMX | This study |
| KKY387 | Same as YPH499 except hrr25Δ::loxP-kanMX-loxP pKK204 | This study |
| KKY401 | Same as KKY387 but TRP1-4xCDRE-LacZ::ura3-52 | This study |
Plasmids
The plasmids used in this study are described in Table 2. To generate pKK191, HRR25 was amplified from genomic DNAby PCR flanked by BamH1 and HindIII sites, and then ligated into pUG36 (CEN URA3 pMET25-yEGFP3; gift from U. Güldener and J.H. Hegemann, Institute for Microbiology, Heinrich-Hiene-University, Dusseldorf, Germany) to create GFP-HRR25. To create pKK194, 3HA-HRR25 flanked by BamH1 and SalI sites was amplified by PCR from pAM4-1 (Murakami et al. 1999) and inserted into pCu423CUP1 (2µ HIS3 pCUP1; Labbâe and Thiele 1999). To create GST-tagged bacterial expression constructs, HRR25 was amplified from genomic DNAby PCR flanked by BamH1 and XhoI sites, and cloned into pGEX-4T-3 (Stratagene) to create pKK196. To create a rapidly degradable, N-degron-tagged version of Hrr25p, 3HA-HRR25 flanked by NdeI and HindIII sites was amplified from pAM4-1 by PCR and ligated into pTS842 (gift from M. Byrnes and T. Stearns, unpubl.). pTS842 contains a Ub-K-LacI cassette, where Ub is ubiquitin, K is arginine, and LacI is the first 23 amino acids of the Escherichia coli LacI gene. This cassette was amplified by PCR from pUB23-arg (Bachmair et al. 1986) and inserted into pTS330 (pRS315; Silorski and Heiter 1989) with GAL4 promoter and ACT1 terminator). For details on the N-degron Ub-K-LacI cassette, see Bachmair et al. (1986).
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| AM4-1 | 2µ pGAL1-3HA-HRR25 | Murakami et al. 1999 |
| AM8-2 | 2µ pGAL1-3HA-HRR25-K38A | Murakami et al. 1999 |
| KK191 | GFP-HRR25 | This study |
| KK194 | 3HA-HRR25 in pCu423CUP1 | This study |
| KK196 | GST-HRR25 in pGEX4T-3 | This study |
| KK204 | 2µ pGAL-3HA-HRR25degron | This study |
| RSP121 | GST-CRZ1 in pGEX4T-3 | Polizotto and Cyert 2001 |
| LMB119 | GAL1p-GST-CRZ1 | L.M. Boustany (unpubl.) |
| LMB127 | 3xGFP-CRZ1 | Boustany and Cyert 2002 |
All PCR products were first ligated into the bacterial vector PCR2.1TOPO, using the TOPO-TAcloning kit (Invitrogen), and then shuttled into the indicated vector.
GST purification of recombinant proteins from E. coli
First, 500 mL BL21/pLys bacterial cells expressing GST-Crz1p or GST-Hrr25p was grown to an O.D.600 of 0.8 and lysed by sonication in 5 mL lysis buffer (50 mM Tris at pH 8, 100 mM NaCl, 2 mM EDTA, 2 mM EGTA, 5 mM DTT) with a standard cocktail of protease inhibitors added (1 mM PMSF, 1 mM benzamidine, 2 µg/mL leupeptin, 2 µ/mL aprotinin). Then, 0.1% Tween-20 and 1.5 M NaCl was added to the lysates, which were then centrifuged at 15,000 rpm for 15 min. The lysates were incubated with 1 mL washed glutathione sepharose beads (Amersham) on a rotator for 2 h at 4°C;. The beads were collected by centrifugation and loaded into a disposable 10-mL column (BioRad). The column was washed with five column volumes of wash buffer (50 mM Tris at pH 8.0, 110 mM KOAc, 2 mM MgCl2, 2 mM DTT, 0.1% Tween-20), two column volumes of wash buffer supplemented with 1 mM ATP, then three column volumes of wash buffer. Proteins were eluted in five 1-mL aliquots in wash buffer with 15 mM reduced glutathione added. Eluted fractions were dialyzed into Buffer 88 (20 mM HEPES at pH 6.8, 150 mM KOAc, 250 mM sorbitol, 2 mM MgOAc, 1 mM DTT, protease inhibitors) using a DispoDialyzer (10,000 MW cutoff; Spectra/Por). Proteins were analyzed by SDS-PAGE along with BSAstandards, and visualized by Coomassie to determine protein concentrations.
Identification of Hrr25p as a Crz1p kinase
GST-Crz1p purified from E. coli was used as a substrate in a yeast protein kinase chip assay essentially as described (Zhu et al. 2000). Protein substrates (of which GST-Crz1 was one) were first arrayed in a 96-well plate and then manually double-printed onto a GPTS-activated glass surface. The proteins were allowed to cross-link to the epoxy group on the glass surface through their primary amine groups for 1 h at room temperature, followed by blocking in 1% BSAin PBS buffer at room temperature for an additional hour. Individual protein kinases purified from yeast as described (Zhu et al. 2000) were added to separate chips to carry out the kinase assays at 30°C for 30 min. After washing with PBS, the chips were exposed to a PhosphorImager screen, and the resulting signals were analyzed by the PhosphorImager.
In vitro phosphorylation of GST-Crz1p in bacterial lysates
Extracts were made from BL21/pLys bacterial cells expressing ZZ-Crz1p, 3HA-Hrr25p, or 3HA-Hrr25pK38A. Five-hundredmilliliter cultures were grown to O.D.600 0.8 and harvested by centrifugation. The cells were disrupted by sonication as described above, without the addition of NaCl. Four-hundred micrograms of each relevant extract was combined with 35 µL of IgG slurry, (Amersham), and the total volume was brought up to 500 µL in binding buffer (50 mM Tris at pH 8, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, protease inhibitors). The reactions were placed on a rotator at 4°C for 1 h. The beads were washed five times with binding buffer + 0.5% Triton X-100 and three times in kinase buffer (50 mM Tris at pH 7.5, 10 mM MgCl2, 1 mM DTT, protease inhibitors), and then resuspended in 10 µL kinase buffer with 10 µM cold ATP and 10 µCi γ32PATP (NEN), and incubated for 30 min at 30°C. The reactions were terminated by the addition of SDS loading buffer, and analyzed by SDS-PAGE and autoradiography. To analyze dephosphorylation of GST-Crz1p by calcineurin, ZZ-Crz1p was phosphorylated in bacterial lysates as above. After phosphorylation, the kinase buffer was removed, and the beads were washed twice in 50 mM Tris at pH 7.5 + 1 mM DTT. The beads were then resuspended in 200 µL of 50 mM Tris at pH 7.5 + 1 mM DTT and supplemented with (1) buffer; (2) 500 U calcineurin (Calbiochem), 2500 U calmodulin (Calbiochem), 20 mM CaCl2; or (3) 500 U calcineurin, 2500 U calmodulin, 100 mM EGTA. The reactions were placed on a rotator at 30°C for 30 min. After incubation, the supernatant was removed and the beads were resuspended in sample buffer. Samples were analyzed by SDS-PAGE and autoradiography.
Quantitative β-galactosidase assays
For HRR25 overexpression assays, ASY832 cells containing indicated plasmids were grown in selective media to an O.D.600 of 0.5, induced with 0.5 mM CuSO4 for 3 h, and then diluted 1:4 into media ± 200 mM CaCl2 for an additional 3 h. For Hrr25p depletion assays, KKY387 cells were grown to log phase in YPGal and diluted to O.D.600 0.2 in YPD or YPGal and grown for 5 h. Next, 10 mM, 20 mM, or 50 mM CaCl2 was added for 1 h. Cells were harvested by centrifugation, washed once in water, and broken open by glass bead lysis (Withee et al. 1997) in breaking buffer (100 mM Tris at pH 8.0, 20% glycerol, 1 mM DTT) with added protease inhibitors. Protein concentration was determined using the Bradford assay (Bio-Rad). β-galactosidase activity was determined as follows: 10-50 µg total extract was combined with 90 µL Z-Buffer (100 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSo4, 0.027% β-mercaptoethanol) and 20 µL of 4 mg/mL ONPG (O-nitrophenyl-β-D-galactopyranoside, Sigma) in a 96-well plate. Each value is an average of two independent cultures, each measured in triplicate. Graphs represent the average of three independent assays.
Immunoblot analysis
Yeast cultures were grown to log phase and harvested. Cells expressing pAM4-1 or pAM8-2 were grown in raffinose until O.D.600 0.5, and then induced with 2% galactose for 4 h. Cells were resuspended in 100 µL Buffer 88 and lysed by glass bead disruption. For IgG pull-downs, 20 µL packed, equilibrated IgG sepharose beads (Amersham) was added to the extracts, and the total volume was brought up to 400 µL in Buffer 88. The reactions were placed on a rotator for 1 h at 4°C and then washed five times in Buffer 88 + 1% NP-40. The beads were resuspended in sample buffer and loaded on a 7% polyacrylamide gel. ZZ-Crz1p was detected using a polyclonal anti-Crz1p antiserum, and an anti-rabbit IgG-coupled HRP secondary antibody (Amersham). HA-Hrr25p was detected with a monoclonal anti-HA16B12 antibody (Covance) and a monoclonal anti-mouse IgG-coupled HRP secondary antibody (Amersham). Immunoblots were developed using ECL (Amersham).
Determination of GST-Crz1p phosphorylation state
For in vitro phosphorylation of GST-Crz1p, 0.1 µg recombinant GST-Crz1p was combined with 0.03 µg recombinant GSTHrr25p in kinase buffer supplemented with 200 µM ATP and protease inhibitors. The reactions were incubated for 40 min at 30°C and terminated by the addition of SDS sample buffer. One-fourth of the reaction was used for analysis. To examine the phosphorylation state of GST-Crz1p in vivo, YPH499 cells expressing GST-CRZ1 (pLMB119; 2µ pGAL4 URA3) were grown to log phase and induced for 4 h with 2% galactose. The culture was divided in two and treated with either 2 µg/mL FK506 or solvent (90% ethanol, 10% Tween-20) for 30 min. Extracts were prepared and GST-Crz1p was isolated using glutathione sepharose. All samples were analyzed by SDS-PAGE and Western blotting using an α-GST antibody.
Phosphorylation of recombinant GST-Crz1p by yeast extract was performed as follows. Extracts were made from KKY387 cells grown in galactose or glucose for the last 5 h of culture time. Then, 250 µg extract was combined with 5 µg recombinant GST-Crz1p immobilized on glutathione sepharose (Amersham), 10µCi γ32P-ATP, an ATP regenerating system [25 U pyruvate kinase (Calbiochem), 4 mM ATP, 5 mM phosphoenolpyruvate], and phosphatase inhibitors (50 mM NaF, 50 mM sodium molybdate, 50 mM EDTA, 50 mM EGTA) in kinase buffer in a final volume of 200 µL. Reactions were incubated on a rotator at 30°C for 40 min, the beads were washed in Buffer 88 + 0.1% Triton X-100, and SDS sample buffer was added. The samples were run on a 5%-12% gradient SDS polyacrylamide gel, dried down, and exposed to film. Quantification was performed using a PhosphorImager (Bio-Rad).
Fluorescence microscopy
Cells expressing green fluorescent protein (GFP) were visualized as described (Stathopoulos-Gerontides et al. 1999) but using a Nikon Eclipse E600 microscope with fluorescence optics and an HB100 mercury lamp. Fluorescein filter sets (Chroma) were used to visualize GFP. Photos were taken using a Hammamatsu digital CCD 47420-95 camera and QED software (QED Imaging). To visualize GFP-Hrr25p, YPH499/pKK191 cells were grown to log phase and examined. Arange of methionine concentrations was used (0-150 mg/L) to confirm that localization of GFP-Hrr25p was not affected by expression level. KKY387 cells were grown to log phase in galactose media, and 2% glucose was added for 3 h where indicated. Then, 50 mM CaCl2 was added as noted. To examine the effects of HRR25 overexpression on GFP-Crz1p localization, KKY271 cells carrying either pCu423CUP1 or pKK206 were diluted to O.D.600 0.25 in media lacking methionine and grown at room temperature for 3 h. Next, 0.5 mM CuSO4 was added for 3 h, and the cells were examined 5 min and 25 min after the addition of 200 mM CaCl2. For each sample, 150-200 cells were counted and scored as nuclear, cytosolic, or nuclear/cytosolic. The average of two independent trials is presented.
Acknowledgments
We thank Victoria Heath and Jagoree Roy for critical reading of the manuscript, and Tim Stearns for suggesting use of the degron tag. We thank the following people for providing plasmids: Akihiko Nakano (HA-HRR25 and HA-HRR25-K38A), Michael Byrnes and Tim Stearns (pTS842), Renee Polizotto (pGEX4T-3-CRZ1), Leila Boustany (GST-CRZ1, GFP-CRZ1), and J.H. Hegemann (pUG36); and Angela Stathopolous-Gerantides for numerous strains and constructs. M.S.C. is supported by NIH research grant GM-48729. K.A.K. is supported by NIH training grant 5T32GM07276. M.S. is supported by NIH research grant GM62480-03. H.Z. is supported by a grant from the Damon Runyon-Walter Winchell Cancer Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1140603.
References
- Aramburu J., Rao, A., and Klee, C.B. 2000. Calcineurin: From structure to function. Curr. Top. Cell. Reg. 36: 237-295. [DOI] [PubMed] [Google Scholar]
- Ausubel F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1987. Current protocols in molecular biology. Wiley, New York.
- Bachmair A., Finley, D., and Varshavsky, A. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179-186. [DOI] [PubMed] [Google Scholar]
- Boustany L.M. and Cyert, M.S. 2002. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes & Dev. 16: 608-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegielska A., Gietzen, K.F., Rivers, A., and Virshup, D.M. 1998. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem. 273: 1357-1364. [DOI] [PubMed] [Google Scholar]
- Crabtree G.R. and Olson, E.N. 2002. NFAT signaling: Choreographing the social lives of cells. Cell 109: 67-79. [DOI] [PubMed] [Google Scholar]
- Cunningham K.W. and Fink, G.R. 1994. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124: 351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M.S. 2003. Calcineurin signalling in Saccharomyces cerevisiae: How yeast go crazy in response to stress. Biochem. Biophys. Res. Comm. (in press). [DOI] [PubMed]
- Cyert M.S. and Thorner, J. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12: 3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M.S., Kunisawa, R., Kaim, D., and Thorner, J. 1991. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. 88: 7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaggio A.J., Lindberg, R.A., Hunter, T., and Hoekstra, M.F. 1992. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc. Natl. Acad. Sci. 89: 7008-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcasanu I.C., Hirata, D., Tsuchiya, E., Nishiyama, F., and Miyakawa, T. 1995. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese, in blocking the entry of ions into the cells. Eur. J. Biochem. 232: 712-717. [PubMed] [Google Scholar]
- Giaever G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Vâeronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andrâe, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387-391. [DOI] [PubMed] [Google Scholar]
- Graef I.A., Chen, F., Chen, L., Kuo, L., and Crabtree, G.R. 2001. Signals transduced by Ca2+/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105: 863-875. [DOI] [PubMed] [Google Scholar]
- Graef I.A., Wang, F., Charron, F., Chen, L., Neilson, J., Tessier-Lavigne, M., and Crabtree, G.R. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 113: 657-670. [DOI] [PubMed] [Google Scholar]
- Guldener U., Heck, S., Fielder, T., Beinhauer, J., and Hegemann, J.H. 1996. Anew efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R., Garciadeblas, B., and Rodrâiguez-Navarro, A. 1991. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 291: 189-191. [DOI] [PubMed] [Google Scholar]
- Ho Y., Mason, S., Kobayashi, R.S., Hoekstra, M., and Andrews, B. 1997. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNAdamage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 94: 581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Hoekstra M.F., Liskay, R.M., Ou, A.C., DeMaggio, A.J., Burbee, D.G., and Heffron, F. 1991. HRR25, a putative protein kinase from budding yeast: Association with repair of damaged DNA. Science 253: 1031-1034. [DOI] [PubMed] [Google Scholar]
- Hoekstra M.F., Dhillon, N., Carmel, G., DeMaggio, A.J., Lindberg, R.A., Hunter, T., and Kuret, J. 1994. Budding and fission yeast casein kinase I isoforms have dual-specificity protein kinase activity. Mol. Biol. Cell 5: 877-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194: 428-453. [DOI] [PubMed] [Google Scholar]
- Kennelly P.J. and Krebs, E.G. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266: 15555-15558. [PubMed] [Google Scholar]
- Kuno T., Tanaka, H., Mukai, H., Chang, C.D., Hiraga, K., Miyakawa, T., and Tanaka, C. 1991. cDNAcloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem. Biophys. Res. Comm. 180: 1159-1163. [DOI] [PubMed] [Google Scholar]
- Labbâe S. and Thiele, D.J. 1999. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306: 145-153. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ishii, S., Tokai, M., Tsutsumi, H., Ohki, O., Akada, R., Tanaka, K., Tsuchiya, E., Fukui, S., and Miyakawa, T. 1991. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol. Gen. Genet. 227: 52-59. [DOI] [PubMed] [Google Scholar]
- Liu F., Virshup, D.M., Nairn, A.C., and Greengard, P. 2002. Mechanism of regulation of casein kinase I activity by group I metabotropic glutamate receptors. J. Biol. Chem. 277: 45393-45399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura, K., Antoch, M.P., Yamazaki, S., Zemenides, P.D., Ralph, M.R., Menaker, M., and Takahashi, J.S. 2000. Positional systemic cloning and functional characterization of the mammalian circadian mutation tau. Science 288: 483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheos D.P., Kingsbury, T.J., Ahsan, U.S., and Cunningham, K.W. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes & Dev. 11: 3445-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P., Morin, N., Baginsky, W., el-Sherbeini, M., Clemas, J.A., Nielsen, J.B., and Foor, F. 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-D-glucan synthase. Mol. Cell. Biol. 15: 5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal I., Rios, G., Mulet, J.M., Serrano, R., and de Larrinoa, I.F. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425: 323-328. [DOI] [PubMed] [Google Scholar]
- Mendoza I., Rubio, F., Rodriguez-Navarro, A., and Pardo, J.M. 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 269: 8792-8796. [PubMed] [Google Scholar]
- Molkentin J.D., Lu, J.R., Antos, C.L., Markham, B., Richardson, J., Robbins, J., Grant, S.R., and Olson, E.N. 1998. Acalcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Geiser, J.R., and Davis, T.N. 1996. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16: 4824-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A., Kimura, K., and Nakano, A. 1999. The inactive form of a yeast casein kinase I suppresses the secretory defect of the sec12 mutant. Implication of negative regulation by the Hrr25 kinase in the vesicle budding from the endoplasmic reticulum. J. Biol. Chem. 274: 3804-3810. [DOI] [PubMed] [Google Scholar]
- Polizotto R.S. and Cyert, M.S. 2001. Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J. Cell Biol. 154: 951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozos T.C., Sekler, I., and Cyert, M.S. 1996. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol. Cell. Biol. 16: 3730-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.C., Hubbard, E.J., Graves, P.R., DePaoli-Roach, A.A., Roach, P.J., Kung, C., Haas, D.W., Hagedorn, C.H., Goebl, M., and Culbertson, M.R. 1992. Yeast casein kinase I homologues: An essential gene pair. Proc. Natl. Acad. Sci. 89: 28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.C., Menold, M.M., Garrett, S., and Culbertson, M.R. 1993. Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol. Cell. Biol. 13: 2870-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H.K., Antebi, A., Fink, G.R., Buckley, C.M., Dorman, T.E., LeVitre, J., Davidow, L.S., Mao, J.I., and Moir, D.T. 1989. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell 58: 133-145. [DOI] [PubMed] [Google Scholar]
- Sherman F. 1991. Getting started with yeast. In: Guide to yeast genetics and molecular biology (eds. C. Guthrie and G.R. Fink), pp. 3-22. Academic Press, San Diego, CA.
- Silorski R.S. and Heiter, P. 1989. Asystem of shuttle vectors and yeast host strains designed for efficient manipulation of DNAin Saccharomyces cerevisiae. Genetics 122: 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A.M. and Cyert, M.S. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes & Dev. 11: 3432-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A., Guo, G.G., and Cyert, M.S. 1999. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes & Dev. 13: 798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancura A., Sessler, A., Leichus, B., and Kuret, J. 1994. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J. Biol. Chem. 269: 19271-19278. [PubMed] [Google Scholar]
- Vielhaber E. and Virshup, D.M. 2001. Casein kinase I: From obscurity to center stage. IUBMB Life. 51: 73-78. [DOI] [PubMed] [Google Scholar]
- Wang X., Hoekstra, M.F., DeMaggio, A.J., Dhillon, N., Vancura, A., Kuret, J., Johnston, G.C., and Singer, R.A. 1996. Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 16: 5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J.L., Mulholland, J., Jeng, R., and Cyer, M.S. 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8: 263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman, K., Gasch, A.P., Li, H.X., Ogawa, N., Botstein, D., Brown, P.O., and Cyert, M.S. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277: 31079-31088. [DOI] [PubMed] [Google Scholar]
- Zhu H., Klemic, J.F., Chang, S., Bertone, P., Casamayor, A., Klemic, K.G., Smith, D., Gerstein, M., Reed, M.A., and Snyder, M. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26: 283-289. [DOI] [PubMed] [Google Scholar]