Abstract
Vesicle trafficking requires membrane fusion, mediated by SNARE proteins, and upstream events that probably include “tethering”, an initial long-range attachment between a vesicle and its target organelle. Among the factors proposed to mediate tethering are a set of multisubunit tethering complexes (MTCs). The Dsl1 complex, with only three subunits, is the simplest known MTC, and is essential for the retrograde traffic of COPI-coated vesicles from the Golgi to the ER. To elucidate structural principles underlying MTC function, we have determined the structure of the Dsl1 complex, revealing a tower containing at its base the binding sites for two ER SNAREs and at its tip a flexible lasso for capturing vesicles. The Dsl1 complex binds to individual SNAREs via their N-terminal regulatory domains and also to assembled SNARE complexes; moreover, it is capable of accelerating SNARE complex assembly. Our results suggest that even the simplest MTC may be capable of orchestrating vesicle capture, uncoating, and fusion.
Keywords: Intracellular trafficking, Dsl1 complex, multisubunit tethering complex, SNARE proteins, x-ray crystallography
Introduction
Within eukaryotic cells, a collection of tethering factors is believed to function in the initial attachment of transport vesicles to their target membranes (Cai et al., 2007; Pfeffer, 1999). Consistent with an ability to bridge membranes over a significant distance, these tethering factors are large and/or elongated. Some of them are long coiled-coil homodimers (Gillingham and Munro, 2003), while others are large hetero-oligomers (multisubunit tethering complexes or MTCs) (Whyte and Munro, 2002). Tethering factors interact with other components of the protein machinery that mediates intracellular trafficking (Cai et al., 2007). For example, most of the known MTCs have been shown to interact with SNAREs, the much smaller, membrane-embedded proteins that assemble to bring the two membranes into close apposition just prior to fusion (Cai et al., 2007). Many tethering factors also interact with Rabs, small Ras-like GTPases that have long been known to play regulatory roles in membrane trafficking. Finally, several MTCs interact with coatomer, the seven-subunit protein complex that forms the coat of COPI transport vesicles. By interacting with vesicle coats, Rab proteins, SNAREs, and possibly membrane lipids, MTCs are well-positioned to orchestrate vesicle capture, docking, and/or fusion. Despite recent progress in elucidating the composition and localization of the MTCs, however, the mechanisms underlying MTC function are not well understood.
Eight MTCs have been identified, each of them implicated in one or more distinct trafficking pathways (Cai et al., 2007; Sztul and Lupashin, 2006; Whyte and Munro, 2002). The known MTCs comprise 3–10 subunits each, with total molecular weights of 250–800 kDa. An essentially complete structure has been reported for only one of these MTCs, TRAPP I (Kim et al., 2006). To gain further insight into tethering factor function, we have been studying the structure of a second, unrelated MTC, the Dsl1 complex (Tripathi et al., 2009). The Dsl1 complex displays weak sequence homology with three other MTCs, the exocyst, COG, and GARP complexes (Koumandou et al., 2007; Whyte and Munro, 2002). The Dsl1 complex is, however, architecturally simpler than these other MTCs. Indeed, the Saccharomyces cerevisiae complex is composed of just three subunits, named Dsl1, Sec39/Dsl3, and Tip20 (Kraynack et al., 2005). All three subunits are encoded by essential genes.
A wealth of data implicates the Dsl1 complex in retrograde trafficking of COPI vesicles from the Golgi to the ER. For example, temperature-sensitive mutations in any of the three subunits lead to defects in Golgi-to-ER trafficking (Andag et al., 2001; Kamena and Spang, 2004; Kraynack et al., 2005; Reilly et al., 2001; Sweet and Pelham, 1993; VanRheenen et al., 2001). Furthermore, temporarily shutting down production of the Dsl1 subunit itself causes a reversible accumulation of COPI coated vesicles (Zink et al., 2009). All three Dsl1 complex subunits are ER-localized, and it seems likely that the complex functions at the surface of the ER to capture Golgi-derived vesicles. ER localization is mediated, at least in part, by direct interactions between the Dsl1 complex and two different ER SNARE proteins, Sec20 and Use1 (Kraynack et al., 2005; Tripathi et al., 2009). Vesicle capture, on the other hand, has been proposed on the basis of genetic and physical evidence to be mediated by a direct interaction between the Dsl1 complex and the COPI coat (Andag et al., 2001; Andag and Schmitt, 2003; Zink et al., 2009).
Here, we report an x-ray crystallography based model of the Dsl1 complex. The architecture of the complex appears well-suited to a role in tethering at a distance, with ER and COPI binding sites separated by as much as 30 nm. The ability of the Dsl1 complex to bind simultaneously to two of the four SNARE proteins proposed to mediate the fusion of Golgi-to-ER trafficking vesicles raised the possibility that the Dsl1 complex might regulate SNARE assembly. Consistent with this, we find that the N-terminal regulatory domains of the ER SNAREs Use1 and Sec20 mediate their interaction with the Dsl1 complex. Moreover, the presence of the Dsl1 complex accelerates SNARE complex formation in vitro, albeit to a modest extent. Finally, we find that the Dsl1 complex binds to fully assembled SNARE complexes. Based on these results, we propose a model in which the Dsl1 complex coordinates vesicle capture and SNARE assembly upstream of membrane fusion.
Results
Crystal Structure of a Dsl1/Sec39 Complex
The Dsl1 complex is composed of three 80–90 kDa subunits: Dsl1, Sec39, and Tip20. Previously, we reported crystal structures collectively representing approximately half of the S. cerevisiae Dsl1 complex, including the Tip20 subunit and the N-terminal domains of the Dsl1 subunit (Tripathi et al., 2009). Here, we present an additional crystal structure comprising the remainder of the Dsl1 complex, including the C-terminal domains of the Dsl1 subunit and most of the Sec39 subunit. We initially crystallized S. cerevisiae Dsl1 residues 340–754 (Dsl1C) bound to Sec39. These crystals were not, however, sufficiently well-ordered to allow structure determination, perhaps because of a Dsl1 region (approximately residues 388–467) predicted to lack well-folded structure (VanRheenen et al., 2001). Using comparative sequence analysis, however, we noticed that Kluyveromyces lactis Dsl1 contains a shorter region of potential flexibility, while still retaining 29% sequence identity with S. cerevisiae Dsl1 (Figure S1A). K. lactis Dsl1 residues 332–686 (Dsl1Clactis), like S. cerevisiae Dsl1C, formed a stable complex with S. cerevisiae Sec39. More importantly, we were able to determine the crystal structure of the Dsl1Clactis–Sec39 complex using MAD phasing and refine it to 2.6 Å resolution.
Both Dsl1Clactis and Sec39 consist mostly of α-helices (Figure 1A). The predicted flexible region of Dsl1Clactis is indeed disordered: no interpretable electron density is observed for a 57-residue segment (residues 367–423). The corresponding segment of S. cerevisiae Dsl1 (residues 378–488) is almost twice as long (111 residues). Importantly, the disordered region of Dsl1 contains the sequence motifs reported to bind to subunits of the coatomer (Andag and Schmitt, 2003; Zink et al., 2009); their inclusion within a flexible “lasso” may allow the Dsl1 complex to capture COPI vesicles over a larger volume than would otherwise be possible. The ordered portion of Dsl1Clactis contains helix bundle domains homologous to those observed in nearly every subunit of a non-TRAPP MTC whose structure has been reported (Cavanaugh et al., 2007; Dong et al., 2005; Hamburger et al., 2006; Moore et al., 2007; Richardson et al., 2009; Sivaram et al., 2006; Wu et al., 2005; see Figure 2C). Sec39, by contrast, displays a novel fold; in particular, its structure is unlike that of any other known tethering complex subunit. Antiparallel α-helices, oriented perpendicular to the long axis of the 18-nm Sec39 rod, are stacked side by side along its entire length. The C-terminal region (residues 520–680) shares moderate structural homology with other proteins containing tandem arrays of antiparallel helices, including protein phosphatase 5 (Das et al., 1998) and the Sec18/NSF co-chaperone Sec17 (Rice and Brunger, 1999).
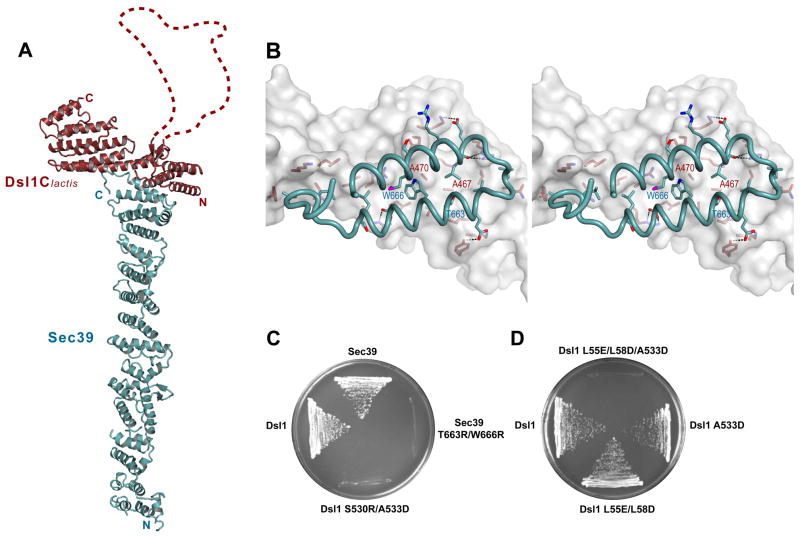
Figure 1. X-ray Crystal Structure of a Dsl1/Sec39 Complex.
(A) Ribbon representation of the complex between Dsl1Clactis (residues 332–686; red) and Sec39 (residues 1–709; blue). Dsl1Clactis residues 367–423, which are apparently disordered, are represented by a dashed red line.
(B) Stereo view of the Dsl1Clactis/Sec39 binding interface. Dsl1Clactis is shown as a surface representation, while Sec39 is shown as a ribbon representation. Side chains are included for residues involved in binding.
(C) Mutations, in either dsl1 or sec39, that disrupted the Dsl1–Sec39 interaction were lethal in yeast. Deletion strains bearing both a CEN-URA plasmid encoding the wild-type protein and a CEN-LEU plasmid encoding the corresponding mutant protein as indicated were grown at 23°C on 5-FOA plates.
(D) dsl1 mutations that disrupt the Dsl1–Sec39 interaction (A533D) and the Dsl1–Tip20 interaction (L55E/L58D) are synthetically lethal.
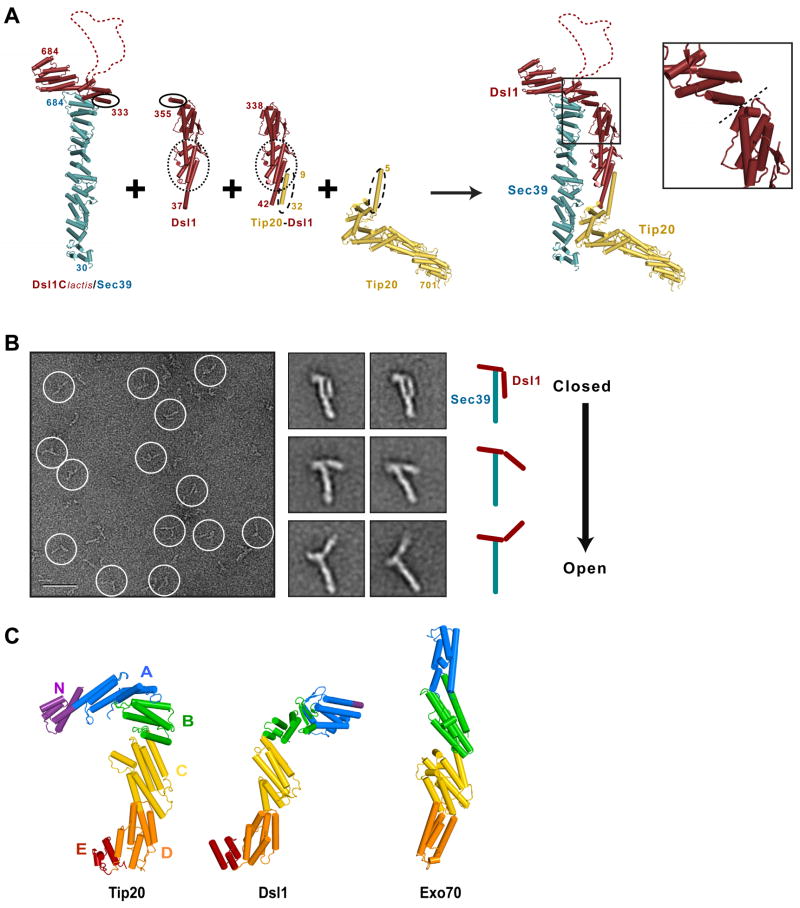
Figure 2. Molecular Architecture of the Yeast Dsl1 Complex.
(A) Modeling of the entire Dsl1 complex using crystal structures of Dsl1Clactis–Sec39, Dsl1 (PDB 3ETU), a Tip20-Dsl1 fusion protein (PDB 3ETV), and Tip20 (PDB 3FHN). Sec39, Dsl1, and Tip20 are colored in blue, red, and yellow, respectively. Regions of overlap were overlaid to yield a model representing the intact Dsl1 complex; these include Dsl1 residues 341 to 355 and Dsl1lactis residues 333–347 (indicated by solid ovals), Dsl1 residues 52 to 205 (dotted ovals), and Tip20 residues 9 to 32 (dashed ovals). Inset: Close-up view of the proposed flexible hinge (dashed line) between domains B and C (see panel C) near the center of the Dsl1 subunit.
(B) Negative stain EM of the Dsl1–Sec39 complex. Left: typical image of a sample stained with uranyl formate (scale bar = 50 nm). Middle: representative class averages displaying different conformations (side length = 36 nm). Right: cartoon representation of the class averages.
(C) Structural alignment of Tip20 (PDB 3FHN), Dsl1 (as modeled in (A)), and Exo70 (PDB 2PFV). Domains C through E were superimposed using DaliLite (Holm and Park, 2000).
The two C-terminal helices of Sec39 mediate its binding to Dsl1C through an interface featuring extensive hydrophobic and polar interactions (Figure 1B). To confirm the physiological relevance of this binding mode, we designed destabilizing substitutions in the center of the binding interface. S. cerevisiae Dsl1C S530R/A533D (equivalent to Dsl1Clactis A467R/A470D) failed to bind Sec39, as judged by gel filtration (Figure S1B). Conversely, introducing the substitutions T663R and W666R into Sec39 abolished its ability to bind Dsl1C.
Next, we carried out complementary experiments in vivo, replacing wild-type S. cerevisiae proteins with their mutant counterparts. Neither Dsl1 S530R/A533D nor Sec39 T663R/W666R was able to rescue a deletion of the corresponding wild-type protein (Figure 1C). Together, these results validate the subunit interface observed in the crystal structure and establish that the Dsl1–Sec39 interaction is required for viability in yeast. Interestingly, the other interaction responsible for the integrity of the Dsl1 complex – between the Dsl1 and Tip20 subunits – could be abolished without causing an obvious growth defect in yeast (Kraynack et al., 2005; Tripathi et al., 2009). We revisited this result by creating a milder, temperature-sensitive mutation in the Dsl1–Sec39 interface (Dsl1–A533D) and found that it is synthetic lethal with disruption of the Dsl1–Tip20 interface (Figure 1D). Thus, the physiological significance of the Dsl1–Tip20 interface is uncovered by a mutation that weakens the Dsl1–Sec39 interface, and all three subunits appear to function cooperatively in the operation of the complex.
Dsl1 Complex Architecture
To generate a model for the entire Dsl1 complex, we superimposed common regions of the Dsl1Clactis–Sec39 crystal structure and the previously-determined Tip20 and Dsl1N structures (Figure 2A). In the resulting model, the Dsl1 complex forms a tower approximately 20 nm in height, with the Sec39 and Tip20 subunits together forming the base of the tower. At the tip of the tower is the flexible lasso located near the middle of the Dsl1 subunit. Sec39 and Tip20 have previously been shown to interact directly with two different ER SNAREs (Tripathi et al., 2009); this interaction, which is further characterized below, likely anchors the base of the tower to the ER membrane.
To further validate the crystallographically-derived structural model, we investigated the Dsl1 complex using negative stain electron microscopy (EM). First, we examined the stable binary complex formed by full-length S. cerevisiae Dsl1 and Sec39 (Figure 2B). In raw images, the Dsl1–Sec39 complex displayed distinct, extended shapes. 7392 particles were manually selected and classified into 200 classes (Figure S2A). The rigid rod-like structure of Sec39 is evident in almost all of the class averages. The larger of the remaining appendages evidently corresponds to the N-terminal region of Dsl1 (residues 1–339; Figure 2B). This interpretation reveals that the relative orientation of the N- and C-terminal halves of the Dsl1 subunit varies dramatically among the class averages, yielding a series of different conformations ranging from a closed conformation that closely matches the model shown in Figure 2A to a much more open conformation (Figure 2B). Although in principle negative staining artifacts might account for some of this variability, we note that the putative hinge region falls at the boundary between Dsl1 domains (Figure 2A, inset) and conclude that the heterogeneity in the averaged images likely reflects intrinsic flexibility near the center of the Dsl1 subunit. Moreover, although the crystallographically-derived model places Sec39 and Tip20 in close proximity, we have not found evidence of a stable, direct Sec39–Tip20 interaction that would prevent flexure around a central hinge (Tripathi et al., 2009). The different conformations we observe might, as discussed below, reflect different stages in a functional cycle; e.g., before and after vesicle tethering.
Since the domain structure of Tip20 closely resembles that of Dsl1 (Figure 2C), we wondered whether Tip20 might also display evidence of flexibility. To investigate this question, we used negative stain EM to examine Tip20 alone (Figure S2B). Most class averages revealed a hooked structure, in close agreement with the previously determined crystal structure. None of these class averages appeared to represent a well-defined extended structure. On a domain-by-domain basis, Tip20 and Dsl1 both resemble the exocyst complex subunit Exo70 (Figure 2C), but Exo70 crystal structures (Dong et al., 2005; Hamburger et al., 2006; Moore et al., 2007) revealed a straight, rod-like conformation with only limited flexibility at the domain B-C interface (Hamburger et al., 2006). Thus, among the known structures of complete (or, in the case of Exo70, nearly complete) MTC subunits, only Dsl1 appears to be flexible. Notably, Exo70 and Tip20 appear to be locked in very different conformations, one straight, the other sharply hooked. Whether interactions with additional factors (e.g., SNAREs, Rabs, phospholipids) might unlock either Exo70 or Tip20 is unknown.
Next, we examined the Tip20–Dsl1–Sec39 sample by negative stain EM, but did not initially obtain useful images; indeed, many of the particles we observed resembled those present in the Dsl1–Sec39 sample described above. Because the Tip20–Dsl1 interaction is weaker than the Dsl1–Sec39 interaction (KD values of 100 and 10 nM, respectively; (Tripathi et al., 2009) and data not shown), we expected the majority of Tip20 molecules to dissociate from the ternary complex at the concentration required to prepare useful EM samples (~40 nM). Therefore, we attempted to stabilize the intact complex by using our earlier crystal structure of a Tip20-Dsl1 fusion protein to engineer a disulfide bond linking Tip20 and Dsl1. The disulfide-containing complex, Tip20(L28C)–Dsl1(L48C)–Sec39, was purified and examined by negative stain EM. Although particle heterogeneity precluded the calculation of meaningful class averages, we did observe individual particles with open and closed conformations consistent with our analysis of the binary Dsl1–Sec39 complex (Figure S2C). These results suggest that the intact Dsl1 complex contains one or more flexible hinges, but do not allow the sites of flexibility (beyond the known Dsl1 hinge) to be localized precisely. We conclude that the intact Dsl1 complex is not locked in a closed conformation, at least in the absence of additional factors.
Dsl1 Complex – SNARE Interactions
Both in vitro and in vivo, the Dsl1 complex interacts with two different ER SNAREs, Use1 and Sec20 (Kraynack et al., 2005; Sweet and Pelham, 1993; Tripathi et al., 2009). To explore the functional significance of these interactions, we first attempted to map the interacting regions more precisely. SNARE proteins are characterized by an approximately 70-residue region called the SNARE motif, usually sandwiched between an N-terminal regulatory domain and a C-terminal transmembrane anchor (Ungar and Hughson, 2003). Four SNARE motifs assemble to form a stable helix bundle capable of driving membrane fusion (see Figure 6). We established previously that the SNARE Use1 binds to Sec39 (Tripathi et al., 2009). Gel filtration experiments showed that the N-terminal half of Sec39 (residues 1–392) is sufficient to bind Use1 (Figure 3A). Conversely, we found that the extreme N-terminus of Use1, residues 1–35, is necessary (Figure 3B) and sufficient (Figure 3C) for stable binding to Sec39. Thus, the interaction between Sec39 and Use1 does not involve the SNARE motif, but rather a short sequence within the N-terminal regulatory region. Consistent with this conclusion, the mammalian ortholog of Sec39 was recently reported to bind the N-terminus of the mammalian ortholog of Use1 (Aoki et al., 2008). In yeast, the replacement of wild-type Use1 with a truncated version lacking the first 35 residues gave rise to a temperature-sensitive growth defect (Figure S3). These results suggest that the Use1–Sec39 interaction is essential at elevated temperature.
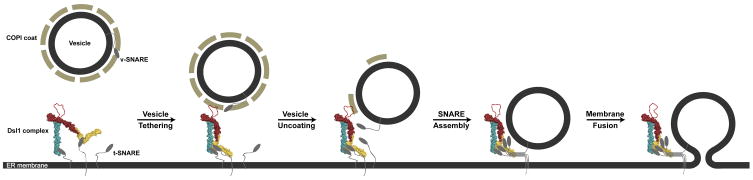
Figure 6. Working Model for Vesicle Tethering via the Dsl1 Complex.
The Dsl1 complex is depicted in a space-filling representation, color-coded as in Figure 2A. Evidence for lasso-COPI interactions has been presented by Schmitt and co-workers (Andag et al., 2001; Andag and Schmitt, 2003; Zink et al., 2009). Although two different conformations of the Dsl1 complex are illustrated, it is not known whether, or in what manner, the conformation and/or flexibility of the complex is affected by its interactions with SNAREs and COPI.
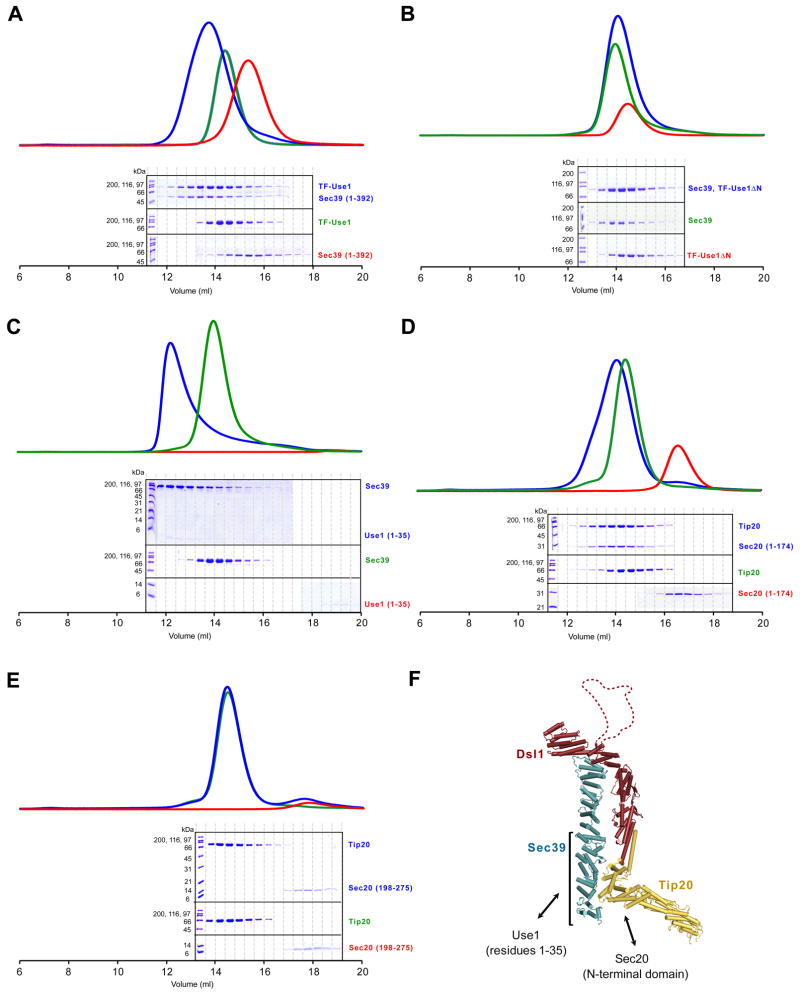
Figure 3. Mapping Interacting Regions Between the Dsl1 complex and the ER SNAREs Use1 and Sec20.
(A) The N-terminal region of Sec39 (residues 1–392) binds TF-Use1 (residues 1–212 fused to trigger factor). Sec39 alone, Use1 alone, or an equimolar mixture were analyzed by gel filtration.
(B) Sec39 does not bind TF-Use1ΔN (residues 36–216 fused to trigger factor).
(C) Sec39 binds the N-terminus of Use1 (residues 1–35). Binding of Sec39 to Use1 residues 1–35, but not to Use1 residues 1–212 (data not shown), appears to induce oligomerization as judged by performance on gel filtration.
(D) Tip20 binds the N-terminal regulatory domain of Sec20 (residue 1–174).
(E) Tip20 does not bind the Sec20 SNARE motif (residues 198–275).
(F) Summary of the interactions between the Dsl1 complex and the SNAREs Use1 and Sec20.
A different subunit of the Dsl1 complex, Tip20, interacts with the ER SNARE Sec20 (Sweet and Pelham, 1993; Tripathi et al., 2009). We found that Tip20 binds the N-terminal regulatory region of Sec20 (residues 1–174), but not the SNARE motif (residues 198–275; Figures 3D and 3E). Conversely, Tip20 residues 82–701, but not residues 357–701, bind Sec20 (Tripathi et al., 2009 and data not shown). Therefore, Tip20 residues 82–356 are necessary for binding to Sec20. Taken together, our results establish that two different Dsl1 complex subunits, Sec39 and Tip20, bind directly to N-terminal regulatory sequences of two different ER SNAREs, Use1 and Sec20 (Figure 3F).
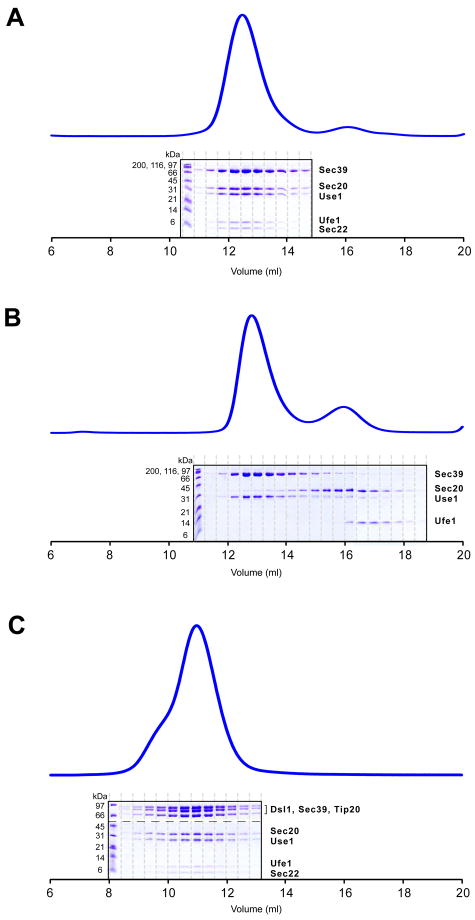
In vivo, SNAREs cycle between unassembled and assembled states. Since the Dsl1 complex binds to two individual SNARE proteins, Use1 and Sec20, we sought to determine whether it is also capable of binding to the assembled SNARE complex. Use1 and Sec20 function in concert with the additional SNARE proteins Ufe1 and Sec22 in mediating retrograde Golgi-to-ER traffic (Burri et al., 2003; Dilcher et al., 2003). To reconstitute a minimal complex containing all four SNAREs, we combined the SNARE motifs of Ufe1 and Sec22 with the full-length cytoplasmic domains of Use1 and Sec20; we retained the regulatory domains of Use1 and Sec20 because they are directly involved in Dsl1 complex binding (see above). We also found that the intact cytoplasmic domain of Use1 was insoluble; therefore, we instead performed SNARE assembly experiments with Use1–Sec39, a stable, soluble complex. Upon mixing, the four SNAREs assembled to form a monodisperse complex (Figure 4A). The yield of assembled SNARE complexes was temperature- and concentration-dependent (data not shown). No assembly was observed if any one of the four SNAREs was omitted from the mixture (Figures 4B and S4A-C); most notably, the three ER SNAREs did not assemble in the absence of the vesicle SNARE, Sec22. We obtained the same results when we used a soluble Use1 fusion protein, generated by tagging the N-terminus with the ribosomal chaperone trigger factor (TF), in place of Use1–Sec39 (data not shown). Finally, to test whether the Dsl1 complex binds to the fully assembled SNARE complex, we mixed four SNAREs and the Dsl1 complex and analyzed the mixture by gel filtration chromatography. Indeed, SNARE complexes assembled in the presence of the Dsl1 complex, and together they formed a stable heptameric complex, Tip20–Dsl1–Sec39–Use1–Sec20–Ufe1–Sec22 (Figure 4C). The heptameric complex was also formed if the Dsl1 complex was mixed with pre-formed SNARE complexes (data not shown). We conclude that the Dsl1 complex is capable of binding both to individual SNAREs and to SNARE complexes, and that the presence of the Dsl1 complex is compatible with SNARE complex assembly. These conclusions are, moreover, consistent with the observation that the Dsl1 complex engages SNARE regulatory domains rather than SNARE motifs, since the latter undergo dramatic conformational changes during SNARE assembly.
Figure 4. SNARE Complexes and Their Interaction with the Dsl1 complex.
(A) Three ER SNAREs, Use1 (residues 1–212, stabilized/solubilized by Sec39), Sec20 (residues 1–275), and Ufe1 (residues 240–320), together with the vesicle SNARE Sec22 (residues 125–188), assemble to form complexes.
(B) The three ER SNAREs do not interact in the absence of the vesicle SNARE, Sec22.
(C) SNARE complexes assemble in the presence of the intact Dsl1 complex. Individual gel filtration fractions were analyzed both by 10% SDS-PAGE gel (to visualize the high molecular weight polypeptides composing the Dsl1 complex) and by tricine SDS-PAGE gel (to visualize SNAREs); in the figure, the gels are separated by a dashed line.
Dsl1 Complex Accelerates SNARE Assembly
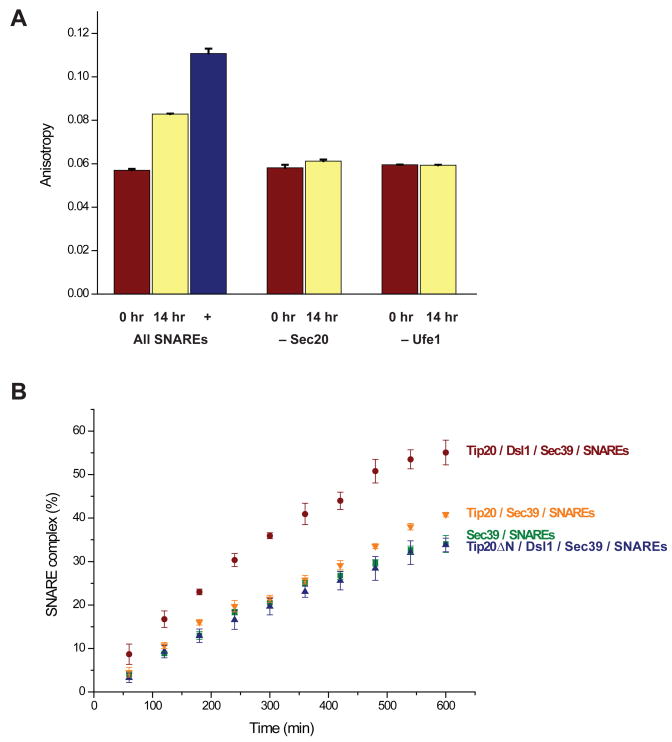
The results so far establish that the Dsl1 complex binds independently to two different ER SNAREs and also to SNARE complexes. To examine whether the Dsl1 complex regulates SNARE assembly, we developed a fluorescence polarization assay that registers the change in anisotropy of fluorescently labeled Sec22 upon SNARE complex formation. As mentioned above, the cytoplasmic domain of Use1 requires for its solubility either a trigger factor tag or Sec39. In preliminary experiments, we found that the trigger factor tag itself had a significant effect on SNARE assembly kinetics (Figure S5). To avoid this complication, we instead used Use1–Sec39 in subsequent experiments. The measured anisotropy increased after prolonged incubation of the four SNARE proteins, indicating that labeled Sec22 was incorporated into a larger molecular species (Figure 5A). Gel filtration experiments confirmed that this species contained all four SNAREs (together with Sec39); the anisotropy of the complex, once purified, remained constant for several days (data not shown). Under the conditions used, about half of the Sec22 assembled into SNARE complexes over the course of 14 hr. Control assembly reactions, in which Sec20 or Ufe1 was omitted, displayed little if any change in anisotropy (Figure 5A).
Figure 5. The Dsl1 Complex Accelerates SNARE Assembly.
(A) Fluorescence polarization experiments were performed using fluorescein-labeled Sec22 as described in the Experimental Procedures. An increase in anisotropy, signifying SNARE complex assembly, was observed after a 14-hr incubation of Use1 (stabilized/solubilized by Sec39), Sec20, Ufe1, and Sec22. The anisotropy of purified SNARE complexes is also shown. Little change in anisotropy, as compared to t=0, was observed if either Sec20 or Ufe1 was omitted from the mixture. Error bars represent SEM for two independent experiments.
(B) SNARE assembly was monitored using fluorescence anisotropy as a function of time. Details are provided in the Experimental Procedures. Error bars represent SEM for three independent experiments.
To evaluate the impact of the Dsl1 complex, we repeated the experiment with different combinations of Dsl1 complex subunits (Figure 5B). The presence of the entire Dsl1 complex accelerated SNARE complex formation, compared to Sec39 alone. When the Dsl1 subunit was omitted, this acceleration was essentially eliminated. Thus the Tip20 subunit, which binds directly to Sec20, did not itself significantly enhance SNARE assembly. To test more directly whether acceleration is dependent on the integrity of the Dsl1 complex, we disrupted the complex specifically by replacing Tip20 with Tip20ΔN (residues 82–701). Tip20ΔN, although it cannot bind the Dsl1 subunit, retains its ability to bind Sec20 (Tripathi et al., 2009). When the interaction of Dsl1 and Tip20 was disrupted, we found that SNARE assembly was no longer accelerated. Therefore, the rate enhancement is dependent on the intact Dsl1 complex. It remains possible that Sec39 alone also affects assembly kinetics. However, the insolubility of Use1 in the absence of either the trigger factor tag, which itself affects assembly, or Sec39 prevented us from addressing this possibility conclusively. Finally, we have not ruled out the possibility that interaction with the t-SNAREs
Discussion
X-ray crystal structures have been used to assemble a model of an intact multisubunit tethering complex of the COG/Dsl1/exocyst/GARP family (Koumandou et al., 2007). The model reveals a tower 20 nm in height with a disordered lasso at its tip. This structure appears to be highly consistent with the inferred role of the Dsl1 complex in COPI vesicle capture (Figure 6). At its base, the Dsl1 complex is anchored to the ER membrane via a bivalent attachment to two different ER SNAREs, Use1 and Sec20. Consistent with our biochemical analyses, co-immunoprecipitation experiments demonstrate that near-stoichiometric quantities of Use1 and Sec20 are recovered in association with the Dsl1 complex (Kraynack et al., 2005). The lasso, on the other hand, contains Trp-containing sequence motifs capable of binding subunits of the coatomer (Andag et al., 2001; Andag and Schmitt, 2003; Zink et al., 2009). The position of these motifs at the distal tip of the complex, within a long (111 residues in S. cerevisiae) disordered segment capable of extending 10 nm or more, seems ideally suited to the purpose of capturing COPI vesicles residing within a wide sweep of three-dimensional space. It has recently been proposed that binding of the vesicle to the Dsl1 complex impacts vesicle uncoating (Zink et al., 2009). Our results imply that tethering, uncoating, SNARE assembly, and membrane fusion may be coordinated by the Dsl1 complex. Our structural model, moreover, provides a foundation for investigating this hypothesis further.
The Dsl1 tower is not monolithic; instead, electron microscopy of negatively stained samples, in conjunction with image classification, indicates that it contains a flexible hinge at the center of the Dsl1 subunit. The linkage between the Dsl1 and Tip20 subunits, mediated by an antiparallel association between N-terminal helices, is probably flexible as well (Tripathi et al., 2009). Thus, although in our structural model the SNARE binding regions of Sec39 and Tip20 are relatively close to one another, the flexible hinge(s) would allow spatial separation of these subunits. The flexibility and/or conformation of one or more of the hinges might be linked to the succession of events that represent a functional cycle. It is striking in this regard that the lasso lies in close proximity to the Dsl1 subunit’s central hinge, suggesting that engagement of the lasso by a COPI vesicle binding could influence this hinge and, in turn, control the relative positioning of Sec39 and Tip20 (Figure 6).
The Dsl1 complex, in addition to binding individual ER SNAREs via their N-terminal regulatory domains, also binds assembled SNARE complexes. In this way, it appears capable of remaining associated with ER SNAREs throughout their functional cycle, as depicted in Figure 6. Moreover, the Dsl1 complex accelerates SNARE complex assembly in vitro. This acceleration, though quantitatively modest, is specific inasmuch as it depends on the integrity of the MTC. One potential mechanism by which an MTC might regulate SNARE assembly is by altering the conformations of the individual SNAREs. This mechanism seems attractive based on previous studies of SNARE assembly, which indicated that regulatory domains in some SNAREs are capable of folding back upon, and protecting from assembly, the SNARE motifs (Munson et al., 2000; Tochio et al., 2001). Tethering factors that engaged these regulatory domains might thereby deprotect the SNARE motifs, accelerating assembly. The addition of Sec39 alone did, in fact, appear to accelerate SNARE assembly (Figure S5). In a related MTC, the opposite result was obtained: an individual exocyst subunit, Sec6, was found to bind the SNARE protein Sec9 and thereby to inhibit the assembly of exocytic SNAREs (Sivaram et al., 2005). Clearly, the regulation of SNARE conformation and assembly by individual MTC subunits remains an open question deserving further exploration.
A second, and perhaps the simplest, mechanism by which the Dsl1 complex might influence SNARE assembly is by serving as a scaffold, binding simultaneously to two of the four SNAREs needed to form a stable complex, and possibly helping to orient them for productive interaction (Figure 6). This mechanism demands that the Dsl1 complex be intact, as suggested by our results, since it uses different subunits to bind different SNAREs. Moreover, the flexibility inherent in the Dsl1 complex might be important, although a priori it is not obvious whether it would assist or impede assembly. It does seem clear, however, that additional factors that affected the conformation and/or flexibility of the Dsl1 complex could greatly enhance (or inhibit) its assembly activity.
In conclusion, we present here a structural model for an intact MTC of the COG/Dsl1/exocyst/GARP family. Two of its three subunits (Dsl1 and Tip20) are homologous to previously reported structures of individual COG and exocyst subunit fragments, while one (Sec39) displays a novel fold. We demonstrate that the integrity of the Dsl1 complex is important both for its function in vivo and for its ability to accelerate SNARE assembly in vitro. Our analysis also reveals what we believe to be the first direct evidence for structural flexibility within an MTC. This flexibility takes two forms. First, a mobile lasso for vesicle capture is presented at the top of the Dsl1 tower. Second, hinge motions within the body of the tower appear to allow for conformational changes during a functional cycle. It will be particularly interesting, going forward, to characterize these conformations, their modulation by other factors, and their specific roles in orchestrating vesicle capture and fusion.
Experimental Procedures
Protein Production
Full-length S. cerevisiae Tip20 (residues 1–701), Dsl1 (residues 1–754), and Sec39 (residues 1–709), and fragments Dsl1C (residues 340–754) and Sec20 (residues 1–275, representing the cytoplasmic region) were purified as described (Tripathi et al., 2009). Sec39 and Dsl1Clactis (K. lactis Dsl1 residues 332–686) were combined for co-expression via pQLinkN (Addgene plasmid 13670) and pQLinkH (Addgene plasmid 13667), respectively (Scheich et al., 2007). Additional expression plasmids for Sec39 residues 1–392, Sec20 residues 1–174, Sec20 residues 198–275, Ufe1 residues 240–320, and Sec22 residues 125–188 were constructed using pProExHTb (Sec20 residues 198–275, Gibco) or pQLinkH (all others). The cytoplasmic domain of Use1 (residues 1–212), as well as an N-terminally truncated variant (residues 36–216), were overproduced with an N-terminal trigger factor tail using pCold TF DNA (Takara Bio). Amino acid substitutions were introduced using QuikChange (Stratagene) site-directed mutagenesis. All proteins were overproduced in Rosetta Escherichia coli cells (Novagen) and purified from cell lysates by Ni2+-affinity chromatography. Except in a few cases (Sec39 residues 1–392, Sec20 residues 1–174, and Sec20 residues 198–275), histidine tags were removed by rTEV digestion. Since removing the TF tag from Use1 caused it to aggregate, tag removal (via Factor Xa) was carried out in the presence of His-tagged (for experiments in Figure 4) or untagged (for experiments in Figure 5) Sec39. Proteins were then further purified by ion exchange (MonoS for Ufe1, MonoQ for all others; GE Healthcare) and size exclusion (S200; GE Healthcare) chromatography. Purified proteins were stored at −80°C in 10 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM Tris[2-carboxyethyl] phosphine (TCEP). The N-terminal peptide of Use1 (residues 1–35) was synthesized by AnaSpec.
Crystallization and Structure Determination
Crystals of selenomethionine-substituted Dsl1Clactis–Sec39 were obtained by vapor diffusion at 4°C using a 2:3 ratio of protein (4 mg ml−1) and well buffer (0.1 M HEPES, pH 7.5, 10% PEG monomethyl ether 5000, 0.1 M MgCl2, 5% dimethyl sulfoxide). Streak seeding was required to produce high quality crystals. Crystals were cryoprotected by brief sequential soaks in well buffer supplemented with 15% and 25% dimethyl sulfoxide. All data were collected at NSLS beamline X29 and processed using the HKL suite (Otwinowski and Minor, 1998).
The Dsl1Clactis–Sec39 structure was determined using multiwavelength anomalous diffraction (MAD) at 2.6 Å resolution. Initial phases were calculated using SHELX (Sheldrick, 2008) and subsequently improved using SHARP (Bricogne et al., 2003). Model building was done in COOT (Emsley and Cowtan, 2004). The model was initially refined with REFMAC (Murshudov, 1997) and subsequently with PHENIX (Adams et al., 2002). Five TLS groups were refined for the model, two for Dsl1 and three for Sec39, in conjunction with individual isotropic B-factor refinement. Two N-terminal helices of Sec39 prior to residue 30 were weakly visible in the electron density map but could not be unambiguously assigned to sequence and were therefore omitted from the model, as was the disordered loop 63–100. Refinement statistics are summarized in Table S1. Ramachandran statistics were calculated using Molprobity (Davis et al., 2007).
Structure-based sequence alignment was guided by DaliLite (Holm and Park, 2000). Molecular graphics were rendered using PyMOL (DeLano, 2002).
Electron Microscopy and Image Processing
Samples were prepared by conventional negative staining with 0.75% (w/v) uranyl formate as described previously (Ohi et al., 2004). Images were collected with a Tecnai T12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates at a nominal magnification of 67,000x and a defocus value of −1.5 μm using low dose procedures. Imaging plates were read out with a scanner (DITABIS Digital Biomedical Imaging System AG, Pforzheim, Germany) using a step size of 15 μm, a gain setting of 20,000, and a laser power setting of 30%; 2 × 2 pixels were averaged to yield a pixel size of 4.5 Å on the specimen level.
A total of 7,392 Sec39–Dsl1 and 9,325 Tip20 particles were interactively selected from the raw images using BOXER, the display program associated with the EMAN software package (Ludtke et al., 1999). Using the SPIDER software package (Frank et al., 1996), the Sec39–Dsl1 particles were windowed into 80 × 80 pixel images and the Tip20 particles into 64 × 64 pixel images respectively. These particles were rotationally and translationally aligned and subjected to 10 cycles of multi-reference alignment. Each round of multi-reference alignment was followed by K-means classification specifying 200 output classes. The references used for the first multi-reference alignment were randomly chosen from the particle images.
Protein Binding Experiments
For mapping the interacting regions of the Dsl1 subunits and SNAREs, binding reactions were prepared by mixing proteins at 6 or 10 μM final concentration in 10 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM TCEP. After incubating at 4°C for 30 min, samples were loaded onto a Superose 6 10/30 column (GE Healthcare). Fractions were analyzed using SDS-PAGE or Tricine-SDS-PAGE gels stained with Coomassie Blue.
For SNARE complex assembly in the absence or presence of the Dsl1 complex, equimolar quantities were mixed to yield 10–30 μM concentrations of each protein in 10 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM TCEP. After overnight incubation at room temperature, samples were loaded onto a Superose 6 10/30 column. Equal volumes from individual 0.4-ml fractions were analyzed as above.
Yeast Methods
Heterozygous diploid deletion strains DSL1/dsl1Δ, SEC39/sec39Δ, and USE1/use1Δ (Invitrogen) were transformed with CEN-URA plasmids (pRS416) containing wild-type copies of the corresponding proteins. Diploid cells were sporulated and dissected to obtain haploid deletion strains complemented by CEN-URA plasmids. CEN-LEU plasmids (pRS415) containing either wild-type or mutant genes were introduced into the appropriate haploid strains. The transformants were selected against CEN-URA plasmids on 5-fluoroorotic acid (5-FOA) plates at 23°C.
Fluorescence Polarization Experiments
Sec22 (residues 125–188, S125C) containing a single Cys residue was labeled with the thiol-reactive 5-iodoacetamidofluorescein (5-IAF, Invitrogen). The labeling reaction proceeded to completion (>90%) as judged by mass spectrometry. Fluorescence anisotropy measurements were performed using a PTI (Photon Technology International) instrument equipped with Glan-Thompson polarizers for selection of vertically and horizontally polarized excitation and emission components. Labeled Sec22 was excited at 490 nm, and the emission was collected at 520 nm with a slit width of ~4 nm. Vertical and horizontal intensities were measured simultaneously.
All experiments were performed at room temperature in 10 mM Tris, pH 8.0, 150 mM NaCl, 0.5 mM TCEP. For experiments in Figure 5B, all components (10 μM final concentration) except fluorescein-labeled Sec22 were mixed. Protease inhibitor cocktail (Sigma-Aldrich) at a 1:250 (v/v) ratio was added as a precaution against protein degradation, after which the fluorescence intensity was recorded for background subtraction. Fluorescence anisotropy was measured at a series of time points after adding labeled Sec22 to a final concentration of 1 μM. The anisotropy value of fully-bound Sec22 was estimated using the corresponding pure complexes isolated by gel filtration. Because protein concentrations were uncertain for the purified complexes, and background fluorescence intensity was less than 0.5% of the total signal, background was not subtracted for purified complexes. No significant difference between emission spectra collected before and after SNARE complex assembly was observed. The percentage of SNARE complex formation was estimated as (rt-r0)/(rbound-r0), where rt, r0, and rbound represent anisotropy values at the indicated time points, at time zero, and of purified complexes, respectively. The reported values are the average of three independent experiments.
For the experiments depicted in Figure 5A, a 100 μl reaction mixture was set up. At indicated time points, samples were diluted to 500 μl, and measured as described above. The reported values are the average of two independent experiments.
Supplementary Material
Acknowledgments
We gratefully acknowledge the staff of the National Synchrotron Light Source X29 beamline for assistance with x-ray data collection; Zong Lin and Hays Rye for assistance and discussion regarding fluorescence anisotropy experiments; Hans Dieter Schmitt for sharing results prior to publication; and Sean Munro, Mary Munson, Anne Spang, and Daniel Ungar for critical comments on the manuscript. This work was supported by a grant to F.M.H. from the National Institutes of Health (GM071574). C.K.Y. acknowledges a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. T.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: Coordinates and structure factors for Dsl1Clactis-Sec39 (RCSB accession code 3K8P) are in the Protein Data Bank.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Andag U, Neumann T, Schmitt HD. The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J Biol Chem. 2001;276:39150–39160. doi: 10.1074/jbc.M105833200. [DOI] [PubMed] [Google Scholar]
- Andag U, Schmitt HD. Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J Biol Chem. 2003;278:51722–51734. doi: 10.1074/jbc.M308740200. [DOI] [PubMed] [Google Scholar]
- Aoki T, Kojima M, Tani K, Tagaya M. Sec22b-dependent assembly of endoplasmic reticulum Q-SNARE proteins. Biochem J. 2008;410:93–100. doi: 10.1042/BJ20071304. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Burri L, Varlamov O, Doege CA, Hofmann K, Beilharz T, Rothman JE, Sollner TH, Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc Natl Acad Sci USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Cavanaugh LF, Chen X, Richardson BC, Ungar D, Pelczer I, Rizo J, Hughson FM. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Dilcher M, Veith B, Chidambaram S, Hartmann E, Schmitt HD, Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. Long coiled-coil proteins and membrane traffic. Biochim Biophys Acta. 2003;1641:71–85. doi: 10.1016/s0167-4889(03)00088-0. [DOI] [PubMed] [Google Scholar]
- Hamburger ZA, Hamburger AE, West AP, Jr, Weis WI. Crystal structure of the S. cerevisiae exocyst component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Kamena F, Spang A. Tip20p prohibits back-fusion of COPII vesicles with the endoplasmic reticulum. Science. 2004;304:286–289. doi: 10.1126/science.1095049. [DOI] [PubMed] [Google Scholar]
- Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynack BA, Chan A, Rosenthal E, Essid M, Umansky B, Waters MG, Schmitt HD. Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol Biol Cell. 2005;16:3963–3677. doi: 10.1091/mbc.E05-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J Mol Biol. 2007;371:410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- Murshudov GN. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1998;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- Reilly BA, Kraynack BA, VanRheenen SM, Waters MG. Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit. Mol Biol Cell. 2001;12:3783–3796. doi: 10.1091/mbc.12.12.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Brunger AT. Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol Cell. 1999;4:85–95. doi: 10.1016/s1097-2765(00)80190-2. [DOI] [PubMed] [Google Scholar]
- Richardson BC, Smith RD, Ungar D, Nakamura A, Jeffrey PD, Lupashin VV, Hughson FM. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci USA. 2009;106:13329–13334. doi: 10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- Sivaram MV, Saporita JA, Furgason ML, Boettcher AJ, Munson M. Dimerization of the exocyst protein Sec6p and its interaction with the t-SNARE Sec9p. Biochemistry. 2005;44:6302–6311. doi: 10.1021/bi048008z. [DOI] [PubMed] [Google Scholar]
- Sweet DJ, Pelham HR. The TIP1 gene of Saccharomyces cerevisiae encodes an 80 kDa cytoplasmic protein that interacts with the cytoplasmic domain of Sec20p. EMBO J. 1993;12:2831–2840. doi: 10.1002/j.1460-2075.1993.tb05944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am J Physiol Cell Physiol. 2006;290:C11–26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- Tochio H, Tsui MM, Banfield DK, Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293:698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16:114–123. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Hughson FM. SNARE protein structure and function. Ann Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Reilly BA, Chamberlain SJ, Waters MG. Dsl1p, an essential protein required for membrane traffic at the endoplasmic reticulum/Golgi interface in yeast. Traffic. 2001;2:212–231. doi: 10.1034/j.1600-0854.2001.020307.x. [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- Zink S, Wenzel D, Wurm CA, Schmitt HD. A link between ER tethering and COP-I vesicle uncoating. Dev Cell. 2009;17:403–416. doi: 10.1016/j.devcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.