Abstract
The “β-catenin destruction complex” is central to canonical Wnt/β-catenin signaling. The scaffolding protein Axin and the tumor suppressor adenomatous polyposis coli protein (APC) are critical components of this complex, required for rapid β-catenin turnover. We determined the crystal structure of a complex between β-catenin and the β-catenin-binding domain of Axin (Axin-CBD). The Axin-CBD forms a helix that occupies the groove formed by the third and fourth armadillo repeats of β-catenin and thus precludes the simultaneous binding of other β-catenin partners in this region. Our biochemical studies demonstrate that, when phosphorylated, the 20-amino acid repeat region of APC competes with Axin for binding to β-catenin. We propose that a key function of APC in the β-catenin destruction complex is to remove phosphorylated β-catenin product from the active site.
Keywords: Wnt, β-catenin, Axin, adenomatous polyposis coli (APC), crystal structure
The canonical Wnt/β-catenin pathway plays a critical role in embryonic development, and its deregulation in adult tissues is associated with various human diseases (for review, see Moon and Kimelman 1998; Wodarz and Nusse 1998; Peifer and Polakis 2000; Moon et al. 2002; Giles et al. 2003). In this pathway, β-catenin mediates the transmission of a Wnt signal into the nucleus and the subsequent activation of target genes. In the absence of a Wnt signal, a cytoplasmic protein complex containing glycogen synthase kinase-3β (GSK-3β), the adenomatous polyposis coli protein (APC), and the scaffolding protein Axin, among others, catalyzes the phosphorylation of β-catenin. Because phosphorylation of β-catenin targets it for degradation by the proteasome, this complex has been termed the “β-catenin destruction complex.” When the pathway is active, binding of Wnt to its receptors leads to the inactivation of the destruction complex and a consequent accumulation of β-catenin. The β-catenin translocates to the nucleus, where it binds to DNA-binding proteins of the Tcf/LEF family. Together they turn on the transcription of Wnt-responsive genes. Although β-catenin levels may also be regulated by other Axin-independent pathways (Kang et al. 2002; Tolwinski et al. 2003), phosphorylation of β-catenin by the β-catenin destruction complex is the central regulatory step of the canonical Wnt/β-catenin signaling pathway.
In the β-catenin destruction complex, GSK-3β phosphorylates the critical residues in the N terminus of β-catenin, contingent upon priming phosphorylation by casein kinase I (CKI; for review, see Polakis 2002). By itself, GSK-3β does not efficiently phosphorylate β-catenin; thus Axin plays a critical role in bringing GSK-3β, CKIα, and β-catenin together to efficiently promote the phosphorylation reaction (Dajani et al. 2003; for review, see Polakis 2002). The importance of Axin in β-catenin destruction is underscored by the presence of mutations in the human AXIN1 gene in certain human cancers that are associated with increased β-catenin levels (Satoh et al. 2000; Webster et al. 2000; Dahmen et al. 2001).
Another essential component of the destruction complex is the tumor suppressor APC. Mutations of APC cause the elevation of cytoplasmic β-catenin levels and are found in ∼85% of colon cancers (for review, see Polakis 2000). The function of APC in the β-catenin destruction complex is connected with Axin, because the overexpression of Axin in APC-mutant cancer cells is sufficient to down-regulate β-catenin levels in these cells (Hart et al. 1998). APC contains repetitive β-catenin interaction motifs, including three 15-amino acid repeats (possibly four; cf. Spink et al. 2001) and seven 20-amino acid repeats. It has been shown that APC plays a role in the transportation of β-catenin from the nucleus to the cytoplasm, where β-catenin is phosphorylated and degraded (for review, see Bienz 2002). Although it has also been proposed that APC may attenuate β-catenin levels by recruiting β-catenin to the β-catenin destruction complex (for review, see Bienz 2002), it remains unclear how APC plays an essential role in the β-catenin destruction complex.
In addition to Axin, APC, GSK-3β, and CKI, many other proteins, such as protein phosphatase 2A (PP2A), have also been found to play a role in the β-catenin destruction complex. A central question now is how these proteins interact to form a molecular machine that efficiently phosphorylates and degrades β-catenin. Specifically, a catalytic machine must be efficient in both substrate recruitment and product release. How does the β-catenin destruction complex keep β-catenin in the complex long enough to be phosphorylated, yet release it quickly enough to maintain the efficiency of phosphorylation?
We have determined the crystal structure of a complex between the armadillo repeat region of β-catenin and the β-catenin-binding domain of Axin, which reveals the structural basis of the β-catenin/Axin interaction. This structure suggests that Axin and the 20-amino acid repeat region of APC compete for binding to β-catenin when they are both involved in the β-catenin destruction complex. Our biochemical studies clearly show that these regions do compete for binding, but only when the 20-amino acid region is phosphorylated. Based on our data, we suggest that APC is required for both the recruitment of β-catenin and the removal of the phosphorylated β-catenin from the active site, which explains the critical role of APC in β-catenin turnover.
Results
Overall structure of the β-catenin/Axin-CBD complex
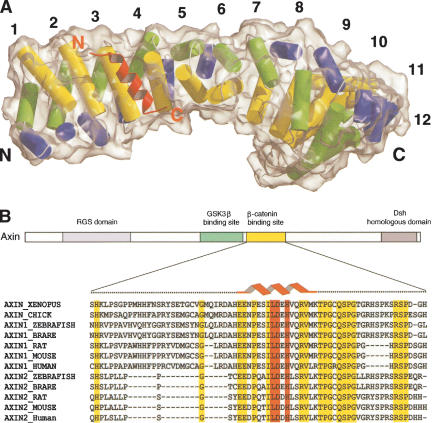
Numerous β-catenin and Axin protein fragments were produced and tested in combination to find a crystallizable β-catenin/Axin complex. We were able to crystallize the complete armadillo repeat region of human β-catenin (residues 133–665) in complex with the β-catenin-binding domain of Xenopus Axin (XAxin-CBD, residues 435–504 of Xenopus Axin). The structure was determined by molecular replacement and refined at 2.2 Å resolution (Fig. 1A). The β-catenin/XAxin-CBD crystal used in this study has one complex per asymmetric unit (Table 1). The final model contains residues 142–665 of β-catenin including the loop in armadillo repeat 10 (residues 550–562). Although we used the whole β-catenin-binding domain of XAxin, only the central region of the XAxin-CBD (residues 466–482) is visible in our electron density map (Fig. 2A). The sequence of the XAxin-CBD visible in the β-catenin/XAxin-CBD crystal structure is well conserved through a broad range of species, with complete conservation of residues required for the interaction (Fig. 1B).
Figure 1.
Overall structure of the β-catenin/XAxin-CBD complex. (A) β-catenin/XAxin-CBD complex structure with the β-catenin molecular surface outlined. Each armadillo repeat of β-catenin, except repeat 7, is composed of three helices that are shown as blue, green, and yellow cylinders, whereas the XAxin-CBD is shown as a red ribbon. The N and C termini are marked for each protein. (B) Primary structure and sequence alignment of Axin. Different functional units of Axin are designated as rectangles. The red helix cartoon represents the XAxin-CBD residues visible in the crystal structure. Residues not visible in the structure are marked in black dashes. Residues highlighted in red are required for the interaction with β-catenin in the β-catenin/XAxin-CBD structure, and other residues conserved among all vertebrate Axin sequences are highlighted in yellow.
Table 1.
Statistics of structure determination of β-catenin/XAxin-CBD complex
| Data collection | |
| Space group | C2 |
| Unit cell dimension | a = 85.141, b = 75.041, c = 101.398, β = 97.473 |
| No. of complex per asym. | 1 |
| Resolution range | 50—2.2 |
| aRmerge | 0.072 (0.529) |
| I/σ(I) | 17.8 (2.7) |
| Completeness (%) | 99.8 (99.9) |
| Multiplicity | 3.7 |
| No. of unique reflections | 37253 |
| Structure Refinement | |
| bRwork/Rfree (%) | 22.1/25.6 |
| Rmsd from ideal | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 1.1 |
| Dihedral angle (°) | 18.4 |
| cRamachandran plot (core, disallowed, %) | 91.3, 0.0 |
| Average B factor | 54.3 |
| No. of protein atoms in the final model | 4118 |
| No. of H2O molecules in the final model | 31 |
Values in parentheses refer to the outer-shell bin.
Rmerge = Σij|(Ii(j) — 〈I(j)|〉 /Σij II(j), where Ii(j) is the intensity of the i-th observation of reflection j. 〈I(j)〉 is the weighted mean of all measurements of j
R = Σj| Fobs(j)| — |Fcalc(j)| /Σj|Fobs(j)|. Rwork and Rfree were calculated with the working and test reflection sets, respectively
As defined in PROCHECK (Laskowski et al. 1993)
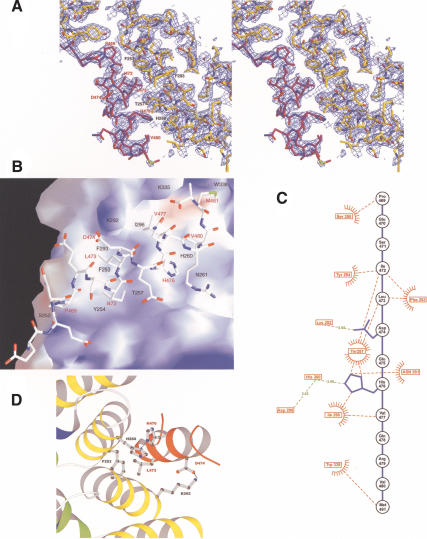
Figure 2.
Critical interactions on the β-catenin/Axin interface. In each case, β-catenin is in yellow and the XAxin-CBD is in red, except in panels B and C. β-Catenin and XAxin-CBD are labeled in black and red, respectively, except in panel C. (A) Stereoview of the 2Fo-Fc electron density map of the XAxin-CBD bound to β-catenin. The map is contoured at 1σ. (B) β-catenin electrostatic surface map of the XAxin-CBD-binding site. The surface of β-catenin is colored according to its relative electrostatic potential, with red representing negative charge and blue representing positive charge. The XAxin-CBD is shown as a stick model. (C) β-Catenin/XAxin-CBD bonding diagram. β-Catenin residues are labeled in red boxes with red text. The backbone and side chains of the XAxin-CBD are shown in blue, and XAxin-CBD residues are labeled in black circles with black text. Hydrogen bonds and charge–charge interactions are designated with green broken lines. A red star-burst together with a red broken line represents hydrophobic interactions. (D) Critical contacts in the β-catenin/XAxin-CBD interface. Residues making critical contacts are shown with side chains in ball-and-stick format.
In the crystal structure, the central region of the XAxin-CBD forms a continuous α-helix that fits into a shallow groove of β-catenin formed by the armadillo repeats (Fig. 1A). The helix of the XAxin-CBD specifically interacts with the third helices of β-catenin armadillo repeats 3 and 4, which are colored in yellow in Figure 1. The β-catenin molecule in the β-catenin/XAxin-CBD complex assumes a superhelical structure similar to the structure of free β-catenin reported previously (Huber et al. 1997), demonstrating that the binding of Axin does not significantly alter the conformation of β-catenin. The rmsd between the Cα positions of the unbound and bound β-catenin is 1.08 Å.
Recognition between β-catenin and Axin-CBD
The β-catenin/Axin interface is rather hydrophobic, with ∼1297 Å2 surface area buried in the complex. Five hydrophobic residues from the XAxin-CBD—Ile 472, Leu 473, Val 477, Val 480, and Met 481—reside on the helix surface complementary to the shallow β-catenin groove. Leu 473 of the Axin-CBD packs against Phe 253 and sits in a shallow hydrophobic pocket formed by Phe 253, Phe 293, and Tyr 254 of β-catenin. Ile 472 of the XAxin-CBD is also part of the hydrophobic core. His 476 and Val 477, in the center of the XAxin-CBD helix, interact with Thr 257 and Ile 296 of β-catenin, whereas Pro 469 and Met 481 at the XAxin-CBD helical termini contact Ser 250 and Trp 338 of β-catenin, respectively (Fig. 2B,C).
In addition to hydrophobic interactions, salt bridges and hydrogen bonds are also critical for the β-catenin/Axin interaction. In particular, the side chain carboxyl group of Asp 474 of the XAxin-CBD is coordinated with the amino group of β-catenin Lys 292 in armadillo repeat 4 through charge–charge interactions, and the side chain of His 476 of the XAxin-CBD forms a hydrogen bond with the side chain of His 260 of β-catenin armadillo repeat 3 that is in turn stabilized by β-catenin Asp 299 (Fig. 2C,D).
Critical contacts between Axin and β-catenin
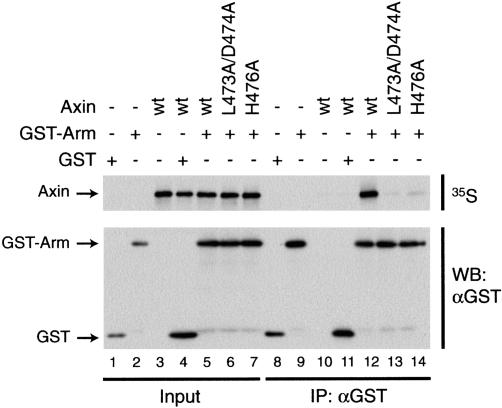
Our crystal structure suggested that XAxin residues Leu 473, Asp 474, and His 476 are important contact points between XAxin and β-catenin (Fig. 2D). To test whether these residues are required for the interaction between the two molecules, we mutated them to alanines and tested the mutant proteins' ability to interact with β-catenin. A double-point mutant form of XAxin (XAxin L473A/D474A) and a single-point mutant form (XAxin H476A) were produced in vitro as 35S-labeled proteins, and incubated with the armadillo repeat region of β-catenin tagged with GST (GST-Arm). When GST-Arm was pulled down with glutathione resin, wild-type XAxin (Fig. 3, lane 12), but neither XAxin L473A/D474A nor XAxin H476A (Fig. 3, lanes 13,14), was able to coprecipitate with GST-Arm. The ability of wild-type XAxin to coprecipitate with GST-Arm is due to the presence of the armadillo repeat region, as GST alone does not coprecipitate XAxin (Fig. 3, lane 11). These results demonstrate that Leu 473, Asp 474, and His 476 are required for the interaction between XAxin and β-catenin.
Figure 3.
L473/D474 and H476 are required for the interaction between Axin and β-catenin. The armadillo repeat region of β-catenin tagged with GST (GST-Arm) was tested for its ability to coprecipitate wild-type XAxin (wt) or XAxin containing point mutations in residues predicted to be important for interacting with β-catenin (L473A/D474A and H476A). (Lanes 1–7) Levels of input proteins prior to pull-down. GST-Arm specifically coprecipitates XAxin (lane 12), but XAxin with a double-point mutation in L473 and D474 (lane 13) or with a single-point mutation in H476 (lane 14) shows greatly decreased interaction with GST-Arm.
Competition between XAxin and APC for β-catenin binding
It is striking that the Axin-CBD occupies the whole binding groove formed by armadillo repeats 3 and 4 of β-catenin with a helical structure, which precludes the simultaneous binding of any other β-catenin partner in this region of the groove. It was proposed that the binding of phosphorylated APC to β-catenin might involve this region of β-catenin (see Discussion; Spink et al. 2001). This raised the possibility that the binding of Axin and APC might be mutually exclusive. To test this possibility, we produced 35S-labeled XAxin in vitro and incubated it with GST-Arm in the presence of competitor peptides derived from APC, one containing two of the 15-amino acid repeat regions (APC-B,C) and another containing two of the 20-amino acid repeat regions (APC-2,3; Fig. 4A). We tested whether phosphorylation of the APC 20-amino acid repeats might enhance their competition with XAxin. As kinases, we used both GSK-3β and CKI, because both have been implicated in the Wnt pathway (for review, see Polakis 2002). Addition of XAxin, which binds both CKI and GSK-3β, to APC causes it to be phosphorylated in Xenopus extracts and SW480 cells (Salic et al. 2000; Rubinfeld et al. 2001), and GSK-3β was previously shown to enhance the binding of β-catenin to APC (Rubinfeld et al. 1996). CKI has been shown to phosphorylate APC, and mutation of potential CKI phosphorylation sites in the 20-amino acid repeats inhibits the function of APC (Rubinfeld et al. 2001). We incubated APC-B,C and APC-2,3 with GSK-3β and CKI and noted a dramatic shift in the mobility of APC-2,3, but not APC-B,C on an SDS gel, indicating multiple phosphorylation sites on APC-2,3 (data not shown). Treatment with CKI alone produced an identical set of shifts (Fig. 4B).
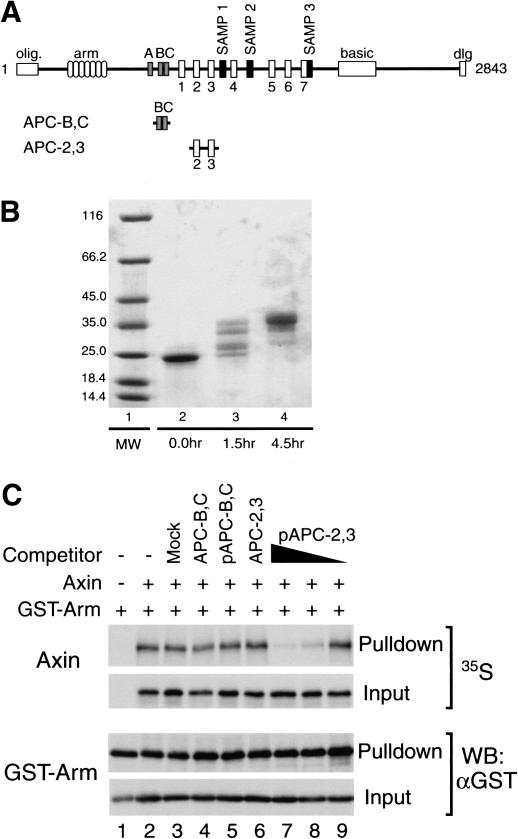
Figure 4.
The 20-amino acid repeat region of APC competes with Axin for binding to β-catenin when phosphorylated. (A) Domain structure of APC. A schematic representation of the APC primary structure shows, from N to C terminus, the oligomerization domain (olig.), armadillo repeats (arm), 15-amino acid β-catenin-binding repeats (A–C), 20-amino acid β-catenin-binding repeats (1–7), Axin binding repeats (SAMP1-3), basic region (basic), and discs large interaction domain (dlg). The human APC (hAPC) peptide fragments used in the competition assay are depicted below. APC-B,C includes the second and third 15-amino acid repeats (amino acids 1133–1189). APC-2,3 includes the second and third 20-amino acid repeats (amino acids 1362–1540). (B) Phosphorylation of APC by CKI. SDS-PAGE was used to monitor the process of the phosphorylation reaction. (Lane 1) Molecular weight marker labeled in kilodaltons. APC-2,3 (unphosphorylated, lane 2) displays multiple band shifts after 1.5-h reaction (lane 3), and a further band shift after another 3-h reaction (lane 4). (C) Competition assay. GST-Arm was tested for its ability to coprecipitate Axin in the presence of various competitors. The 20-amino acid repeat fragment APC-2,3 once phosphorylated (lanes 7–9, 0.3 μg pAPC-2,3, 10- and 100-fold dilutions) can specifically and dose-dependently block coprecipitation of Axin by GST-Arm. The unphosphorylated APC-2,3 (lane 6, 0.3 μg APC-2,3), the 15-amino acid repeat fragment APC-B,C whether unphosphorylated (lane 4, 0.3 μg APC-B,C) or kinase-treated (lane 5, 0.3 μg pAPC-B,C), and a mock phosphorylation reaction containing no peptide (lane 3), all do not block Axin binding to β-catenin.
Figure 4C shows that the phosphorylated 20-amino acid repeat peptide (pAPC-2,3) specifically and dose-dependently competes with XAxin for binding to β-catenin (Fig. 4C, lanes 7–9), whereas high levels of the unphosphorylated 20-amino acid repeat peptide (APC-2,3; Fig. 4C, lane 6), the unphosphorylated 15-amino acid repeat peptide (APC-B,C; Fig. 4C, lane 4), and the kinase-treated 15-amino acid peptide (pAPC-B,C; Fig. 4C, lane 5) do not compete for binding to β-catenin. These results demonstrate that when phosphorylated by CKI, the 20-amino acid repeat region of APC is capable of competing with XAxin for binding to β-catenin. Identical results were obtained with pAPC-2,3 phosphorylated with both CKI and GSK-3β (data not shown).
Phosphorylation of β-catenin does not affect its binding to XAxin
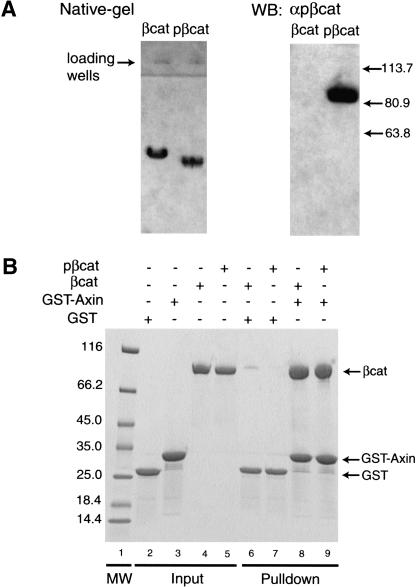
We wondered whether the phosphorylation of the N-terminal domain of β-catenin by GSK-3β could prevent XAxin from binding to β-catenin, which would provide a mechanism coupling the phosphorylation of β-catenin to its release from the destruction complex. Therefore we compared the XAxin binding affinity of unphosphorylated and phosphorylated full-length β-catenin. Full-length β-catenin was purified and extensively phosphorylated by CKI and GSK-3β and then repurified by anion exchange chromatography. The phosphorylated β-catenin demonstrated a peak shift in the anion exchange chromatography and a band shift on a native PHAST gel compared with unphosphorylated β-catenin (Fig. 5A). A Western blot with a phospho-β-catenin (pS33/pS37/pT41)-specific antibody confirmed that the β-catenin sample was phosphorylated at S33/S37/T41 and is thus likely to be fully phosphorylated (Fig. 5A). As shown in Figure 5B, the GST-tagged β-catenin-binding domain of XAxin (GST-Axin) pulled down similar amounts of unphosphorylated and phosphorylated β-catenin. Thus the phosphorylation of β-catenin did not alter its binding to XAxin. These results suggest that N-terminal phosphorylation of β-catenin is not responsible for its release from the destruction complex.
Figure 5.
The phosphorylation of β-catenin by CKI and GSK-3β does not affect its binding to Axin. (A) Native electrophoresis and Western blot of full-length β-catenin (βcat; 85kD) and full-length β-catenin treated with CKI and GSK-3β (pβcat). (Left) Coomassie blue-stained native gel. (Right) Western blot probed with a phospho-β-catenin (Ser 33/37/Thr 41) antibody, with relative molecular weight marked in kilodaltons on its right. (B) GST-pull-down binding assay. βcat and pβcat were compared in their ability to bind GST-tagged XAxin-CBD (GST-Axin). GST was used as a control. (Lanes 2–5) Input proteins. (Lanes 8,9) GST-Axin pulls down similar amounts of βcat and pβcat. (Lane 1) A molecular weight marker is labeled in kilodaltons on the left.
Discussion
Structural basis of β-catenin/Axin interactions
Formation of the β-catenin/Axin complex is essential for β-catenin turnover and is thus a critical step in canonical Wnt/β-catenin signaling (for review, see Polakis 2002). Previous studies using truncation mutants have shown that a short conserved region of Axin (Axin-CBD), which is C-terminal to the GSK-3β-binding site, is necessary and sufficient for specific β-catenin/Axin interaction (Fig. 1B; Behrens et al. 1998; Hart et al. 1998; Ikeda et al. 1998; Itoh et al. 1998; Kishida et al. 1998; Nakamura et al. 1998; Sakanaka et al. 1998; Yamamoto et al. 1998). In our crystal structure, only a short alpha helical portion of the XAxin-CBD is visible in the electron density map. As SDS electrophoresis of the dissolved crystal confirms that the flanking amino acids of the XAxin-CBD are still present in our crystals, these residues are likely to be flexible in the complex. Potentially, these regions are also flexible in full-length Axin in order to allow β-catenin to interact with other proteins bound to Axin such as GSK-3β (see below).
Importantly, the helical region of the Axin-CBD sequence is the most conserved region in the β-catenin-binding domain of Axin. Specifically, three residues in the XAxin-CBD that make critical contacts with β-catenin, that is, Leu 473, Asp 474, and His 476, are conserved among all vertebrate Axins (Axin1 and Axin2/Conductin; Fig. 1B). Together with the mutational analysis of the interaction between full-length XAxin and β-catenin, our results suggest that these conserved residues are critical in the Axin family for their recognition of β-catenin. Our crystal structure is completely consistent with previous biochemical studies. Site-directed mutagenesis studies have shown that Phe 253, His 260, and Lys 292 of β-catenin are critical for β-catenin/Axin (Conductin) interactions (Graham et al. 2000; von Kries et al. 2000). In our crystal structure, three critical Axin residues, Leu 473, Asp 474, and His 476, interact specifically with these three β-catenin residues, respectively (Fig. 2D).
The Axin-CBD helix runs along the armadillo repeats of β-catenin in a parallel fashion; that is, the N- to C-terminal axis of the Axin-CBD helix is roughly parallel to that of the superhelix formed by the armadillo repeats (Fig. 1A). One possible consequence of the parallel orientation between β-catenin armadillo repeats and the Axin-CBD is to place the N-terminal domain of β-catenin in close proximity to GSK-3β, as the GSK-3β-binding site of Axin is just N-terminal to the β-catenin-binding site (Fig. 1B). This arrangement might facilitate the phosphorylation of β-catenin's N terminus by GSK-3β. However, the significance of this orientation remains to be tested, because it is unclear whether GSK-3β and β-catenin have a relatively fixed spatial configuration in the β-catenin destruction complex. Axin nucleates the destruction complex as a scaffold for GSK-3β and β-catenin. The key Axin structural elements required for the GSK-3β/Axin and β-catenin/Axin interactions are two alpha helices separated by an ∼65-residue-long linker region that is conserved among Axin1, but not Axin2, proteins (Fig. 1B; Dajani et al. 2003). In crystal structures of the individual complexes, the linker is disordered. It will be interesting to test whether it becomes ordered when both GSK-3β and β-catenin are bound to Axin simultaneously.
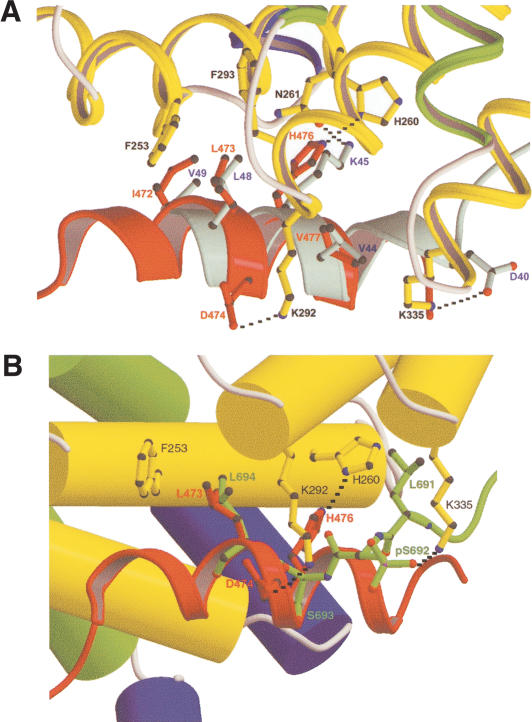
The XAxin-CBD binds to β-catenin in the same region as the C-terminal helix of the Tcf catenin-binding domain (Tcf-CBD) and region IV of the phosphorylated E-cadherin cytosolic domain (phospho-E-cadherin) through strikingly similar interactions (Fig. 6; Graham et al. 2000, 2001; Huber and Weis 2001), although the XAxin-CBD runs in the β-catenin groove with an orientation opposite to that of the Tcf-CBD and phospho-E-cadherin. The position of three side chains (I472, L473, and His 476) in the Axin-CBD helix plays a key role in determining this unusual orientation of the Axin-CBD helix (Fig. 6). It is also clear from a structural point of view that the binding of Tcf or phospho-E-cadherin competes with the binding of Axin to β-catenin. In agreement with the structural data, a previous study showed that Tcf3 can inhibit β-catenin turnover by preventing Axin from binding β-catenin (Lee et al. 2001).
Figure 6.
Structural comparison. (A) Binding mode comparison between the C-terminal helix of the hTcf4-CBD and the XAxin-CBD. The armadillo repeat regions from β-catenin/XAxin-CBD and β-catenin/hTcf4 complexes were superimposed. Note that these two helices bind to β-catenin in almost completely opposite directions. The hTcf4 helix is colored in cyan and labeled in blue. Important residues are shown in ball-and-stick format. The hydrogen bonding and charge–charge interactions are designated with black broken lines. (B) Structural comparison of the XAxin-CBD and phospho-E-cadherin bound to β-catenin. β-Catenin is shown in solid cylinders colored as in Figure 1A with the residues labeled in black. XAxin-CBD is shown as a red ribbon, and E-cadherin is shown as a green ball-and-stick figure superimposed onto the β-catenin/XAxin-CBD complex. The side chain of β-catenin residue Lys 335 has been modified to the conformation in the β-catenin/phospho-E-cadherin complex to show the charge–charge interaction with the phosphoserine.
Phosphorylated APC 20-amino acid repeats compete with the Axin-CBD for β-catenin binding
The tumor suppressor APC is required for β-catenin turnover, but its role in facilitating β-catenin phosphorylation remains unclear. The central region of APC contains three 15-amino acid repeats and seven 20-amino acid repeats that bind β-catenin. Each of these repeats may bind to one molecule of β-catenin (Tickenbrock et al. 2003). Three SAMP repeats, interspersed among the 20-amino acid repeats, mediate APC's interaction with Axin (Fig. 4A). It remains mysterious why there are two different types of β-catenin binding repeats in the APC sequence. The importance of these different repeats is supported by their presence in APCs from a wide range of species. One possibility is that the 15-amino acid repeats and 20-amino acid repeats play different roles in the regulation of β-catenin by APC.
The crystal structure of β-catenin in complex with an APC 15-amino acid repeat revealed that the APC 15-amino acid repeats bind in the long positively charged groove formed by armadillo repeats 5–8 of β-catenin (Spink et al. 2001), a site that is also used by the central region of the hTcf4-CBD, but not the Axin-CBD helix. Although the β-catenin-binding site for the 20-amino acid repeats has not been revealed by direct structural studies, the binding site has been mapped by mutational analysis in a yeast two-hybrid analysis and shown to overlap the region that binds the 15-amino acid repeats (von Kries et al. 2000). In support of this, the unphosphorylated 15-amino acid and 20-amino acid repeats compete with each other for binding to β-catenin (Spink et al. 2001).
Each of the 20-amino acid repeats and region IV of E-cadherin contain a conserved SxxxSxSxL sequence, which includes GSK-3β and CKI consensus phosphorylation sites (Rubinfeld et al. 2001). Phosphorylation of E-cadherin by CKII and GSK-3β dramatically increases its affinity for β-catenin (Spink et al. 2001). APC 20-amino acid repeats may also increase their affinity for β-catenin when phosphorylated, because conversion of one of the serines in the CKI phosphorylation site of rat APC 20-amino acid repeat 3 to aspartate, to mimic a phosphoserine, increased the affinity of this repeat for β-catenin 24-fold (Tickenbrock et al. 2003). Based on our previously published mutagenesis data (Graham et al. 2000) and homology between the APC 20-amino acid repeats and region IV of the E-cadherin cytoplasmic domain, it was suggested that phosphorylation of the 20-amino acid repeats might increase their affinity for β-catenin and cause them to change their binding site to armadillo repeats 3 and 4 (Spink et al. 2001). If this model is correct, our structure suggests that the phosphorylated 20-amino acid repeats of APC, but neither the unphosphorylated 20-amino acid repeats nor the 15-amino acid repeats, should compete with Axin for binding to β-catenin.
We tested this possibility and found that phosphorylation of the 20-amino acid repeats by GSK-3β and CKI, the two kinases found in the destruction complex, allowed APC to compete with Axin for binding to β-catenin. The unphosphorylated 20-amino acid repeats did not compete with Axin for β-catenin binding, consistent with the idea that phosphorylation of these repeats enhances their affinity and/or changes their binding sites on β-catenin. In contrast, the 15-amino acid repeats were not phosphorylated by either GSK-3β or CKI, nor did they prevent Axin from binding to β-catenin. These results support the hypothesis that the 15-amino acid and 20-amino acid repeats have different roles in the destruction complex.
Our results suggest that the two regions of APC contact two separate regions of β-catenin, one in the Axin-binding region (repeats 3 and 4, bound by the phosphorylated 20-amino acid repeats) and the other more C-terminal (repeats 5–8, bound by 15-amino acid repeats). Bipartite binding could be important for two reasons. First, the binding of the 15-amino acid repeats (or unphosphorylated 20-amino acid repeats) could help to position certain 20-amino acid repeats close to the Axin/GSK-3β active site, promoting the phosphorylation of these 20-amino acid repeats and thus allowing the 20-amino acid repeats to compete with Axin for β-catenin binding. Second, APC could remain tethered to β-catenin by the 15-amino acid repeats, whereas the binding of the 20-amino acid repeats to β-catenin could be dynamically regulated by phosphorylation. Thus, upon phosphorylation, one phosphorylated APC 20-amino acid repeat and one 15-amino acid repeat could bind simultaneously to different regions of the same β-catenin molecule with high affinity (armadillo repeats 3 and 4, and 5–8, respectively) and effectively compete with Axin for binding to β-catenin. But when this 20-amino acid repeat is dephosphorylated, only one 15-amino acid (or 20-amino acid) repeat may remain bound to the β-catenin molecule with a moderate affinity in the μM range (Spink et al. 2001; Tickenbrock et al. 2003).
A mechanistic model of the β-catenin destruction complex
A critical feature of the β-catenin destruction complex is its ability to rapidly phosphorylate β-catenin molecules as they are synthesized, targeting them for degradation. An essential aspect of efficient phosphorylation is the release of phosphorylated β-catenin from the complex. One way that phosphorylated β-catenin might be released from the destruction complex is if the phosphorylation of the N terminus of β-catenin by GSK-3β were to trigger the release of β-catenin from the β-catenin-binding domain of Axin, perhaps through a conformational change mediated by the phosphorylated β-catenin N terminus. However, Figure 6 shows that the Axin-CBD interacts with both unphosphorylated and CKI/GSK-3β-phosphorylated β-catenin with similar affinity. Evidently, a different mechanism for the release of reaction product (phospho-β-catenin) from the substrate binding site is required to explain how the destruction complex functions as an efficient enzyme, promoting rapid phosphorylation and turnover of β-catenin.
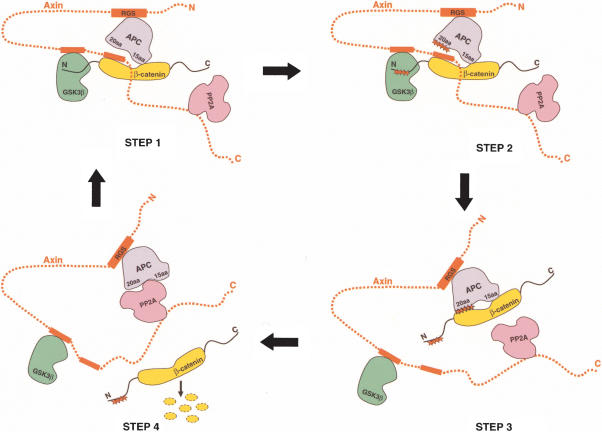
Based on our studies, we suggest that phosphorylation of the APC 20-amino acid repeats could be a key regulatory step in the release of phosphorylated β-catenin. Different β-catenin-binding sites for the 15-amino acid and 20-amino acid repeats of APC could explain how the β-catenin destruction complex catalyzes efficient phosphorylation of β-catenin molecules (Fig. 7). In step 1, Axin recruits unphosphorylated β-catenin to the destruction complex. Axin contains binding sites for all known key players of the destruction complex including GSK-3β, APC, and β-catenin, and can assemble them into a functional multiprotein complex. Axin binds β-catenin at repeats 3 and 4, as shown in our structure. The GSK-3β-binding site of Axin is N-terminal to the β-catenin-binding site, conveniently positioning GSK-3β near the N terminus of β-catenin, which can then be phosphorylated. APC is recruited to the complex through an interaction between its SAMP repeats and the RGS domain of Axin. Both the 15-amino acid and unphosphorylated 20-amino acid repeats of APC may play a role in recruiting free β-catenin to the β-catenin destruction complex at this stage by binding to armadillo repeats 5–8 of β-catenin (Spink et al. 2001). Step 2 is the phosphorylation step. Positioned inside the multiprotein complex, the N-terminal regulatory phosphorylation sites of β-catenin are phosphorylated by CKI and GSK-3β sequentially (for review, see Polakis 2002). We suggest that the 20-amino acid repeats of APC may also be phosphorylated by CKI and GSK-3β. In support of this, Salic et al. (2000) demonstrated that the binding of Axin to APC in Xenopus extracts caused a dramatic increase in the phosphorylation of APC. In step 3, the phosphorylated APC 20-amino acid repeats compete β-catenin away from the Axin-binding site, as seen in our in vitro assay, while the 15-amino acid repeats stay bound to β-catenin. This opens up Axin's β-catenin-binding site for the next β-catenin substrate molecule. In step 4, after the phosphorylated 20-amino acid repeats displace β-catenin from Axin, they could then move away from the GSK-3β active site, which allows APC to be dephosphorylated. The dephosphorylation of APC would reduce the binding affinity between APC and β-catenin, allowing β-catenin to be released to the ubiquitin ligase system, and switch the 20-amino acid repeats of APC back to their recruitment role (Fig. 7).
Figure 7.
A working model for the β-catenin destruction complex. The components in the β-catenin destruction complex are colored as follows: Axin (red); β-catenin (yellow); GSK-3β (green); APC (purple); PP2A (pink). Axin's binding regions for β-catenin, GSK-3β, and APC are shown as red rectangles. The 20-amino acid and 15-amino acid repeats of APC are labeled. Phosphorylation at the β-catenin N terminus or the APC 20-amino acid repeats is designated by red stars. Step 1 illustrates the recruitment of β-catenin with the help of APC. Step 2 shows phosphorylation of β-catenin by GSK-3β (after priming phosphorylation by CKI, which binds Axin but is not shown in the model). Step 3 illustrates the release of phosphorylated β-catenin product from Axin by the phosphorylated APC 20-amino acid repeats. Step 4 illustrates the dephosphorylation of the APC 20-amino acid repeats by PP2A and the release of phosphorylated β-catenin, which is subsequently degraded via the ubiquitin ligase–proteasome system. For simplicity, CKI and other regulatory proteins are not shown in the model.
A candidate for dephosphorylation of APC 20-amino acid repeats is serine/threonine protein phosphatase 2A (PP2A). PP2A, interacting with both Axin and APC directly, is an integral component of the β-catenin destruction complex, and it is clear that PP2A promotes the turnover of β-catenin (Hsu et al. 1999; Seeling et al. 1999; Ikeda et al. 2000; Ratcliffe et al. 2000; Li et al. 2001; Yamamoto et al. 2001). The PP2A-specific inhibitor okadaic acid inhibits β-catenin turnover (Seeling et al. 1999; Li et al. 2001). In addition, APC is required for PP2A function in β-catenin turnover (Seeling et al. 1999). PP2A can dephosphorylate APC in vitro and possibly in vivo (Seeling et al. 1999; Ikeda et al. 2000; Yamamoto et al. 2001). These observations cannot be explained by the recruitment model of APC, in which the dephosphorylation of APC by PP2A would be expected to reduce the rate of β-catenin turnover. In our model, PP2A aids β-catenin turnover by promoting the release of phosphorylated β-catenin product from the β-catenin destruction complex. A second important role of PP2A is to switch APC back to its β-catenin recruitment mode, because the phosphorylated APC 20-amino acid repeat precludes a new β-catenin molecule from binding to Axin.
In our model, APC plays key roles, both in recruitment of unphosphorylated β-catenin substrate and release of phosphorylated β-catenin product. Although the phosphorylation of APC may promote the nuclear export of β-catenin because of the enhanced binding (Rosin-Arbesfeld et al. 2000), in the β-catenin destruction complex, the dephosphorylation and phosphorylation of APC 20-amino acid repeats switches them between a β-catenin recruiting role and a phospho-β-catenin removal role. In addition, our model explains the necessity of PP2A for efficient turnover of β-catenin. Although APC is required for the release of phosphorylated β-catenin product from the active site, PP2A is needed for the release of phosphorylated β-catenin product from APC, and to prepare APC for the next reaction cycle.
Materials and methods
Expression, purification, and crystallization of the β-catenin/XAxin-CBD complex
Recombinant GST-tagged forms of the armadillo repeat region of β-catenin (residues 133–665) and the XAxin-CBD (residues 435–504) with a TEV cleavage site following the GST-tag were affinity-purified on glutathione resin (Amersham Pharmacia). The GST-tags were cleaved; the proteins were repurified on glutathione resin to remove the cleaved GST, and passed over a Q-sepharose HP column (Amersham Pharmacia). To complex the proteins, a 1:2 molar ratio of β-catenin and XAxin-CBD were incubated together at 4°C for 1 h, and excessive XAxin-CBD was removed by centrifuge filtration. Final complex concentration was 2 mg/mL in 20 mM Tris at pH 8.5, 0.15 M NaCl, and 2 mM dithiothreitol (DTT).
Initial crystals were obtained by the hanging drop vapor diffusion method at 20°C from the Hampton Screen I. Optimized growth conditions produced single crystals in 2–3 d with a typical size of 0.1 × 0.1 × 0.2 mm by mixing 1 μL of the protein complex stock solution and 1 μL of the reservoir solution, which consisted of 0.1 M sodium citrate at pH 5.6, 15% isopropanol (v/v), 7% PEG 4000–6000 (w/v), and 5 mM DTT. Crystals were frozen in a cryoprotection solution containing 15 mM Tris at pH 8.5, 80 mM NaCl, 80 mM sodium citrate at pH 5.6, 15% isopropanol, 22% PEG 6000, and 15% ethylene glycol.
Data collection, structure determination, and structural analysis
Data sets for the β-catenin/XAxin-CBD complex crystals were collected on the Advanced Light Source Synchrotron beamline 5.0.1 (λ = 1.0000 Å), and processed and scaled with the DENZO/SCALEPACK program packages (Otwinowski and Minor 1997). A 2.2 Å data set was used in the structure determination and the refinement (Table 1).
The structure was solved by molecular replacement in AmoRe (Navaza 1994). The best solution (correlation coefficient 61.0% and R-factor 40.4%) was obtained using the β-catenin structure in the β-catenin/E-cadherin complex (PDB entry 1I7X) as the search model. The model was refined with CNS programs (Brunger et al. 1998) using a maximum likelihood target. Model building was conducted in Xtalview (McRee 1999). A test set of 5.0% of the reflections and a 1σ cutoff of all the data between 50.0 and 2.2 Å were used throughout the refinement. The initial model of the XAxin-CBD was built in after the first round of rigid body refinement, positional refinement, B factor refinement, and simulated annealing. The overall complex model was refined and rebuilt until R-factors converged. The stereochemical quality of structural models was monitored using Procheck (Laskowski et al. 1993).
Structural superposition of β-catenin/XAxin-CBD, β-catenin/hTcf4-CBD (PDB entry: 1JDH) and β-catenin/phospho-E-cadherin (PDB entry: 1I7W) were performed based on the Cα's in the armadillo repeats 2–5 of β-catenin (residues 181–349) with Swisspdb Viewer (Guex and Peitsch 1997). After superposition, the rmsd was 0.68 Å between β-catenin/XAxin-CBD and β-catenin/phospho-E-cadherin structures, and 0.51 Å between β-catenin/XAxin-CBD and β-catenin/hTcf4-CBD structures. The following programs were used to generate the figures in this paper: GRASP (Nicholls et al. 1991), Molscript (Kraulis 1991), and RASTER3D (Merritt and Murphy 1994).
The coordinates of the β-catenin/Axin structure have been deposited in the Protein Data Bank with the ID code 1QZ7.
Expression, purification, and phosphorylation of APC fragments and full-length β-catenin
Human APC fragments were purified as described for β-catenin and the XAxin-CBD. APC-B,C contains residues 1133–1189, and APC-2,3 contains residues 1362–1540. Proteins were stored in 20 mM Tris at pH 8.5, 0.2 M NaCl, and 2 mM DTT.
APC-2,3 was either phosphorylated by CKI-δ (New England BioLabs) alone for 4.5 h, or sequentially with CKI-δ for 1.5 h and then CKI-δ plus GSK-3β (New England BioLabs) for another 3 h using the vendor's suggested reaction conditions. EDTA was added to quench the reaction. Phosphorylation was monitored by SDS-PAGE.
Full-length Xenopus β-catenin (residues 1–781) used for the Axin binding experiments was purified using a construct kindly provided by Drs. Ken-Ichi Takemaru and Randall Moon, as described for the armadillo repeat region above, except that a His-tagged construct was initially purified on a Ni-NTA column. Phosphorylation was with CKI-δ for 3 h followed by GSK-3β for 3 h. Following phosphorylation, kinases were removed by purification on a Q-column. Pure full-length β-catenin was stored in 20 mM Tris at pH 7.5, 0.25 M NaCl, and 2 mM DTT.
Native electrophoresis followed by Western blotting using a rabbit anti-phospho-β-catenin (pSer 33/pSer 37/pThr 41) antibody (New England BioLabs) and horseradish peroxidase (HRP) anti-rabbit secondary antibody (Zymed) was used to detect the phosphorylation of β-catenin.
In vitro binding assays
Wild-type XAxin for in vitro binding studies was produced using CS2+Xaxin-myc (Hedgepeth et al. 1999). Point mutants were introduced into XAxin-myc by the QuikChange procedure (Stratagene) using the primers GAGGAAAATCCGGAATC TATTgcGGcTGAACACGTGCAGCGTG for L473A/D474A (plasmid: XAxin41), and GGAATCTATTCTGGATGAAgcC GTGCAGCGTGTTATGAAAACCC for H476A (plasmid: XAxin55), where the lowercase letters show the altered bases. 35S-labeled Axin was produced using the TNT SP6 Quick Coupled Transcription/Translation system (Promega). Approximately 0.3 μg of recombinant GST-Arm was incubated with 8 μL of TNT-produced 35S-labeled Axin (either wild-type or mutant) at 4°C, with nutation, in a total volume of 24 μL. Incubation buffer was 20 mM Tris at pH 8.5, 150 mM NaCl, and 2 mM DTT. One-microliter or 4-μL samples were retained for analysis of input by autoradiography and Western blotting, respectively. GST-Arm was precipitated by incubation with glutathione resin (Amersham Pharmacia) for 1 h at 4°C with nutation. Unbound protein was removed by washing with 10 mM Tris at pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, and 1% deoxycholate. Proteins were analyzed by SDS-PAGE followed by autoradiography or Western blotting using goat anti-GST primary antibodies (Amersham Pharmacia) and HRP anti-goat secondary antibodies (Zymed). Competitions were performed essentially the same way, except that an additional incubation was performed in the presence of the competitor peptide with nutation at 4°C.
Unphosphorylated (βcat), phosphorylated full-length Xenopus β-catenin (pβcat), and XAxin-CBD used in the in vitro binding assay were prepared as described above. Approximately 20 μg βcat or 20 μg pβcat were incubated with 4 μg GST-tagged XAxin-CBD (or GST as a control) corresponding to a molar ratio of 2:1 (full-length β-catenin vs. GST-XAxin-CBD) for 30 min at 20°C in 20 mM Tris at pH 7.5, 0.15 M NaCl, 0.1% Triton X-100, and 2 mM DTT binding buffer. The protein complexes were isolated on glutathione resin after a 30-min incubation at 20°C and wash with the binding buffer, and analyzed by SDS-PAGE.
Acknowledgments
We thank the staff at ALS beamline 5.0.2 for assistance with the synchrotron data collection. We thank Drs. Ken-Ichi Takemaru and Randall Moon for a Xenopus β-catenin construct. We also thank Carole Weaver for her valuable comments on the manuscript. This work was supported by NIH grants CA90351 to W.X. and HD27262 to D.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1142603.
References
- Behrens J., Jerchow, B.A., Wurtele, M., Grimm, J., Asbrand, C., Wirtz, R., Kuhl, M., Wedlich, D., and Birchmeier, W. 1998. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science 280: 596-599. [DOI] [PubMed] [Google Scholar]
- Bienz M. 2002. The subcellular destinations of APC proteins. Nat. Rev. Mol. Cell Biol. 3: 328-338. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54: 905-921. [DOI] [PubMed] [Google Scholar]
- Dahmen R.P., Koch, A., Denkhaus, D., Tonn, J.C., Sorensen, N., Berthold, F., Behrens, J., Birchmeier, W., Wiestler, O.D., and Pietsch, T. 2001. Deletions of AXIN1, a component of the WNT/wingless pathway, in sporadic medulloblastomas. Cancer Res. 61: 7039-7043. [PubMed] [Google Scholar]
- Dajani R., Fraser, E., Roe, S.M., Yeo, M., Good, V.M., Thompson, V., Dale, T.C., and Pearl, L.H. 2003. Structural basis for recruitment of glycogen synthase kinase 3β to the axin-APC scaffold complex. EMBO J. 22: 494-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles R.H., van Es, J.H., and Clevers, H. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653: 1-24. [DOI] [PubMed] [Google Scholar]
- Graham T.A., Weaver, C., Mao, F., Kimelman, D., and Xu, W. 2000. Crystal structure of a β-catenin/Tcf complex. Cell 103: 885-896. [DOI] [PubMed] [Google Scholar]
- Graham T.A., Ferkey, D.M., Mao, F., Kimelman, D., and Xu, W. 2001. Tcf4 can specifically recognize β-catenin using alternative conformations. Nat. Struct. Biol. 8: 1048-1052. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714-2723. [DOI] [PubMed] [Google Scholar]
- Hart M.J., de los Santos, R., Albert, I.N., Rubinfeld, B., and Polakis, P. 1998. Downregulation of β-catenin by human Axin and its association with the APC tumor suppressor, β-catenin and GSK3β. Curr. Biol. 8: 573-581. [DOI] [PubMed] [Google Scholar]
- Hedgepeth C.M., Deardorff, M.A., and Klein, P.S. 1999. Xenopus axin interacts with glycogen synthase kinase-3 beta and is expressed in the anterior midbrain. Mech. Dev. 80: 147-151. [DOI] [PubMed] [Google Scholar]
- Hsu W., Zeng, L., and Costantini, F. 1999. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J. Biol. Chem. 274: 3439-3445. [DOI] [PubMed] [Google Scholar]
- Huber A.H. and Weis, W.I. 2001. The Structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105: 391-402. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Nelson, W.J., and Weis, W.I. 1997. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90: 871-882. [DOI] [PubMed] [Google Scholar]
- Ikeda S., Kishida, S., Yamamoto, H., Murai, H., Koyama, S., and Kikuchi, A. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 17: 1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kishida, M., Matsuura, Y., Usui, H., and Kikuchi, A. 2000. GSK-3β-dependent phosphorylation of adenomatous polyposis coli gene product can be modulated by β-catenin and protein phosphatase 2A complexed with Axin. Oncogene 19: 537-545. [DOI] [PubMed] [Google Scholar]
- Itoh K., Krupnik, V.E., and Sokol, S.Y. 1998. Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and β-catenin. Curr. Biol. 8: 591-594. [DOI] [PubMed] [Google Scholar]
- Kang D.E., Soriano, S., Xia, X., Eberhart, C.G., De Strooper, B., Zheng, H., and Koo, E.H. 2002. Presenilin couples the paired phosphorylation of β-catenin independent of axin: Implications for β-catenin activation in tumorigenesis. Cell 110: 751-762. [DOI] [PubMed] [Google Scholar]
- Kishida S., Yamamoto, H., Ikeda, S., Kishida, M., Sakamoto, I., Koyama, S., and Kikuchi, A. 1998. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of β-catenin. J. Biol. Chem. 273: 10823-10826. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr. 24: 946-950. [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283-291. [Google Scholar]
- Lee E., Salic, A., and Kirschner, M.W. 2001. Physiological regulation of β-catenin stability by Tcf3 and CK1 epsilon. J. Cell Biol. 154: 983-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yost, H.J., Virshup, D.M., and Seeling, J.M. 2001. Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in Xenopus. EMBO J. 20: 4122-4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156-165. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy, M.E.P. 1994. RASTER3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50: 869-873. [DOI] [PubMed] [Google Scholar]
- Moon R.T. and Kimelman, D. 1998. From cortical rotation to organizer gene expression: Toward a molecular explanation of axis specification in Xenopus. Bioessays 20: 536-545. [DOI] [PubMed] [Google Scholar]
- Moon R.T., Bowerman, B., Boutros, M., and Perrimon, N. 2002. The promise and perils of Wnt signaling through β-catenin. Science 296: 1644-1646. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Hamada, F., Ishidate, T., Anai, K., Kawahara, K., Toyoshima, K., and Akiyama, T. 1998. Axin, an inhibitor of the Wnt signalling pathway, interacts with β-catenin, GSK-3β and APC and reduces the β-catenin level. Genes Cells 3: 395-403. [DOI] [PubMed] [Google Scholar]
- Navaza J. 1994. AMoRe: An automated package for molecular replacement. Acta Crystallogr. A 50: 157-163. [Google Scholar]
- Nicholls A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Gen. 11: 282-296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307-326. [DOI] [PubMed] [Google Scholar]
- Peifer M. and Polakis, P. 2000. Wnt signaling in oncogenesis and embryogenesis—A look outside the nucleus. Science 287: 1606-1609. [DOI] [PubMed] [Google Scholar]
- Polakis P. 2000. Wnt signaling and cancer. Genes & Dev. 14: 1837-1851. [PubMed] [Google Scholar]
- ____. 2002. Casein kinase 1: A Wnt'er of disconnect. Curr. Biol. 12: 499-501. [DOI] [PubMed] [Google Scholar]
- Ratcliffe M.J., Itoh, K., and Sokol, S.Y. 2000. A positive role for the PP2A catalytic subunit in Wnt signal transduction. J. Biol. Chem. 275: 35680-35683. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Townsley, F., and Bienz, M. 2000. The APC tumour suppressor has a nuclear export function. Nature 406: 1009-1012. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S., and Polakis, P. 1996. Binding of GSK3β to the APC–β-catenin complex and regulation of complex assembly. Science 272: 1023-1026. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Tice, D.A., and Polakis, P. 2001. Axin-dependent phosphorylation of the adenomatous polyposis coli protein mediated by casein kinase 1ε. J. Biol. Chem. 276: 39037-39045. [DOI] [PubMed] [Google Scholar]
- Sakanaka C., Weiss, J.B., and Williams, L.T. 1998. Bridging of β-catenin and glycogen synthase kinase-3β by axin and inhibition of β-catenin-mediated transcription. Proc. Natl. Acad. Sci. 95: 3020-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Lee, E., Mayer, L., and Kirschner, M.W. 2000. Control of β-catenin stability: Reconstitution of the cytoplasmic steps of the wnt pathway in Xenopus egg extracts. Mol. Cell. 5: 523-532. [DOI] [PubMed] [Google Scholar]
- Satoh S., Daigo, Y., Furukawa, Y., Kato, T., Miwa, N., Nishiwaki, T., Kawasoe, T., Ishiguro, H., Fujita, M., Tokino, T., et al. 2000. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 24: 245-250. [DOI] [PubMed] [Google Scholar]
- Seeling J.M., Miller, J.R., Gil, R., Moon, R.T., White, R., and Virshup, D.M. 1999. Regulation of β-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 283: 2089-2091. [DOI] [PubMed] [Google Scholar]
- Spink E.K., Fridman, S.G., and Weis, W.I. 2001. Molecular mechanisms of β-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC–β-catenin complex. EMBO J. 20: 6203-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickenbrock L., Kossmeier, K., Rehmann, H., Herrmann, C., and Muller, O. 2003. Differences between the interaction of β-catenin with nonphosphorylated and single-mimicked phosphorylated 20-amino acid residue repeats of the APC protein. J. Mol. Biol. 327: 359-367. [DOI] [PubMed] [Google Scholar]
- Tolwinski N.S., Wehrli, M., Rives, A., Erdeniz, N., DiNardo, S., and Wieschaus, E. 2003. Wg/Wnt signal can be transmitted through arrow/LRP5,6 and Axin independently of Zw3/Gsk3β activity. Dev. Cell 4: 407-418. [DOI] [PubMed] [Google Scholar]
- von Kries J.P., Winbeck, G., Asbrand, C., Schwarz-Romond, T., Sochnikova, N., Dell'Oro, A., Behrens, J., and Birchmeier, W. 2000. Hot spots in β-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol. 7: 800-807. [DOI] [PubMed] [Google Scholar]
- Webster M.T., Rozycka, M., Sara, E., Davis, E., Smalley, M., Young, N., Dale, T.C., and Wooster, R. 2000. Sequence variants of the axin gene in breast, colon, and other cancers: An analysis of mutations that interfere with GSK3 binding. Genes Chromosomes Cancer 28: 443-453. [PubMed] [Google Scholar]
- Wodarz A. and Nusse, R. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14: 59-88. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Kishida, S., Uochi, T., Ikeda, S., Koyama, S., Asashima, M., and Kikuchi, A. 1998. Axil, a member of the Axin family, interacts with both glycogen synthase kinase 3β and β-catenin and inhibits axis formation of Xenopus embryos. Mol. Cell. Biol. 18: 2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Hinoi, T., Michiue, T., Fukui, A., Usui, H., Janssens, V., Van Hoof, C., Goris, J., Asashima, M., and Kikuchi, A. 2001. Inhibition of the Wnt signaling pathway by the PR61 subunit of protein phosphatase 2A. J. Biol. Chem. 276: 26875-26882. [DOI] [PubMed] [Google Scholar]