Summary
A major goal in cell biology is to understand the functional organization of macromolecular complexes in vivo. Electron microscopy is helping cell biologists to achieve this goal, thanks to its ability to resolve structural details in the nanometer range. While issues related to specimen preparation, imaging, and image interpretation make this approach to cell architecture difficult, recent improvements in methods, equipment, and software have facilitated the study of both important macromolecular complexes and comparatively large volumes from cellular specimens. Here, we describe recent progress in electron microscopy of cells and the ways in which the relevant methodologies are helping to elucidate cell architecture.
Keywords: cryo-electron tomography, rapid freezing, freeze-substitution, 3-D reconstruction, resolution
Introduction
Light microscopy (LM) of living samples has made tremendous contributions to our understanding of macromolecular organization in cells, thanks to improvements in specific stains, optics, and cameras that can now record low levels of light with a good signal-to-noise ratio (SNR). Nonetheless, the spatial resolution of these methods imposes limitations on the data available, and the unavoidable motions of small components in living materials impose more, making alternative imaging methods necessary. Electron microscopy (EM) provides almost three orders of magnitude better spatial resolution than LM, permitting another flowering of observation, data, and discovery [1]. However, EM too has limitations, like the need for samples to withstand high vacuum and damage from the electron beam. Fortunately, technologies have recently emerged or been popularized that allow EM to make significant contributions to our understanding of cellular organization. Rapid freezing immobilizes cells and molecular structures in amorphous ice, a “vitrified” state that preserves native biological structure, allowing a high resolution “snapshot” of physiological conditions. Cryo-electron microscopy (cryo-EM) has long been popular for 3-D structural studies of isolated macromolecules [2], but modern electron guns with improved coherence and cameras with improved detective quantum efficiency have helped to improve image SNR and make cryo-imaging of cells more practical.
It is difficult, however, to freeze samples so quickly that only vitreous ice is formed. The crystallization of water damages cellular structure, and to avoid this one must either drop the cell’s temperature at ~104 °C/sec or inhibit ice crystal formation with solutes or a physical effect, like high pressure. Moreover, the frozen sample, like any other EM specimen, must be thin enough to minimize multiple scattering of beam electrons and to make inelastic scattering rare. This means that for view by EM, most cells must be sliced into thin “sections”, be they frozen or not. Frozen-hydrated samples are particularly sensitive to the electron beam, so they must be imaged with sufficiently few electrons that biological structure is preserved at a resolution suitable for the questions being addressed. Moreover, since cryo-EM images are low in electron contrast, they usually display low SNR; visible image detail is therefore limited not by the microscope but by the available data about the electron scattering that occurs at each volume element (voxel) in the specimen. Averaging of multiple images of equivalent structures is necessary to improve the SNR and allow the intrinsic resolution of EM to be realized in visible detail.
An alternative way to handle frozen samples is to maintain low temperatures while the vitrified cellular water is dissolved by an organic solvent containing solutes that will cross-link macromolecules (freeze-substitution fixation), making them hardier to subsequent treatments, like embedding in plastic. These samples are comparatively easy to cut into thin sections, but they commonly require stain to improve their contrast. This increases image SNR, but one is now imaging stain, not the macromolecules themselves, and there is concern about the congruence of the sample with a living cell. Nonetheless, these methods have been evaluated by comparison with images from LM, freeze-fracture EM, and x-ray scattering, documenting their reliability to a resolution of ~5 nm [3]. They are therefore now in widespread use.
An additional problem is that biological structures are intrinsically three-dimensional (3D). Electron images, like all lens-generated images, are projections into two dimensions, which superimposes densities along the direction of projection, discarding valuable information. If, however, samples are imaged from multiple angles, these projections can be combined by any of several algorithms to generate accurate 3D images, a process called tomography. This approach can provide information about both cellular and macromolecular structures, so it has become the imaging method of choice for many biological specimens.
The combination of these technologies defines modern EM. Below we provide examples to demonstrate the value of both cryo-EM of frozen-hydrated cells and cell parts and of samples prepared by rapid freezing, then freeze-substitution fixation (RF/FSF). We conclude with our opinions about the ways in which these approaches to cell structure can most profitably be merged into a suite of technologies that will connect the living cell, as seen by LM, with the organization of its component macromolecules.
A. Cryo-Electron Tomography
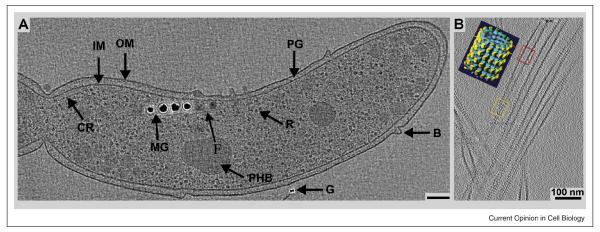
The excellent preservation of cellular structures by rapid freezing and cryo-electron tomography (cryo-ET) is illustrated in Figure 1. This bacterial cells was thin enough to be imaged in toto; multiple tilted views have allowed a 3D reconstruction, which is sampled here as a single, 10 nm slice at the midplane of the cell. The visible detail is impressive, since tomograms allow one to extract thin slices from the reconstructed volume, removing the material above and below the selected slice; details of cell structure can thus be seen directly. The result is analogous to confocal light microscopy, which can display narrow planes of image data from a cell’s fluorescence without the out-of-focus fluorescence that plagues conventional, full-field imaging. In this tomogram one can see individual ribosomes, both cell membranes, together with blebs on the cell surface, and a slender cytoskeletal fiber that connects adjacent “magnetosome”, the small particles of iron whose ordered arrangement makes a detector that allows these organisms to navigate relative to the earth’s magnetic fields.
Fig. 1.
Slices through a cryo-electron tomography volumes. A: Magnetospirillum magneticum sp AMB-1: adapted from [7•] with permission. General features of AMB-1 cells are highlighted in this 12-nm thick slice from a cryo-ET reconstruction: outer membrane (OM), inner membrane (IM), peptidoglycan layer (PG), ribosomes (R), outer membrane bleb (B), chemoreceptor bundle (CR), poly-b-hydroxybutyrate granule (PHB), gold fiduciary marker (G), intracellular fiber (F) and the magnetosome chain (MG). (B: Tomographically reconstructed microtubule-Eg5 kinesin complexes compared to a helical reconstruction (inset: Eg5 motor domain shown as yellow densities). Red and orange frames mark the location of the tomographic slice with respect to the total microtubule volume. Red cuts at the lumen of the tube, and orange cuts along the outer surface). Both tomograms demonstrate molecular preservations to ~3nm resolution. AMB-1 images reveal that magnetosomes are invaginations of the inner cell membrane. Image B: Julia Cope, Hoenger lab, CU-Boulder). Scale bars = 100 nm.
Some regions of even eukaryotic cells are thin enough for this kind of direct imaging and 3-D reconstruction (ca. 300 - 500 nm). Filopodia [4•] and the edges of naturally flat cells, like fibroblasts [5] and Dictyostelium, have been imaged as whole-mounts after growth directly on EM-grids. In these cases cryo-ET has been particularly powerful for elucidating the complex inner structure of cytoplasm; it reveals the organization of cytoplasmic membranes and cytoskeletal fiber, both in 3D and with unprecedented detail. This possibility has opened a new approach for structural investigations into small eukariotic cells (Ostreococcus tauri) [6] and bacteria (e.g. [7• -11]). Typically, these cryo-images or tomograms allow direct visualization of detail at 6-8 nm on cellular specimens (Fig. 1a) and approximately to 3 nm on very thin specimens (Fig. 1B); this resolution defines small structural features, like individual kinesin motor domains, without the use of averaging procedures. The structures preserved by cryo-fixation include even finer details, but seeing them requires averaging to improve image SNR. This may be achieved either by conventional methods based on 2-D projections, or now by averaging 3-D volumes picked from tomograms [12••] (Fig. 2).
Fig. 2.
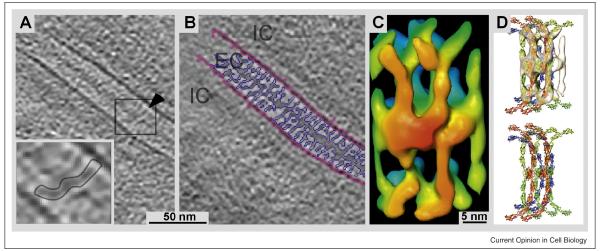
Cryo-ET reconstruction of cadherin molecules bridging the extracellular space in a desmosome. This tomogram was obtained from a cryo-section cut from a fully vitrified specimen. A: Due to the absence of chemical fixation and/or any substitution processes cryo-ET reveals molecular details at high resolution. The inset outlines the density of a spanning molecular bridge. B: Image as in A with the membranes marked in pink and the connecting cadherins-based density in purple. C: 3-D volume obtained from an average of subtomograms. D: Molecular model with the X-ray structures of C-cadherin fitted into the volume shown in C. Figure adapted from [12••] (with permission).
Cryo-ET has also contributed to our understanding of specimens like microtubules and actin complexes, particularly in situations where single-particle averaging or symmetry-based methods, like helical reconstruction, would have failed, Examples include the randomly distributed densities observed inside microtubules [13,14] and reconstruction of the seam in a microtubule lattice (Hoenger group, unpublished), the Actin-Arp2/3 complex [15], intermediate filaments [16], Clathrin-coated pits [17•], and nuclear pore complexes [18]. Fig. 1B shows 3-D reconstructions of several microtubules, most of which exhibit a lattice seam; they are therefore unlike the volume shown in the inset that came from a truly helical, 15-protofilament microtubule, to which helical methods could be applied.
Similar imaging of thicker samples requires that they be sectioned before they are imaged. While microtomy of frozen-hydrated samples was initiated in the 1970s [19] and improved in the eighties [20], the technology did not become popular until recently. The Manella team [21] and the Dubochet lab [22] have collected beautiful data from vitrified, unstained/unfixed thin-sections. Ways to make this demanding technique easier are now emerging [23-26], and the potential of vitrified sectioning for molecular analysis is shown in the molecular arrangement of cadherins in desmosomes produced by a combination of vitrified sectioning and averaging of subvolumes extracted from the initial tomogram [12] (Figure 2). Essentially the same image enhancement strategy has been used to study the structure of plunge-frozen axonemes [27••]; detailed descriptions of the power of tomographic subvolume averaging are given in [28] and [29].
B. EM of Fixed and Embedded Samples
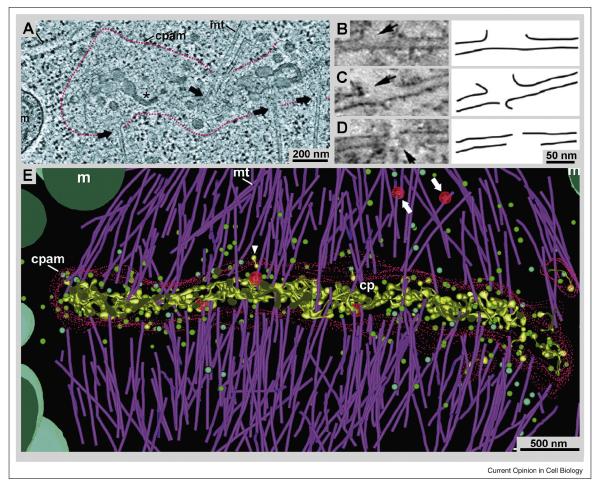
Tomographic imaging of sections 50 – 400 nm thick, cut from RF/FSF samples, has provided informative 3D views of many biological samples [30]. Thicker samples have also been imaged, but at some cost in resolution and often to characterize the distribution of specific stains [31]. These reconstructions can be made either with free software (e.g., IMOD <bio3d.colorado.edu>) or with commercial packages from the makers of EMs or suitable digital cameras. Slices of the 3D images reveal impressive cellular detail (Fig 3), because the viewer can alter the position, orientation, and thickness of the plane of sampling until a scientific question is answered. This capability is particularly valuable for a complex mixture of membranes and fibers, like that found in the cell plate that forms at the midplane of a plant cell during cytokinesis (Fig. 3A). Here, the density of vesicles, microtubules, and matix components had confounded morphologists using conventional EM. ET, on the other hand, allows such a fine sampling of detail that minute structural features, like aberrations in the walls of single microtubules can be found and characterized (Fig. 3B-D). The combination of this kind of detail with the overview provided by 3D imaging has allowed biologists to determine the relative position of many cytoplasmic constituents, providing data to build highly informative models of cellular substructure (Fig. 3E) [32].
Fig. 3.
Tomographic 3-D reconstructions from ~300-nm thick plastic sections, showing both the fine details and the global view of cellular structure that are visible in this kind of ET. A: 20 nm slice from a tomogram of the plant cell cytokinetic organelle; the “phragmoplast”, and its associated cell plate assembly matrix (cpam) are outlined in red. For details see [48•]. Figure adapted with permission. B – D: ~2 nm slices from a tomogram of a mitotic spindle showing that RF/FSF preserves molecular details, such as defects in microtubule structure caused by katanin [49•]. Arrows indicate breaks in the microtubule wall that are absent when katanin is deleted. E: 3-D model of phragmoplast microtubules, linking their plus-ends to the cpam. These models demonstrate the complexity of 3-D data that can be analyzed by tomography and tomographic modeling methods [48] (with permission).
To make such a display (Fig. 3E), multiple slices were viewed seriatim, and features of interest on each section were abstracted as graphic objects, resulting in a “model” of cellular substructure that facilitates the analysis of connectivities, surface areas, volumes, proximities, and visible textures. The advantages of this approach are the reliability of cell preservation achieved by rapid freezing, the likely preservation of this structure by freeze-substitution fixation and embedding, the high SNR of stained images, the ability to image in 3D, and the quantitative approach to structure that is facilitated by having all the information in a computer. Space resolution of these samples is 4 – 8 nm, depending on the preparation and the conditions of imaging/reconstructing. Equally important, the amount of sample imaged can be large. While most electronic cameras have only ~4 × 106 useful pixels, bigger cameras have been constructed [33], and montages from conventional cameras are possible [32]. When such imaging is coupled with serial sections, considerable cellular volume can be reconstructed, e.g., an entire fission yeast cell [34•]. By combining lower resolution images of individual areas with large-scale montaging, it is realistic to think about reconstructing entire mammalian cells [35], but the use of 2 nm voxels to represent a sample with ~5 nm resolution means that a cuboidal mammalian cell, 20 μm on a side, will require 1012 voxels, a daunting amount of information to manage, process, and analyze.
The good news, though, is that parts of any tomogram may be viewed and “segmented”, i.e., broken down into the parts that are relevant for the current scientific question, and the resulting graphic model is often smaller and simpler than the initial 3D data (Fig. 3E). Even more important, available software packages can extract numerical information from such reconstructions, so quantitative relationships can be derived from the data.
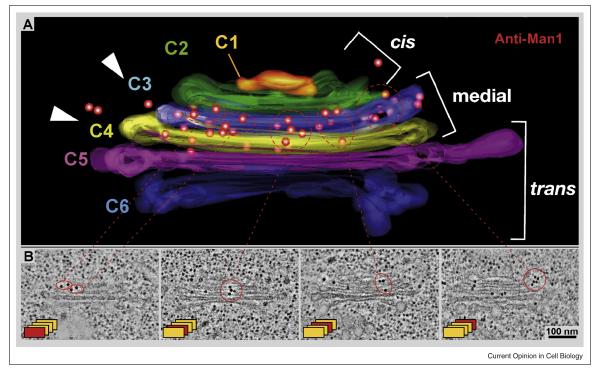
EM has long been combined with protein labeling to locate specific macromolecules in cells. Melding these methods with ET, however, presents problems. High quality fixation often inactivates epitopes, and more gentle fixations can fail to preserve biological structure at the required resolution. Several investigators have developed partial solutions to these problems. One school has accepted the limitations of room temperature aldehyde fixation, so they impregnate fixed samples with sucrose, then cut cryo-sections, and image them by ET. Some membrane structures are well preserved by this method, but others are not, and most cytoskeletal fibers are essentially invisible. Another school insists on RF/FSF, whereupon the sample is in an organic solvent and cannot be treated directly with antibody. However, these samples can be infiltrated with hydrophilic plastic at temperatures as low as −55°C, the plastic polymerized with UV light, and thereafter one can warm to room temperature, cut sections, apply antibodies, and stain. Now antibody labeling is limited to the section’s surface, but the preservation of both antigenicity and cell structure are excellent, even for cytoskeletal structures. Moreover, ET reveals structures that are not labeled, in addition to those that are, placing the immune labeling in the context of 3-D organelle structure. For example, this approach has allowed the group led by Andrew Staehelin to localize an isoform of α-mannosidase to two medial Golgi cisternae with high accuracy and confidence (Fig 4).
Fig. 4.
3D localization of antigens by the combination of ET with immuno-labeling. Arabidopsis meristems were prepared by RF/FSF, sliced ~300 nm, labeled with antibodies to α-1,2 mannosidase 1, and imaged as tilt series for ET [50•]. C1 – 6 label successive cisternae in the cis, medial, and trans parts of this Golgi complex, and the position of the antigen is marked by 15 nm gold particles through indirect immuno-localization. (With permission).
Another method for protein localization employs a small, membrane-permeating, arsenical ligand that binds a fluorophore to a tetra-sulfhydryl structure, which can be engineered into a protein of interest. The fluorophore converts energy from incident photons into free radicals that can oxidize diaminobenzidine (DAB) into an osmiophilic precipitate that is EM-visible [36••]. While this method has not yet been made suitable for RF/FSF samples, it has produced valuable data. Likewise, some fluorescent dyes can be photobleached to precipitate DAB into an EM-visible form, labeling aspects of intracellular membrane traffic [37]. Photo-conversion of the green fluorescent protein (GFP) has also been demonstrated [38], but the utility of this approach seems to be limited to situations where the labeled protein is abundant and localized within a membrane. Clearly, it would be of great value to have an EM equivalent of GFP, but although several labs have pursued this goal, no widely used method has yet appeared.
C) Perspective on the Use of Frozen-Hydrated and Low Temperature-Fixed samples for the Characterization of Macromolecules in cells
From the above it is clear that each of these approaches to sample preparation for EM has strengths and limitations. Cryo-ET has great advantages over other kinds of cryo-EM for the study of large, flexible, and non-periodic structures, like cells and organelles, because it provides 3D information without symmetry-based reconstruction methods. Tomography is now available to many investigators and is quite easy to do. Tilt-series at room-temperature can be recorded almost automatically within 30 min [39], and analogous data for vitrified samples usually take <80 min. New procedures for the alignment of tilted views should increase the accuracy of reconstructions, particularly when focused on small volumes [40,41]. New, cartridge-based tilting stages that shield specimens from perturbations have also been implemented. However, getting sufficient contrast from cryo-specimens currently requires under-focusing the objective lens by 2-6 μm. Imperfections in electronic lenses (particularly spherical aberration) allow defocusing to enhance phase contrast, which is the major source of information in cryo-micrographs. This trick, however, affects high-resolution components of the image by an unfavorable modulation of the contrast-transfer-function. Hence, visualizing structures smaller than 2.5nm requires a correction for the details of lens imaging, a process that is particularly tricky on tilted images. This problem could be avoided by the use of “phase plates”, analogous to phase imaging in the LM; then lower defocus values could be used [42,43]. Furthermore, improved lens technology, such as in the FEI-Titan series, and direct electron detectors (e.g. CMOS detectors [44, 32]), as well as imaging filters and aberrations correctors [45] promise to generate improved cryo-ET data in the near future.
Cryo-EM of frozen-hydrated samples is capable in principle of yielding resolutions of ~ 0.2 nm, but very large numbers of identical images would have to be averaged to approach this goal. Achieving even1-2 nm resolution requires 10,000 – 20,000 particles to make a 3-D map. Cryo-ET is further limited by inaccuracies in the alignment of serial tilts, but this approach should still be able to achieve <2nm, good enough for direct comparisons with atomic resolution structures. Reaching this resolution will again require the averaging of many identical samples to get sufficient SNR, so effective and efficient methods for image processing are necessary. Moreover, macromolecular complexes in cells are commonly machines that move through multiple states, each with its own structure, so in practice even more samples must be imaged, allowing classifications of multiple states as well as their alignments. This kind of work has been accomplished for muscle myosin [46••], but it is both demanding on the investigator and consumptive of computer time. Nonetheless, it represents an important frontier for future work in cellular imaging. Several computer programs are now available to help with this work, such as “particle estimation by electron tomography” (PEET) in the IMOD package or software from the Frangakis [12], Subramaniam [28] and Baumeister labs [47]. This endeavor is likely to be a domain where cryoEM has its biggest impact on solving cellular structures to molecular detail.
Viewing large expanses of cellular material, on the other hand, is not the greatest strength of cryoEM. Even when excellent sections have been cut, they are rather thin, and serial cryo-sectioning is still in its infancy. Serial tomography of plastic sections, on the other hand, is now routine in several labs, so imaging large cellular volumes at a resolution of ~5 nm is straightforward. Granted, there is the potential for artifact, as a result of RF/FSF and the use of stains, but if the resolution goal is not too demanding, these methods, properly employed, are likely to be very reliable. They also offer the possibility of macromolecular recognition through labeling, and there are several novel approaches to this kind of imaging now being developed in different labs; a “GFP for EM” may become a reality in the not far distant future. Thus, high resolution imaging of cellular detail is most likely to come from cryoET, while for work where information at 5 nm or bigger can answer the question, RF/FSF samples are likely to be preferable. If labeling methods become perfected, the value of the latter kind of imaging will increase even more, but regardless of such progress, it will be the intelligent combination of these methodologies that is likely to help elucidate cell architecture in macromolecular terms.
Acknowledgements
The authors’ laboratory has been supported by RR000592 from the NIH/NCRR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McIntosh JR, Nicastro D, Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Dubochet J, McDowall AW, Menge B, Schmid EN, Lickfeld KG. Electron microscopy of frozen-hydrated bacteria. J Bacteriol. 1983;155:381–390. doi: 10.1128/jb.155.1.381-390.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh JR. Electron microscopy of cells: a new beginning for a new century. J Cell Biol. 2001;153:F25–32. doi: 10.1083/jcb.153.6.f25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, Gerisch G. Organization of actin networks in intact filopodia. Curr Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022.A combination of cryo-ET and image processing reveals important aspects of actin organization in these slender projections from living cells.
- 5.Koning RI, Zovko S, Bárcena M, Oostergetel GT, Koerten HK, Galjart N, Koster AJ, Mommaas A Mieke. Cryo electron tomography of vitrified fibroblasts: microtubule plus ends in situ. J Struct Biol. 2008;161:459–468. doi: 10.1016/j.jsb.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Henderson GP, Gan L, Jensen GJ. 3-D ultrastructure of O. tauri: electron cryotomography of an entire eukaryotic cell. PLoS ONE. 2007;2:e749. doi: 10.1371/journal.pone.0000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231.This cryo-ET study of a small bacterium demonstrates the way in which tomographic imaging of a frozen-hydrated cell can reveal internal details of structure that clarify how the macromolecules in the cell are organized to do their various jobs.
- 8.Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008m;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol. 2008;67:1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol. 2008;163:10–17. doi: 10.1016/j.jsb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borgnia MJ, Subramaniam S, Milne JL. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J Bacteriol. 2008;190:2588–2596. doi: 10.1128/JB.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Amoudi A, Díez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994.Samples of skin were rapidly frozen to vitrify cellular water, then sliced in a cryo-microtome, producing specimens that were thin enough for cryo-ET. 3D reconstructions, combined with image averaging and computer graphics have revealed the organization of a protein that is important for cell-cell adhesion.
- 13.Sui H, Downing KH. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 2006;442:475–478. doi: 10.1038/nature04816. [DOI] [PubMed] [Google Scholar]
- 14.Cyrklaff M, Kudryashev M, Leis A, Leonard K, Baumeister W, Menard R, Meissner M, Frischknecht F. Cryoelectron tomography reveals periodic material at the inner side of subpellicular microtubules in apicomplexan parasites. J Exp Med. 2007;204:1281–1287. doi: 10.1084/jem.20062405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldie KN, Wedig T, Mitra AK, Aebi U, Herrmann H, Hoenger A. Dissecting the 3-D structure of vimentin intermediate filaments by cryo-electron tomography. J Struct Biol. 2007;158:378–385. doi: 10.1016/j.jsb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Boll W, Kirchhausen T, Harrison SC, Walz T. Cryo-electron tomography of clathrin-coated vesicles: structural implications for coat assembly. J Mol Biol. 2007;365:892–899. doi: 10.1016/j.jmb.2006.10.036.The use of cryo-ET, rather than a symmetry-based mode of 3D reconstruction, allows these investigators to visualize the naturally occurring variation in the structure of clathrin cages and coated vesicles.
- 18.Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 19.Christensen AK. Frozen thin sections of fresh tissue for electron microscopy, with a description of pancreas and liver. J. Cell Biol. 1971;51:772–804. doi: 10.1083/jcb.51.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowall AW, Chang JJ, Freeman R, Lepault J, Walter CA, Dubochet J. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. J Microsc. 1983;131:1–9. doi: 10.1111/j.1365-2818.1983.tb04225.x. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh CE, Leith A, Mannella CA, Frank J, Marko M. Towards high-resolution three-dimensional imaging of native mammalian tissue: electron tomography of frozen-hydrated rat liver sections. J Struct Biol. 2006;153:1–13. doi: 10.1016/j.jsb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlén LP, Richter K, Blanc NS, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladinsky MS, Pierson JM, McIntosh JR. Vitreous cryo-sectioning of cells facilitated by a micromanipulator. J Microsc. 2006;224:129–134. doi: 10.1111/j.1365-2818.2006.01674.x. [DOI] [PubMed] [Google Scholar]
- 24.Bouchet-Marquis C, Zuber B, Glynn AM, Eltsov M, Grabenbauer M, Goldie KN, Thomas D, Frangakis AS, Dubochet J, Chrétien D. Visualization of cell microtubules in their native state. Biol Cell. 2007;99:45–53. doi: 10.1042/BC20060081. [DOI] [PubMed] [Google Scholar]
- 25.Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J Bacteriol. 2007;189:4485–4493. doi: 10.1128/JB.01948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruska M, Medalia O, Baumeister W, Leis A. Electron tomography of vitreous sections from cultured mammalian cells. J Struct Biol. 2008;161:384–392. doi: 10.1016/j.jsb.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618.Cryo-ET of isolated axonemes, the microtubule component of flagella, shows how the doublet microtubules of this structure and its many enzymes and connections are organized to make a functional biological machine.
- 28.Bartesaghi A, Sprechmann P, Liu J, Randall G, Sapiro G, Subramaniam S. Classification and 3D averaging with missing wedge correction in biological electron tomography. J Struct Biol. 2008;162:436–50. doi: 10.1016/j.jsb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Förster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron subtomograms using constrained correlation. J Struct Biol. 2008;161:276–86. doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 30.McIntosh JR, editor. Methods in Cell Biology. Vol. 79. Academic Press; 2008. Cellular Electron Microscopy; p. 850. [Google Scholar]
- 31.Bouwer JC, Mackey MR, Lawrence A, Deerinck TJ, Jones YZ, Terada M, Martone ME, Peltier S, Ellisman MH. Automated most-probable loss tomography of thick selectively stained biological specimens with quantitative measurement of resolution improvement. J Struct Biol. 2004;148:297–306. doi: 10.1016/j.jsb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA. Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell. 2001;13:2033–2051. doi: 10.1105/TPC.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin L, Milazzo AC, Kleinfelder S, Li S, Leblanc P, Duttweiler F, Bouwer JC, Peltier ST, Ellisman MH, Xuong NH. Applications of direct detection device in transmission electron microscopy. J Struct Biol. 2008;161:352–358. doi: 10.1016/j.jsb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Höög JL, Schwartz C, Noon AT, O’Toole ET, Mastronarde DN, McIntosh JR, Antony C. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev Cell. 2007;12:349–361. doi: 10.1016/j.devcel.2007.01.020.Tomographic reconstructions of multiple serial sections, all linked into a single 3D model, allow the study of an entire eukaryotic cell. Here both microtubules and various organelles are displayed for visualization and study.
- 35.Noske AB, Costin AJ, Morgan GP, Marsh BJ. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J Struct Biol. 2008;161:298–313. doi: 10.1016/j.jsb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci U S A. 2006;103:17777–17782. doi: 10.1073/pnas.0608509103.A single labeling reagent is used to permit both visualization in the light microscope and subsequent localization by EM. This kind of clever tagging of important molecules is likely to lead the way towards a broader understanding of macromolecular organization in cells.
- 37.Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh JR, Howell KE. HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J Cell Biol. 1994;127:29–38. doi: 10.1083/jcb.127.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Structr. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Castaño-Díez D, Al-Amoudi A, Glynn AM, Seybert A, Frangakis AS. Fiducial-less alignment of cryo-sections. J Struct Biol. 2007;159:413–423. doi: 10.1016/j.jsb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Kaneko Y, Danev R, Nagayama K, Nakamoto H. Intact carboxysomes in a cyanobacterial cell visualized by hilbert differential contrast transmission electron microscopy. J Bacteriol. 2006;188:805–808. doi: 10.1128/JB.188.2.805-808.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danev R, Nagayama K. Single particle analysis based on Zernike phase contrast transmission electron microscopy. J Struct Biol. 2008;161:211–218. doi: 10.1016/j.jsb.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Deptuch G, Besson A, Rehak P, Szelezniak M, Wall J, Winter M, Zhu Y. Direct electron imaging in electron microscopy with monolithic active pixel sensors. Ultramicroscopy. 2007;107:674–684. doi: 10.1016/j.ultramic.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Haider M, Müller H, Uhlemann S, Zach J, Loebau U, Hoeschen R. Prerequisites for a Cc/Cs-corrected ultrahigh-resolution TEM. Ultramicroscopy. 2008;108:167–178. doi: 10.1016/j.ultramic.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Wu S, Reedy MC, Winkler H, Lucaveche C, Cheng Y, Reedy MK, Taylor KA. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J Mol Biol. 2006;362:844–860. doi: 10.1016/j.jmb.2006.07.084.3D reconstructions of a highly organized insect muscle allow the identification of functional intermediates in the behavior of myosin cross-bridges. Here, both classification and averaging are used to improve the SNR of different states in the mechanochemical cross-bridge cycle.
- 47.Förster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron subtomograms using constrained correlation. J Struct Biol. 2008;161:276–286. doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Segui-Simarro JM, Austin JR, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749.ET of serial sections of RF/FSF material allows the assembly of informative models that show the relationships between phragmoplast microtubules and the vesicles that assemble to form the cell plate that will separate two dividing plant cells
- 49.Srayko MT, O’Toole E, Hyman AA, Muller-Reichert T. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol. 2006;16:1944–1949. doi: 10.1016/j.cub.2006.08.029.ET of RF/FSF nematode embryos provides both the overview to see spindle microtubule organization (not shown here) and the details of katanin action as it helps to sever these polymers.
- 50.Staehelin LA, Kang BH. Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol. 2008;147:1454–1468. doi: 10.1104/pp.108.120618.ET of sections cut from hydrophilic plastic-embedded plant cells allow the application of antibodies for labeling of specific enzymes in the Golgi complex. Although antibody binding is limited to the surface of each section, the use of ET allows the tracking of membranes and the assembly of a 3D model in which the position of the label can be visualized.